Abstract
Phlebotomus papatasi sand flies are among the primary vectors of Leishmania major parasites from Morocco to the Indian subcontinent and from southern Europe to central and eastern Africa. Antibody-based immunity to sand fly salivary gland proteins in human populations remains a complex contextual problem that is not yet fully understood. We profiled the immunoreactivities of plasma antibodies to sand fly salivary gland sonicates (SGSs) from 229 human blood donors residing in different regions of sand fly endemicity throughout Jordan and Egypt as well as 69 US military personnel, who were differentially exposed to P. papatasi bites and L. major infections in Iraq. Compared with plasma from control region donors, antibodies were significantly immunoreactive to five salivary proteins (12, 26, 30, 38, and 44 kDa) among Jordanian and Egyptian donors, with immunoglobulin G4 being the dominant anti-SGS isotype. US personnel were significantly immunoreactive to only two salivary proteins (38 and 14 kDa). Using k-means clustering, donors were segregated into four clusters distinguished by unique immunoreactivity profiles to varying combinations of the significantly immunogenic salivary proteins. SGS-induced cellular proliferation was diminished among donors residing in sand fly-endemic regions. These data provide a clearer picture of human immune responses to sand fly vector salivary constituents.
Introduction
The causative agents of leishmaniasis are protozoan parasites of the genus Leishmania. All species within the genus share a common lifecycle that involves an extracellular infective stage present in the sand fly vector followed by an intracellular stage of replication within a vertebrate host. Estimates of annual global disease incidence are approximately 0.2–0.4 and 0.7–1.2 million cases of active visceral and cutaneous leishmaniasis, respectively.1 Leishmaniasis is primarily found in tropical and subtropical regions, where vector-competent sand fly species of the family Psychodidae reside. Medically relevant genera of the sand fly subfamily Phlebotominae are distinguished by distinct geographical regions of endemicity: Phlebotomus in the Old World (the Mediterranean, Middle East, and Africa) and Lutzomyia in the New World (Central and South America).2
Currently, no efficacious vaccine exists to combat infection or transmission of Leishmania. In lieu of a vaccine directed at the parasites, attention has focused on another aspect of the disease cycle: the sand fly vector. One hypothesis is that, through pre-exposure to sand fly vector salivary components, individuals are immunized to establish a robust immunological memory against vector salivary factors. This response may generate a hostile microenvironment for parasite transmission on reexposure. For every species of Leishmania parasite, only a few sand fly species are capable of vectoring a specific Leishmania species,3 resulting in a very restrictive and unique vector–parasite exposure profile. Key to the downstream development of a vaccine is the careful examination of host immunological responses to vector salivary proteins among persons naturally pre-exposed to sand fly bites.4,5
Many studies have sought to investigate the serological markers of hosts, both animal and human, against sand fly species,6–14 although the problem remains a highly complex and contextual matter warranting deeper novel analysis. To better characterize acquired human host immune responses to sand fly saliva, we collected peripheral blood samples from 229 people who reside in Middle Eastern regions (Egypt and Jordan) of variable endemic disease ecologies for cutaneous leishmaniasis and its predominant Old World vector, the sand fly P. papatasi. Additionally, samples were collected from 69 US military personnel, differentially exposed to P. papatasi bites and L. major infections, to aid in comparisons with immunologically naïve individuals. Our work sought to understand host population variation with reference to antivector saliva immunity and identify salivary proteins for later investigation as potential exposure markers.
Methods
Study sites and human peripheral blood donors.
Six sites in the Middle East and North Africa (MENA) were chosen for collection of 229 human peripheral blood samples during late August and early September of 2007. Table 1 shows regional donor profiles. Geospatial coordinates for these regions were as previously described.15 Each site was selected based on ecological characteristics in Jordan16–23 and Egypt24–29 pertaining to active cases of leishmaniasis transmission and the presence of P. papatasi vectors. These sites included Cairo (CA), Egypt and Amman (AM), Jordan (a lack of sand fly vectors); Aswan (AW), Egypt and Malka (MA), Jordan (regions of endemic P. papatasi populations but that lack clinical reports of L. major infections); and North Sinai (NS), Egypt and Swaymeh (SW), Jordan (regions of endemic P. papatasi population as well as regions with reports of active L. major transmission). Blood samples from regional Egyptian and Jordanian donors were drawn from healthy adults, none of whom presented with active lesions. Peripheral blood samples were similarly collected from 69 US military personnel at Walter Reed Army Medical Center, Washington, DC. US donors were interviewed for histories of suspected sand fly bite exposure or presented with clinically confirmed cases of cutaneous leishmaniasis but were untreated before blood sampling. All donors were ≥ 18 years of age, and the number of male and female participants was approximately equal. All donors signed documents of informed consent per local and US laws, and samples were stripped of all personal identifying information before processing.
