Abstract
Seoul virus, an Old World hantavirus, is maintained in brown rats and causes a mild form of hemorrhagic fever with renal syndrome (HFRS) in humans. We captured rodents in New Orleans, Louisiana and tested them for the presence of Old World hantaviruses by reverse transcription polymerase chain reaction (RT-PCR) with sequencing, cell culture, and electron microscopy; 6 (3.4%) of 178 rodents captured—all brown rats—were positive for a Seoul virus variant previously coined Tchoupitoulas virus, which was noted in rodents in New Orleans in the 1980s. The finding of Tchoupitoulas virus in New Orleans over 25 years since its first discovery suggests stable endemicity in the city. Although the degree to which this virus causes human infection and disease remains unknown, repeated demonstration of Seoul virus in rodent populations, recent cases of laboratory-confirmed HFRS in some US cities, and a possible link with hypertensive renal disease warrant additional investigation in both rodents and humans.
Introduction
Hantaviruses are enveloped, tripartite, single-stranded, negative-sense RNA viruses belonging to the Bunyaviridae family, genus Hantavirus.1 Hantaviruses are maintained in rodents and shrews, usually with a tight pairing between the specific virus and host species. The Hantavirus genus is taxonomically divided into Old (i.e., Eurasia) and New World (i.e., the Americas) groups that are associated with distinct diseases: hemorrhagic fever with renal syndrome (HFRS) and hantavirus pulmonary syndrome, respectively. At least seven pathogenic Old World hantaviruses have been recognized. One Old World virus, Seoul, can be found worldwide, typically in urban environments, a finding which is attributed to the unintentional transportation of its reservoir, the brown or Norway rat (Rattus norvegicus), around the world on ships beginning in the seafaring middle ages.2 Seoul virus infections typically result in a milder form of HFRS, usually without overt hemorrhage.1,3
Brown rats and potentially, Old World hantaviruses are found in almost all cities in the world, with high population densities in port cities. New Orleans, Louisiana, located at the Mississippi River outlet to the Gulf of Mexico, is one of North America's oldest and busiest port cities. In the 1980s, Tsai and others4 reported the results of studies of hantaviruses in various US cities; over 30% of 79 rodents trapped along the waterfront in New Orleans were positive for antibodies to Old World hantaviruses, and a Seoul virus variant, which they coined Tchoupitoulas virus, was isolated from a brown rat by reverse transcription polymerase chain reaction (RT-PCR).4,5 No additional investigation of hantaviruses has been conducted in New Orleans. Because of the potential risk to public health from hantavirus-infected rodents, we sought to determine if Tchoupitoulas or other hantaviruses are still present in rodents in the city.
Materials and Methods
This study was approved by the Tulane University Institutional Animal Care and Use Committee and Biosafety Committee. Potentially infectious materials were handled at biosafety level 3 at the Regional Biosafety Laboratory facilities at the Tulane National Primate Research Center.
Study site selection.
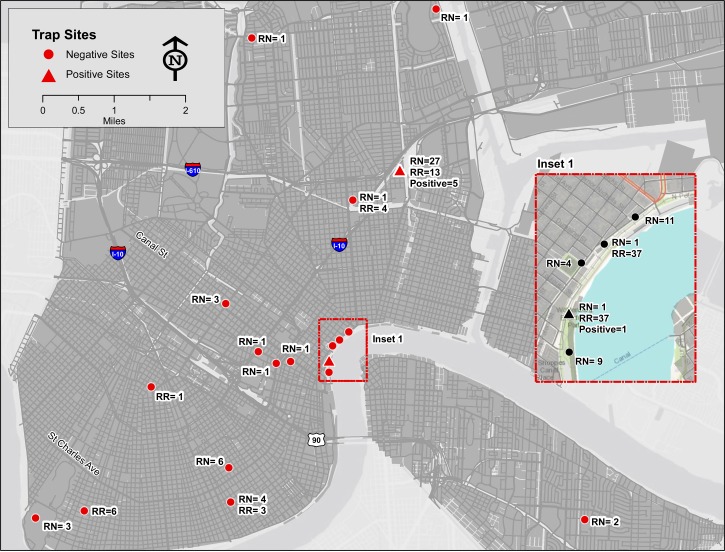
Rodent trapping was conducted periodically between July of 2008 and April of 2010. Rodent trap sites were a convenience sample based on routine rodent control activities of the New Orleans Mosquito, Termite, and Rodent Control Board responding to trouble calls or as part of routine rodent management services. Trap areas included wharf areas bordering the Mississippi River and confluent canals, external areas of private residences, and areas of high human traffic (Figure 1). Global Positioning System (GPS) coordinates of all trap sites were taken.