Table 1.
Summary of human peripheral blood donors
| Abbreviation | Country | P. papatasi endemicity* | L. major transmission reports* | Number of blood donors | |
|---|---|---|---|---|---|
| MENA sites† | |||||
| Cairo | CA | Egypt | − | − | 37 |
| Aswan | AW | Egypt | + | − | 40 |
| North Sinai | NS | Egypt | + | + | 34 |
| Amman | AM | Jordan | − | − | 37 |
| Malka | MA | Jordan | + | − | 41 |
| Swaymeh | SW | Jordan | + | + | 40 |
| US military‡ | P. papatasi bite exposure§ | L. major infection§ | |||
| Control | United States | − | − | 29 | |
| Bitten | United States | + | − | 17 | |
| Infected | United States | + | + | 23 | |
Marks indicate the presence (+) or absence (−) of endemic P. papatasi populations and known cases of active L. major transmission in the region.
MENA locations in which human peripheral blood donors resided.
The condition of US military personnel stationed in the United States (control) or deployed in Iraq (bitten and infected) during 2007.
Marks indicate yes (+) or no (−) as to whether US military personnel reported exposure to sand fly bites or there was a confirmed diagnosis of cutaneous leishmaniasis while deployed in Iraq.
Human host blood collection.
Ten to fifteen milliliters blood were collected in yellow top Vacutainer blood collection tubes with ACD solution A anticoagulant (BD Biosciences, San Jose, CA). Tubes were kept at 4°C and centrifuged at 3,000 rpm for 10 minutes. The aqueous top plasma layer was removed and stored at −20°C. Remaining mixed leukocytes and red blood cells were segregated using Lymphocyte Separation Media (Cellgro, Manassas, VA) following the manufacturer's protocol. Remaining cells were suspended in red blood cell lysis buffer (0.15 M ammonium chloride, 10 mM potassium bicarbonate, and 0.1 mM (ethylenedinitrilo)tetraacetic acid [EDTA]), incubated for 5 minutes on ice, washed with 1× phosphate buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4), and resuspended in standard RPMI-1640 media (Cellgro) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine (referred to as RPMI-C). Mixed cells in RPMI-C media were combined 1:1 in freezing media (20% dimethyl sulfoxide [DMSO], 20% FBS, and 60% RPMI-C), slow frozen overnight at −80°C in an isopropanol chamber, transferred to dry ice for transport, and ultimately transferred to liquid nitrogen for long-term storage before use in cell proliferative assays.
Sand fly colony.
P. papatasi sand flies used in this study were derived from the Israeli strain maintained at the Laboratory of Malaria and Vector Research, National Institutes of Health (Rockville, MD) and the University of Notre Dame (Notre Dame, IN) as previously described.15
Salivary gland sonicates.
Salivary glands from 1- to 2-week-old unfed female P. papatasi sand flies were dissected intact under 1× PBS and flash frozen at −80°C for storage. To procure salivary gland protein sonicates (SGSs), salivary glands were subjected to three cycles of rapid freeze–thaw (−80°C for 2 minutes and then a 37°C water bath for 1 minute), centrifuged for 1 minute at 14,000 rpm, sonicated in a room temperature water bath for 1 minute at 80% power using an Ultrasonic Processor (Cole Parmer, Vernon Hills, IL), and then, filter-sterilized by centrifugation at 14,000 rpm for 1 minute through a 0.22-μm Ultrafree centrifugal filter unit (Millipore, Billerica, MA). Total protein quantities were assessed by standard curve to bovine serum albumin dilutions using the Protein A280 feature of a NanoDrop 1000 microvolume UV-Vis spectrophotometer (Thermo Scientific, Waltham, MA). A whole-salivary gland equivalent concentration was considered approximately 1.5 μg based on estimates of complete lobe pairs from sugar-fed female Lu. longipalpis salivary gland protein concentrations.30
Mass spectrometry and protein sequence alignments.