Figure 1.
Rodent trap sites in New Orleans. RN = Rattus norvengicus; RR = Rattus rattus.
Capture and processing of rodents.
Tomahawk live capture traps (Tomahawk Live Trap, Hazelhurst, WI) were placed along building borders, near burrows, or at sites of reported rodent activity. Approximately 25 traps were set per night. Traps were baited with peanut butter-coated wafers, set out before sunset, and then, checked at daybreak the next day. Captured rodents were brought in their traps to a processing center at the New Orleans Mosquito, Termite, and Rodent Control Board, where they were assigned a unique identification number and euthanized using vaporized isofluorane. The rodent species (based on morphometric analysis), sex, weight, total length (snout to tail), tail length, and presence of ectoparasites were recorded. Necropsies were performed to collect samples of blood, lung, kidney, pancreas, liver, and spleen as previously described.6 Samples were stored at −80°C until laboratory analysis.
RT-PCR and genetic characterization of viruses.
Lung tissue from all rodents was homogenized and tested by RT-PCR using a primer set that amplifies a 281-base pair region of the small (S) segment of all Old World hantaviruses.7 Amplicons of the correct size were then gel-extracted, cloned into the pCR4-TOPO vector (Invitrogen, Carlsbad, CA), and sent for commercial sequencing (GENEWIZ, Inc., Southfield, NJ). Resulting sequences were then aligned using ClustalX8 and analyzed using MrBayes (Bayesian)9 or MEGA5.2 (www.megasoftware.net) neighbor-joining, maximum likelihood, and maximum parsimony software10 for comparison with known hantavirus sequences in the National Center for Biological Information database.
Virus isolation.
Virus isolation from selected RT-PCR–positive rodents was carried out using a modification of the method outlined by Elliott and others.11 Briefly, gravity-settled lung homogenates were prepared and blind passaged two times on subconfluent Vero E6 cells. Virus presence was validated by RT-PCR of culture supernatant, immunofluorescence assay, and electron microscopy of infected cells.
Immunofluorescence assay.
Vero E6 cells grown in eight-well glass chamber slides were mock-infected or infected for 24 hours and then fixed with 4% paraformaldehyde for 10 minutes at 4°C. Cells were then permeabolized with 0.05% Triton-X for 10 minutes and blocked with 5% bovine serum albumin and 10% goat serum in phosphate-buffered saline (PBS). Cells were sequentially stained with mouse anti-Seoul virus nucleocapsid monoclonal antibody (PROGEN Biotechnik GmbH, Heidelberg, Germany) and a goat anti-mouse Alexa-fluor 488 secondary antibody (Invitrogen, Carlsbad, CA), and then, they were visualized on an Olympus IX51 Inverted Microscope (Center Valley, PA).
Transmission electron microscopy.
Specimens were fixed overnight at 4°C with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4). Samples were post-fixed for 1 hour with 0.5% osmium tetroxide/0.8% potassium ferricyanide, 1 hour with 1% tannic acid, and overnight with 1% uranyl acetate at 4°C. Samples were dehydrated with a graded ethanol series and embedded in Spurr's resin. Thin sections were cut with a Leica UCT ultramicrotome (Vienna, Austria) and stained with 1% uranyl acetate and Reynold's lead citrate before viewing at 120 kV on an FEI Tecnai BT Spirit transmission electron microscope (Hillsboro, OR). Digital images were acquired with an AMT digital camera system (AMT, Chazy, NY) and processed using Adobe Photoshop CS5 (Adobe Systems Inc, San Jose, CA).
Results
Trap success and species distribution.
From a total of 1,932 trap nights, 178 animals were captured (trap success = 9.2%), including 135 (75.8%) brown rats and 43 (24.2%) roof rats (R. rattus). The ratio of males to females was 2:1 for both species. Six (3.4%) rodents, all brown rats, were RT-PCR–positive (four female adults, one male adult, and one female juvenile). Positive rodents were noted at two trap sites (one site along the Mississippi River and the second site in a residential neighborhood approximately 2 miles to the north) on two separate occasions 6 months apart (Figure 1).
RT-PCR and phylogenetic analysis.
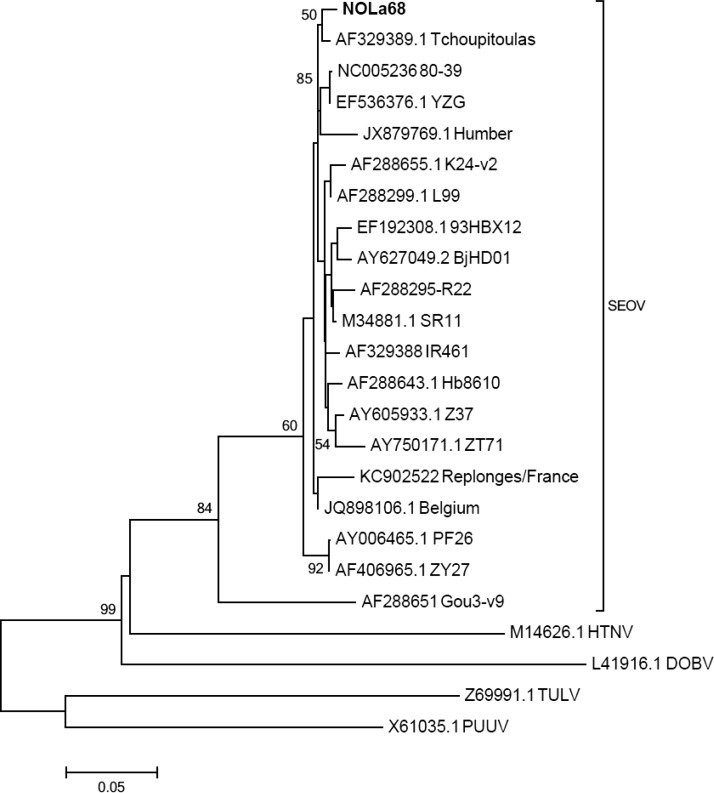
Sequence analysis of selected RT-PCR–positive rodents revealed 99% homology with the previously identified Tchoupitoulas variant of Seoul virus.4 Given the almost identical sequences of the 281-base pair amplicons produced, a representative sequence was used for phylogenetic comparison (Figure 2). Phylogenetic analysis confirmed that all rodent-derived hantavirus sequences from this study belonged to the same branch as the previously identified Tchoupitoulas virus within the Seoul virus clade. Congruency between Bayesian and bootstrapped neighbor-joining, maximum parsimony, and maximum likelihood analyses of Clustal alignments validated the phylogenetic assignment (data not shown).
Figure 2.
Maximum likelihood phylogenetic tree showing relatedness of a representative hantavirus (NOLa68; bold) indentified from rodent samples collected in New Orleans, Louisiana, with other Old World hantavirus sequences available from GenBank. The tree compares partial S-segment sequences (nucleotide positions 973–1253 of accession number M14626.1) and was constructed using the Tamura three-parameter model with γ-distributed rate heterogeneity and a proportion of invariant sites (T92 + G + I). Each branch shows the GenBank accession number followed by common virus identifier. The scale bar indicates nucleotide substitutions per site. GenBank accession numbers of sequences derived from this study are KF683313, KF683314, and KF683315. DOBV = Dobrava virus; HTNV = Hantaan virus; PUUV = Puumala virus; SEOV = Seoul virus; TULV = Tula virus.
Virus isolation, immunofluorescence assay, and electron microscopy.