Twenty salivary gland protein equivalents from P. papatasi SGS in 1× PBS were boiled at 85°C for 5 minutes with Tris-glycine sodium dodecyl sulfate (SDS) sample buffer and NuPAGE reducing agent (Novex, Carlsbad, CA). Samples were applied to a pre-cast 4–20% stacking Tris-glycine gel (Novex) with a PageRuler pre-stained protein ladder (Fermentas, Pittsburgh, PA) on the side and electrophoresed at 125 V for 90 minutes. Salivary proteins were run in single dimension electrophoresis to allow cross-comparison with immunoblots. The gel was bathed in fixation solution (50% methanol and 10% acetic acid) overnight and then stained for 4 hours in 1% Coomassie Blue R-250 (Bio-Rad, Hercules, CA). The gel was destained overnight in destaining solution (5% methanol and 7.5% acetic acid) and stored in 7% acetic acid for imaging on a gel doc Universal Hood II (Bio-Rad, Hercules, CA). Ten intensely stained protein gel bands were cut precisely using clean, sterile scalpels. Gel slices were prepared by trypsin digestion for mass spectrometry (MS) as previously described.31 Samples were desalted by Ziptip (Millipore, Billerica, MA) before liquid chromatography (LC) /MS/MS analysis performed on a 5500QTrap operating in ion-trap mode (AB Sciex, Farmingham, MA) connected to an Eksigent (AB Sciex) Nano Ultra UHPLC system. Samples were acquired as described.32 Briefly, each gel band was analyzed in technical duplicate and separated over a gradient from 2% to 35% acetonitrile (0.1% formic acid) on a 100-μm × 100-mm C18BEH column (Waters, Milford, MA) running at 600 nL/min for 1 hour. A TOP8 data-dependent method was used for spectral acquisition. Peak lists were subjected to database search against all available Phlebotomus and Lutzomyia spp. protein sequences available through the US National Center for Biotechnology Information (NCBI)33 combined with a list of common contaminants (e.g., trypsin, keratin, etc.). False discovery rates (FDRs) were determined by the target–decoy approach.34 The Paragon algorithm within the Protein Pilot software suite (AB Sciex, Farmingham, Massachusetts, USA) was used for database searches,32 and proteins within the 95% FDR interval were chosen for additional analysis. FDR descriptions for combined protein, peptide, and spectral level analyses are included in Supplemental Figure 1. Alignment analyses of published protein sequences were conducted using Clustal Omega35 or LAlign36 web-based programs with default settings.
Immunoblots.
Western immunoblots to examine human plasma donor antibodies reactive against SGS were conducted using a modified version of a previously reported protocol.37 Briefly, 30 SGSs were boiled and separated on pre-cast 10% Tris-glycine SDS–polyacrylamide gel electrophoresis (PAGE) gels (Novex) with a PageRuler pre-stained protein ladder (Fermentas) on the side and transferred to nitrocellulose membranes using an XCell II blot module (Novex). Membranes were blocked for 1 hour at room temperature (RT) in blocking buffer (0.1% Tween 20/1× tris-buffered saline and 5% dried milk), washed three times in 1× TBS, and set in a Mini-Protean II multiscreen apparatus (Bio-Rad). Blood plasma from human donors was diluted 1:100 in blocking buffer and exposed to the membranes. A single common donor was exposed to Israeli colony P. papatasi bites 24 hours before blood collection, which served as a normalizing control on each blot. Membranes and plasma were incubated overnight at 4°C, washed three times in 1× TBS, and then, exposed to a secondary alkaline phosphatase-conjugated goat anti-human Fc antibody (Zymed, South San Francisco, CA) diluted 1:5,000 in blocking buffer. Membranes were developed with colorimetric substrate One Step NPT-BCIP (Thermo Scientific). Membranes were imaged using a gel doc Universal Hood II (Bio-Rad). White balance for all images was normalized to a single immunoblot (Figure 1A) using Photoshop CS5 software (Adobe, San Jose, CA) to equalize background intensity. Because of non-specific colorimetric artifacts in the lanes of many immunoblots, the use of automated image analysis software, such as ImageJ, to score the plasma antibody reactivity to SGS protein bands proved impossible, because a universal baseline was not applicable to all lanes, even in a single immunoblot. Three people manually scored each blot, and they were independent observers blinded to the regional identities of the plasma donors applied to each immunoblot. Observers cataloged the banding pattern and intensity of each blot based on a numeric scale (0 = no bands at a given kilodalton value; 1 = low-intensity band; 2 = moderate-intensity band; 3 = high-intensity band) relative to the positive control plasma lane appearance. The scores were combined based on a uniform rule set favoring the majority opinion at each kilodalton range. The combined score calculations were checked and verified by each investigator independently, providing the most unbiased dataset possible.