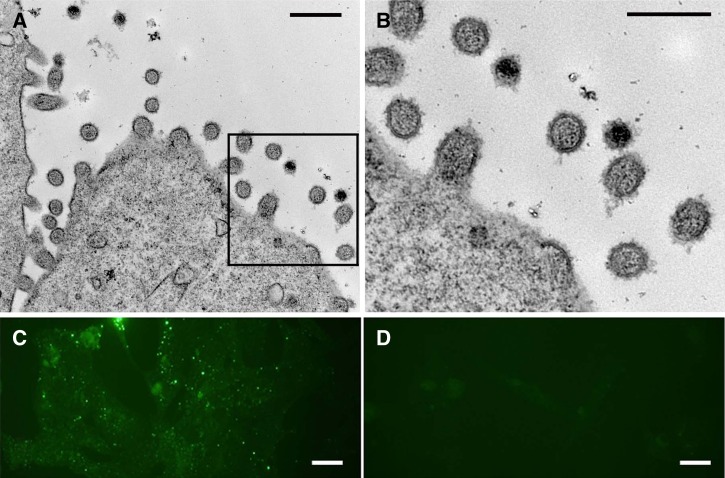
RT-PCR on Vero E6 cell culture supernatant produced the expected 281-base pair product amplified from infected tissues (data not shown). Immunofluorescence assay testing of cells revealed classical punctate, cytoplasmic staining of hantavirus nucleocapsid protein. Electron microscopy showed classic hantavirus morphology of pleomorphic, ovoid virions ranging from 80 to 120 nm in diameter (Figure 3).
Figure 3.
(A and B) Electron micrographs showing classic hantavirus particles ranging from 80 to 120 nm in diameter budding from the surface of Vero cells. (C) Immunofluorescence assay showing characteristic punctuate, cytoplasmic staining of infected Vero E6 cells stained with anti-Seoul virus nucleocapsid antibody. (D) Mock-infected Vero E6 cells. Scale bars: A and B, 250 nm; C and D, 20 μm.
Discussion
Our results show that the Tchoupitoulas variant of Seoul virus is still present in the brown rat population in New Orleans, although at low prevalence. The finding of this same virus over 25 years since its first discovery suggests a stable endemicity in the city. It remains unknown whether human infection and disease caused by Tchoupitoulas virus have occurred in New Orleans, although the 1982 report by Tsai and others5 on the testing of 57 rodent and mosquito control personnel, virology laboratory workers, and sanitarians in New Orleans for anti-Hantaan virus immunoglobulin G (IgG) antibody by immunofluorescence assay showed no positives.5 No systematic testing has been done since that time, and no routine surveillance for hantavirus infection or HFRS exists in the city. Additionally, clinical laboratory testing for hantaviruses is not routinely available.
In addition to New Orleans, serologic evidence of Seoul or related Old World hantaviruses has been reported in rodents in Baltimore, Maryland12–17; Houston, Texas13,18,19; Philadelphia, Pennsylvania13,18,19; Cincinnati, Ohio; Columbus, Ohio; Los Angeles, California20; New York, New York; San Francisco, California; Seattle, Washington; Tacoma, Washington; Hilo, Hawaii4; and areas of North Carolina,12 Maryland, West Virginia,21 Minnesota, California,21,22 Alaska,21 and Mississippi,23 with hantavirus isolates and/or PCR amplification available from rodents in Houston and Philadelphia.18 The distribution of infected rodents may be extremely localized,14 which we noted in our study.
Serologic evidence of human exposure to Old World hantaviruses has been shown in Baltimore,17,24–27 Los Angeles,20 and various counties of Colorado, Maryland, North Carolina, and West Virginia.28 Nevertheless, only five cases of HFRS caused by Seoul virus infection have been reported in the United States: one case each in Maryland29 and southern Texas30 and three retrospectively identified in Baltimore, Maryland.3 Only the Maryland case was definitively confirmed through PCR. Only the case from southern Texas was fatal, with the patient showing significant pulmonary involvement, which is usually associated with New World hantaviruses. No specific exposure to rodent excreta could be shown in these five cases. In addition to acute HFRS, an association of Old World hantavirus infection and chronic hypertensive renal disease has been suggested,3,31,32 although no long-term sequelae were noted in a controlled trial of persons with past HFRS caused by Hantaan virus.33
The degree to which Old World hantaviruses present a threat to public health in the United States remains murky and probably varies considerably by city and region. It may, indeed, be that human infection is rare, considering that infections with rodent-borne viruses seem to be rare, even when rodent contact is frequently reported.25 However, the milder clinical illness associated with HFRS caused by Seoul virus may often impede recognition, especially considering that most physicians in the United States are likely unfamiliar with this condition. The situation is further compounded by the absence of formal surveillance or readily available laboratory diagnostics. Nevertheless, the repeated demonstration of Seoul virus in rodent populations, recent cases of laboratory-confirmed HFRS caused by Seoul virus, and the possible link with chronic hypertensive renal disease warrant additional enzootic investigation and human surveillance of Old World hantaviruses in New Orleans and other US cities.
ACKNOWLEDGMENTS
The authors thank Nell Bond, Greg Glass, Mary Greene, James Mills, Andrew Ruiz, and Corrie Ntiforo for technical assistance.
Footnotes
Financial support: Funding for this research was provided by a Tulane University Research Enhancement grant.
Authors' addresses: Robert W. Cross, Bradley Waffa, Lina M. Moses, Andrew Bennett, Thomas G. Voss, and Daniel G. Bausch, Health Science Center, Tulane University, New Orleans, LA, E-mails: rwcross@utmb.edu, bjwaffa@ncsu.edu, linammoses@gmail.com, Andrew.j.bennett@gmail.com, thomas.voss@sri.com, and dbausch@tulane.edu. Ashley Freeman and Claudia Riegel, New Orleans Mosquito, Termite, and Rodent Control Board, New Orleans, LA, E-mails: Anfree1005@yahoo.com and criegel@cityofno.com. David Safronetz, Elizabeth R. Fischer, and Heinz Feldmann, Rocky Mountain Laboratories, Hamilton, MT, E-mails: safronetzd@niaid.nih.gov, efischer@niaid.nih.gov, and feldmannh@niaid.nih.gov.
References
- 1.Jonsson CB, Figueiredo LT, Vapalahti O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin Microbiol Rev. 2010;23:412–441. doi: 10.1128/CMR.00062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin XD, Guo WP, Wang W, Zou Y, Hao ZY, Zhou DJ, Dong X, Qu YG, Li MH, Tian HF, Wen JF, Plyusnin A, Xu J, Zhang YZ. Migration of norway rats resulted in the worldwide distribution of seoul hantavirus today. J Virol. 2012;86:972–981. doi: 10.1128/JVI.00725-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glass GE, Watson AJ, LeDuc JW, Childs JE. Domestic cases of hemorrhagic fever with renal syndrome in the United States. Nephron. 1994;68:48–51. doi: 10.1159/000188086. [DOI] [PubMed] [Google Scholar]
- 4.Tsai TF, Bauer SP, Sasso DR, Whitfield SG, McCormick JB, Caraway TC, McFarland L, Bradford H, Kurata T. Serological and virological evidence of a Hantaan virus-related enzootic in the United States. J Infect Dis. 1985;152:126–136. doi: 10.1093/infdis/152.1.126. [DOI] [PubMed] [Google Scholar]
- 5.Tsai TF, Bauer SP, Sasso DR, McCormick JB, Bradford H, Caraway CT, McFarland LM, Medrano O, Soulie G. Preliminary evidence that Hantaan or a closely related virus is enzootic in domestic rodents. N Engl J Med. 1982;307:623–625. doi: 10.1056/NEJM198209023071013. [DOI] [PubMed] [Google Scholar]
- 6.Mills J, Yates T, Childs J, Parmenter R, Ksiazek T, Rollin P, Peters C. Guidelines for working with rodents potentially infected with hantavirus. J Mammal. 1995;76:716–722. [Google Scholar]
- 7.Arthur RR, Lofts RS, Gomez J, Glass GE, Leduc JW, Childs JE. Grouping of hantaviruses by small (S) genome segment polymerase chain reaction and amplification of viral RNA from wild-caught rats. Am J Trop Med Hyg. 1992;47:210–224. doi: 10.4269/ajtmh.1992.47.210. [DOI] [PubMed] [Google Scholar]
- 8.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 9.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 10.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott LH, Ksiazek TG, Rollin PE, Spiropoulou CF, Morzunov S, Monroe M, Goldsmith CS, Humphrey CD, Zaki SR, Krebs JW, Maupin GO, Gage KL, Childs JE, Nichol ST, Peters CJ. Isolation of the causative agent of hantavirus pulmonary syndrome. Am J Trop Med Hyg. 1994;51:102–108. doi: 10.4269/ajtmh.1994.51.102. [DOI] [PubMed] [Google Scholar]