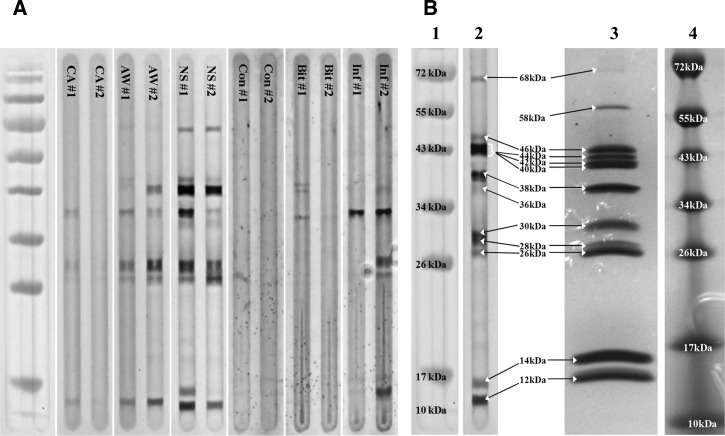
Figure 1.
Characterization of whole P. papatasi salivary gland protein extract sonicates and human host antibody reactions. (A) Immunoblot examples of plasma donors from three regions in Egypt and differentially exposed US military personnel with antibody specificity to P. papatasi salivary proteins. Lanes are designated by regional plasma donor codes (CA, AW, and NS) or differential exposure codes (Con = Control, Bit = Bitten, and Inf = Infected). (B) Comparison of immunoblot example and whole P. papatasi salivary gland protein sonicate. Lane 2 displays an immunoblot image of a North Sinai plasma donor with broad antibody specificities to P. papatasi salivary gland proteins. Lane 3 displays a Coomassie Brilliant Blue R-250-stained P. papatasi whole-salivary gland protein sonicate run on a denaturing SDS-PAGE gel. Lanes 1 and 4 are PageRuler protein MW ladders (Fermentas).
Enzyme-Linked Immunosorbent Assay.
Isolated SGSs in 1× PBS were applied at 1.5 μg per well onto a MaxiSorp flat-bottom 96-well plate (Nunc, Penfield, NY) and incubated overnight at 4°C. Some wells on each plate were coated instead with purified human immunoglobulin G (IgG) or IgE (Abcam, Cambridge, MA) at known concentrations for 1:100, 1:1,000, and 1:10,000 dilutions, which served as standards for calculations of human donor anti-SGS antibody concentrations. IgG standards (Abcam) were composed of a mixture of all subclasses (IgG1, -2, -3, and -4) at known percentages of the total. Plates were washed with 0.05% Tween-20/1× PBS and blocked with 5% bovine serum albumin (BSA) in 1× PBS for 1 hour at RT. Human plasma in blocking solution was applied at a 1:100 dilution and incubated overnight at 4°C. Mouse secondary biotinylated anti-human antibodies, specific for IgG1, -2, -3, and -4 and IgE isotypes (Sigma-Aldrich, Saint Louis, MO), were applied at a 1:100 dilution in blocking buffer, covered from light, and incubated overnight at 4°C. Plates were incubated with streptavidin-conjugated alkaline phosphatase (Sigma-Aldrich) diluted 1:500 in TBS-MgCl2/1% BSA solution for 2 hours at RT and covered from light. After a final wash, para-nitrophenyl phosphate (pNPP) substrate buffer (Sigma-Aldrich) was applied, and the plates were incubated for 30 minutes at RT and read at 405 nm on a SpectraMax M2 microplate reader (Molecular Devices, Sunnyvale, CA). IgG and IgE standards were used to calculate donor anti-SGS antibody concentrations by standard curve fitting.
Proliferation assays.
Monocytes from cryopreserved mixed leukocyte samples of human donors were collected by adherence to six-well cell culture plates (Costar, Manassas, VA) after quick thaws (1 minute in a 37°C water bath), washing in RPMI-C media, and a 2-hour incubation at 37°C under 5% CO2. Cells were stimulated every other day for 1 week with recombinant human granulocyte/macrophage colony-stimulating factor (GM-CSF; 400 U per 1 × 106 monocytes) and IL-4 (160 U per 1 × 106 monocytes) to generate monocyte-derived dendritic cells (MDDCs). Cells were harvested by repeated washing with 0.5 mM EDTA in 1× PBS, replated in a 96-well flat-bottom cell culture plate at a concentration of 4 × 104 MDDCs per well in RPMI-C media, and then, allowed to rest overnight at 37°C under 5% CO2. Cells were stimulated with SGS overnight at a concentration of one salivary gland equivalent (1.5 μg) per well. Negative control wells received only RPMI-C media, whereas positive control wells received 400 ng phytohemagglutinin (PHA; Sigma-Aldrich) per well. Mixed leukocytes from the same donors were coincubated with stimulated MDDCs at a concentration of 8 × 104 per well at 37°C under 5% CO2 for 4 days; 1 μCi tritiated thymidine (Perkin-Elmer, Waltham, MA) in RPMI-C was added to each well for the final 24 hours of coculture. Mixed cultures were harvested using a scintillation harvester and filter mat paper (Brandel, Gaithersburg, MD) per the manufacturer's protocol. Scintillation fluid allowed β-particle decay readings on a Wallac MicroBeta TriLux microplate liquid scintillation counter (Perkin-Elmer). Final values are reported as the stimulation index—the ratio of β-counts (3H-thymidine uptake) between unstimulated and SGS pre-stimulated MDDCs and mixed leukocyte cocultures.
Luminex cytokine quantification.
Culture supernatants from the proliferation reactions were collected just before 3H-thymidine addition. Supernatants were prepared for quantitative cytokine and chemokine fluorescence-based analysis using a Human Cytokine LINCOplex Premixed 96-well kit (Millipore, Billerica, MA) following the manufacturer's protocols. Plates were read on a Luminex 200 machine, and the mean fluorescence intensity (MFI) was calculated following the manufacturer's protocols for internal normalization. Final MFI values for each of 21 cytokines or chemokines in supernatants from each culture exposed in vitro to SGS were subsequently normalized to supernatant values of paired negative control cultures left unexposed to SGS.
Clustering analysis.
K-means clustering was performed using Cluster v.3.0.38 Data were formatted as sample (plasma donor) and gene (SGS protein kilodaltons) tables to generate k = 4 clusters using dataset means measured by Euclidean distance. Visualization and data export were achieved with Java Treeview v.1.1.4.39
Statistics.
Using GraphPad Prism v.5.0 (Graphpad Software, San Diego, CA), combined immunoblot scoring data were analyzed by one-way analysis of variance (ANOVA) and Bonferroni multiple comparison post-tests performed at each SGS protein kilodalton for every regional plasma donor group at a 95% confidence interval. A simple calculation was used to describe immunogenicity of each salivary protein: (probability of donor plasma antibody specificity to a particular salivary protein within a regional sample group) × (average band intensity among that regional group) = arbitrary units (a.u.) of immunogenicity between zero and two. The same software was used to calculate Pearson correlation by two-tailed distribution using percentages of individuals from each single region that clustered into the four k-means clusters as well as Student's t test comparing regional groups mixed leukocyte proliferation data.
Results
Host antibody reactivity to vector salivary proteins.
To better understand human anti-sand fly salivary antibody presence based on differential exposure to wild P. papatasi sand flies, six sites were selected as collection points for human host peripheral blood samples. Each regional site, three sites in Egypt and three sites in Jordan, was categorized into different P. papatasi and leishmaniasis ecotopes (Table 1). Additionally, US military personnel represented responses by individuals immunologically naive to newly encountered sand fly saliva. Plasma from a total of 111 Egyptian or 118 Jordanian residents and 69 US military personnel was tested against PAGE-separated SGS from inbred female P. papatasi by Western immunoblot (Figure 1A and Supplemental Figure 2). Concurrently, PAGE-separated SGS revealed 11 heavily expressed salivary proteins (∼12, 14, 26, 28, 30, 38, 40, 42, 44, 46, and 58 kDa), with 1 modestly expressed protein of ∼68 kDa (Figure 1B). Human donor blood plasma antibodies displayed differential specificities for those proteins (Figure 1). Images of immunoblots were scored by signal intensity of plasma antibodies specific for P. papatasi salivary proteins at each approximate molecular weight (MW). All final immunoblot scores at each MW for Egyptian and Jordanian donors were compared with those scores of donors residing in control regions, where no sand fly vectors were reported to be present. US military donor scores were compared with those scores of control donors who denied any travel to P. papatasi-endemic regions. Antibody specificities for five salivary proteins (∼12, 26, 30, 38, and 44 kDa) were found to be significantly different from control regions within one or both Middle Eastern nations, whereas antibody specificity was significantly different compared with the control group for only two salivary proteins (∼14 and 38 kDa) among Iraq-based US donors infected with Leishmania (P < 0.05) (Table 2). Notably, among Middle Eastern donors, there seemed to be very little, if any, specificity for salivary proteins of ∼15 or 21 kDa (data not shown) and some minor differential recognition of an ∼36 kDa protein that was not observed to be a majorly expressed protein in the P. papatasi laboratory colony SGS (Figure 1B). However, a salivary protein at ∼14 kDa was significantly differentially recognized by antibodies from US military donors.