- 12.Easterbrook JD, Shields T, Klein SL, Glass GE. Norway rat population in Baltimore, Maryland, 2004. Vector Borne Zoonotic Dis. 2005;5:296–299. doi: 10.1089/vbz.2005.5.296. [DOI] [PubMed] [Google Scholar]
- 13.LeDuc JW, Smith GA, Childs JE, Pinheiro FP, Maiztegui JI, Niklasson B, Antoniades A, Robinson DM, Khin M, Shortridge KF, Wooster MT, Elwell MR, Ilbery PLT, Koech D, Rosa EST, Rosen L. Global survey of antibody to Hantaan-related viruses among peridomestic rodents. Bull World Health Organ. 1986;64:139–144. [PMC free article] [PubMed] [Google Scholar]
- 14.Childs JE, Korch GW, Smith GA, Terry AD, Leduc JW. Geographical distribution and age related prevalence of antibody to Hantaan-like virus in rat populations of Baltimore, Maryland, USA. Am J Trop Med Hyg. 1985;34:385–387. doi: 10.4269/ajtmh.1985.34.385. [DOI] [PubMed] [Google Scholar]
- 15.Easterbrook JD, Kaplan JB, Vanasco NB, Reeves WK, Purcell RH, Kosoy MY, Glass GE, Watson J, Klein SL. A survey of zoonotic pathogens carried by Norway rats in Baltimore, Maryland, USA. Epidemiol Infect. 2007;135:1192–1199. doi: 10.1017/S0950268806007746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korch GW, Childs JE, Glass GE, Rossi CA, LeDuc JW. Serologic evidence of hantaviral infections within small mammal communities of Baltimore, Maryland: spatial and temporal patterns and host range. Am J Trop Med Hyg. 1989;41:230–240. doi: 10.4269/ajtmh.1989.41.230. [DOI] [PubMed] [Google Scholar]
- 17.Gibbs CJ, Jr, Takenaka A, Franko M, Gajdusek DC, Griffin MD, Chields J, Korch GW, Wartzok D. Seroepidemiology of Hantaan virus. Lancet. 1982;2:1406–1407. doi: 10.1016/s0140-6736(82)91310-1. [DOI] [PubMed] [Google Scholar]
- 18.LeDuc JW, Smith GA, Johnson KM. Hantaan-like viruses from domestic rats captured in the United States. Am J Trop Med Hyg. 1984;33:992–998. doi: 10.4269/ajtmh.1984.33.992. [DOI] [PubMed] [Google Scholar]
- 19.LeDuc JW, Smith GA, Bagley LR, Hasty SE, Johnson KM. Hantaan virus or an antigenically similar virus in Rattus populations (Letter to the Editor) N Engl J Med. 1982;307:624. [Google Scholar]
- 20.Smith HM, Reporter R, Rood MP, Linscott AJ, Mascola LM, Hogrefe W, Purcell RH. Prevalence study of antibody to ratborne pathogens and other agents among patients using a free clinic in downtown Los Angeles. J Infect Dis. 2002;186:1673–1676. doi: 10.1086/345377. [DOI] [PubMed] [Google Scholar]
- 21.Lee P-W, Yanagihara R, Franko MC, Amyx HL, Gibbs CJ, Gajdusek DC. Korean hemorrhagic fever virus or similar antigenic agents in wild-rodent populations in the United States (Letter to the Editor) N Engl J Med. 1982;307:624–625. [Google Scholar]
- 22.Yanagihara R, Daum CA, Lee PW, Baek LJ, Amyx HL, Gajdusek DC, Gibbs CJ., Jr Serological survey of Prospect Hill virus infection in indigenous wild rodents in the USA. Trans R Soc Trop Med Hyg. 1987;81:42–45. doi: 10.1016/0035-9203(87)90275-6. [DOI] [PubMed] [Google Scholar]