Table 2.
Characteristics of salivary proteins with significant differences in host antibody specificity profiles
| Salivary proteins (estimated MW) | |||||
|---|---|---|---|---|---|
| 12 kDa | 26 kDa | 30 kDa | 38 kDa | 44 kDa | |
| MENA* | |||||
| Percentage positive sera† | |||||
| Egypt | |||||
| CA (−/−) | 43.24 | 27.03 | 37.84 | 45.95 | 8.11 |
| AW (+/−) | 90.00‡ | 70.00‡ | 90.00‡ | 95.00‡ | 42.50‡ |
| NS (+/+) | 79.41‡ | 55.88‡ | 88.24‡ | 79.41‡ | 41.18‡ |
| Jordan | |||||
| AM (−/−) | 56.76 | 21.62 | 59.46 | 70.27 | 13.51 |
| MA (+/−) | 58.54 | 21.95 | 78.05‡ | 73.17 | 12.20 |
| SW (+/+) | 77.50‡ | 35.00 | 90.00‡ | 60.00 | 45.00‡ |
| Average blot intensity§ | |||||
| Egypt | |||||
| CA (−/−) | 0.51 ± 0.08 | 0.38 ± 0.13 | 0.62 ± 0.15 | 0.59 ± 0.12 | 0.14 ± 0.11 |
| AW (+/−) | 1.31 ± 0.15‡ | 0.90 ± 0.11‡ | 1.83 ± 0.16‡ | 1.33 ± 0.11‡ | 0.70 ± 0.12‡ |
| NS (+/+) | 1.26 ± 0.18‡ | 0.96 ± 0.17‡ | 2.09 ± 0.18‡ | 1.38 ± 0.17‡ | 0.84 ± 0.15‡ |
| Jordan | |||||
| AM (−/−) | 0.66 ± 0.10 | 0.24 ± 0.14 | 0.86 ± 0.15 | 0.97 ± 0.08 | 0.22 ± 0.11 |
| MA (+/−) | 0.76 ± 0.06 | 0.29 ± 0.15 | 1.37 ± 0.16‡ | 1.15 ± 0.10 | 0.13 ± 0.12 |
| SW (+/+) | 1.08 ± 0.14‡ | 0.41 ± 0.15 | 1.53 ± 0.15‡ | 0.94 ± 0.10 | 0.75 ± 0.12‡ |
| Immunogenicity (a.u.)¶ | |||||
| Egypt | |||||
| CA (−/−) | 0.22 | 0.10 | 0.24 | 0.27 | 0.01 |
| AW (+/−) | 1.18‡ | 0.63‡ | 1.64‡ | 1.26‡ | 0.30‡ |
| NS (+/+) | 1.00‡ | 0.53‡ | 1.84‡ | 1.10‡ | 0.35‡ |
| Jordan | |||||
| AM (−/−) | 0.38 | 0.05 | 0.51 | 0.68 | 0.03 |
| MA (+/−) | 0.44 | 0.06 | 1.07‡ | 0.84 | 0.02 |
| SW (+/+) | 0.83‡ | 0.14 | 1.37‡ | 0.56 | 0.34‡ |
| 12 kDa | 14 kDa | 26 kDa | 30 kDa | 38 kDa | |
| US military* | |||||
| Percentage positive sera† | |||||
| United States | |||||
| Control | 48.28 | 51.72 | 44.83 | 48.28 | 48.28 |
| Bitten | 23.53 | 52.94 | 64.71 | 29.41 | 76.47 |
| Infected | 34.78 | 13.04‡ | 30.43 | 21.74 | 73.91‡ |
| Average blot intensity§ | |||||
| United States | |||||
| Control | 0.52 ± 0.57 | 0.53 ± 0.53 | 0.50 ± 0.60 | 0.64 ± 0.77 | 0.48 ± 0.51 |
| Bitten | 0.24 ± 0.44 | 0.71 ± 0.85 | 0.74 ± 0.62 | 0.29 ± 0.47 | 1.00 ± 0.71 |
| Infected | 0.52 ± 0.85 | 0.30 ± 0.82‡ | 0.48 ± 0.79 | 0.30 ± 0.70 | 1.70 ± 1.19‡ |
| Immunogenicity (arbitrary units)¶ | |||||
| United States | |||||
| Control | 0.25 | 0.28 | 0.22 | 0.31 | 0.23 |
| Bitten | 0.06 | 0.37 | 0.48 | 0.09 | 0.76 |
| Infected | 0.18 | 0.04‡ | 0.15 | 0.07 | 1.25‡ |
Collection site abbreviations and conditional marks correspond to those abbreviations and marks presented in Table 1 (e.g., +/− refers to a region with known endemic P. papatasi populations but no reports of L. major infections or for US military personnel, donor reported history of sand fly bites but there is no confirmed diagnoses of cutaneous leishmaniasis).
Percentage of serum donors from the given location with antibody recognition of salivary proteins at the estimated MW.
Those regions with significant (P ≤ 0.05) differences in antibody reactivity compared with national negative control regions or US personnel who were not bitten by sand flies.
Western immunoblots of human sera against P. papatasi SGS were scored manually by three independent observers using a scale: 0 = no reaction; 1 = weak reaction; 2 = moderate reaction; 3 = intense reaction (all judged relative to a constant positive control).
Immunogenicity here is calculated as (probability of donor serum antibody specificity to a particular salivary protein within a regional sample group) × (average band intensity among that regional group) = a.u. of immunogenicity.
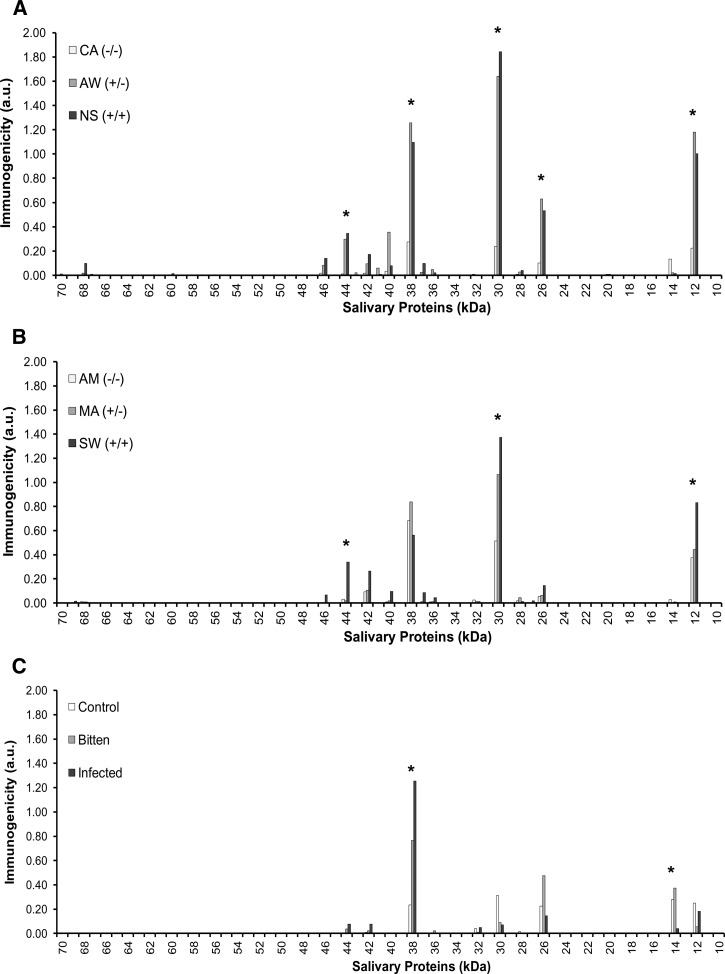
Antibody specificity for salivary proteins can be summarized using a novel calculation of immunogenicity (Figure 2). Regarding Egyptian and Jordanian donors, plasma displayed the greatest immunogenicity against salivary proteins at ∼12, 30, and 38 kDa, regardless of geographic origin. Greater relative difference to control regions was more readily apparent among plasma from Egyptian donors. Jordanian donor plasma from Amman, a control region, exhibited much higher immunogenicity to those three salivary proteins compared with donor plasma from Cairo, the Egyptian control region (Figure 2 and Table 1). This increased antibody specificity for three highly immunogenic salivary proteins, in part, accounts for the lack of statistical significance in testing differences between intraregional profiles for Jordanian donors. The percentage of individuals reacting to the five significant salivary proteins was greater among individuals residing in AW compared with MA (Table 2). However, between NS and SW, the percent of persons reacting to salivary proteins of ∼12, 30, and 44 kDa was similar, although more Egyptian donors displayed antibodies against salivary proteins of ∼26 and 38 kDa.