- 23.Forthal DN, Bauer SP, McCormick JB. Antibody to hemorrhagic fever with renal syndrome viruses (Hantaviruses) in the United States. Am J Epidemiol. 1987;126:1210–1213. doi: 10.1093/oxfordjournals.aje.a114760. [DOI] [PubMed] [Google Scholar]
- 24.Auwaerter PG, Oldach D, Mundy LM, Burton A, Warner ML, Vance E, Moore RD, Rossi CA. Hantavirus serologies in patients hospitalized with community-acquired pneumonia. J Infect Dis. 1996;173:237–239. doi: 10.1093/infdis/173.1.237. [DOI] [PubMed] [Google Scholar]
- 25.Childs JE, Glass GE, Ksiazek TG, Rossi CA, Oro JG, Leduc JW. Human-rodent contact and infection with lymphocytic choriomeningitis and Seoul viruses in an inner-city population. Am J Trop Med Hyg. 1991;44:117–121. doi: 10.4269/ajtmh.1991.44.117. [DOI] [PubMed] [Google Scholar]
- 26.Diglisic G, Rossi CA, Doti A, Walshe DK. Seroprevalence study of Hantavirus infection in the community based population. Md Med J. 1999;48:303–306. [PubMed] [Google Scholar]
- 27.Khabbaz RF, Ksiazek TG, Caiaffa WT, Rollin PE, Taylor E, Vlahov D. Seoul hantavirus seropositivity among injecting drug users in Baltimore. J Infect Dis. 1994;170:1636–1637. doi: 10.1093/infdis/170.6.1636. [DOI] [PubMed] [Google Scholar]
- 28.Yanagihara R, Chin CT, Weiss MB, Gajdusek DC, Diwan AR, Poland JB, Kleeman KT, Wilfert CM, Meiklejohn G, Glezen WP. Serological evidence of Hantaan virus infection in the United States. Am J Trop Med Hyg. 1985;34:396–399. doi: 10.4269/ajtmh.1985.34.396. [DOI] [PubMed] [Google Scholar]
- 29.Woods C, Palekar R, Kim P, Blythe D, de Senarclens O, Feldman K, Farnon EC, Rollin PE, Albarino CG, Nichol ST, Smith M. Domestically acquired seoul virus causing hemorrhagic fever with renal syndrome-Maryland, 2008. Clin Infect Dis. 2009;49:e109–e112. doi: 10.1086/644742. [DOI] [PubMed] [Google Scholar]
- 30.Roig IL, Musher DM, Tweardy DJ. Severe pulmonary involvement in a case attributed to domestically acquired Seoul hantavirus in the United States. Clin Infect Dis. 2012;54:91–94. doi: 10.1093/cid/cir748. [DOI] [PubMed] [Google Scholar]
- 31.Glass GE, Watson AJ, LeDuc JW, Kelen GD, Quinn TC, Childs JE. Infection with a ratborne hantavirus in US residents is consistently associated with hypertensive renal disease. J Infect Dis. 1993;167:614–620. doi: 10.1093/infdis/167.3.614. [DOI] [PubMed] [Google Scholar]
- 32.LeDuc JW, Childs JE, Glass GE. The Hantaviruses, etiologic agents of hemorrhagic fever with renal syndrome: a possible cause of hypertension and chronic renal disease in the United States. Annu Rev Public Health. 1992;13:79–98. doi: 10.1146/annurev.pu.13.050192.000455. [DOI] [PubMed] [Google Scholar]
- 33.Mathes RW, Page WF, Crawford HM, McBean AM, Miller RN. Long-term sequelae of hemorrhagic fever with renal syndrome attributable to hantaan virus in Korean War veterans. Mil Med. 2005;170:315–319. doi: 10.7205/milmed.170.4.315. [DOI] [PubMed] [Google Scholar]