Figure 2.
Immunogenicity to P. papatasi salivary gland proteins among plasma donors. (A) Immunogenicity of all P. papatasi salivary gland proteins by Egyptian blood plasma donor antibody specificities from three regions (CA, AW, and NS) using consensus estimates of MW on immunoblot images from three independent observers. (B) Immunogenicity of all P. papatasi salivary gland proteins by Jordanian blood plasma donor antibody specificities from three regions (AM, MA, and SW). Ecological conditions in the regional key are the same as those conditions outlined in Table 1. (C) Immunogenicity of all P. papatasi salivary gland proteins of Iraq-based US personnel and blood plasma donor antibody specificities from individuals differentially exposed to sand fly bites and Leishmania parasites. Exposure criteria are the same as those criteria outlined in Table 1. Immunogenicity is calculated as (probability of donor plasma antibody reaction of protein among a regional group) × (average of band intensity among that regional group) = a.u. *Significant difference(s) in host plasma antibody specificity for protein(s) at these MWs between sand fly-endemic and control regional sites or between sand fly bite-exposed military personnel and unexposed control donors (P ≤ 0.05).
Considering the immunogenicity of salivary proteins to US military donors, values for the ∼38 kDa salivary protein were substantially greater than those values of all other proteins (Figure 2). Although 64% of bite-exposed individuals displayed specificity for the ∼26 kDa salivary protein (Table 2), signal intensity and immunogenicity were not significantly different from signal intensity and immunogenicity of control donor plasma. Although the immunogenicity of an ∼14 kDa protein was low relative to the immunogenicity of an ∼38 kDa protein, a statistically significant difference between Leishmania-infected donors and controls was found (P < 0.05, Student's t test).
Despite significant relative differences of raw immunoblot scores to the control region for Egyptian donors, immunogenicity values for salivary proteins at ∼26 and 44 kDa were low compared with those values of ∼12, 30, and 38 kDa proteins. Both the 26 and 44 kDa proteins were found in close relative proximity to one or more proteins of similar MW (Figure 1B). Because each donor plasma profile displayed differential specificity for majorly expressed salivary proteins, manual scoring may have been imprecise for proteins at those two MWs. Therefore, we took additional steps to more thoroughly characterize both the majorly expressed salivary proteins and the global single human donor plasma antibody specificity profiles.
Sand fly salivary protein identification.
To more thoroughly differentiate and characterize abundantly expressed P. papatasi salivary proteins, 10 bands were excised from SGS segregated by denaturing PAGE (∼12, 14, 26, 28, 30, 38, 40, 42, 44, and 46 kDa). MS analyses were performed on the purified proteins after trypsin digestion. Resultant spectra were compared against all available NCBI protein database sequences for Phlebotomus and Lutzomyia spp.33 Table 3 outlines the top protein matches, ordered by rank of best local match to our database, for spectra of each protein band. All top-ranked matches were against published protein sequences from the P. papatasi Israeli colony strain. Despite seemingly clear and unique in-gel MW differences for proteins in the 26 and 28 kDa bands as well as those proteins in the 40, 42, 44, and 46 kDa bands, multiple high-confidence protein matches were found for each analyzed band. Moreover, given the high homology in primary sequence for PpSP28 and PpSP30 (top matches for the ∼26 and 28 kDa bands) as well as between PpSP42 and PpSP44 (top matches for the ∼40, 42, 44, and 46 kDa bands), identification was based predominantly on spectral matches to domains exclusively unique to each protein. Within the range of ∼26–30 kDa, we distinguished three primary salivary proteins that corresponded to differential donor antibody specificities (Figure 1B), where MS matches to the ∼26 and 28 kDa proteins were equally confident between PpSP28 and PpSP30. The band at ∼30 kDa matched best to a 29 kDa salivary protein precursor (gi∣76589378).
Table 3.
Identification of majorly expressed P. papatasi salivary proteins by MS
| P. papatasi salivary protein (estimated MW)*/rank | Unused | Sequence coverage† (%) | Accession number | Protein name | Species | No. of unique peptides | Confidence of identification‡ | Local FDR for protein (%) |
|---|---|---|---|---|---|---|---|---|
| 12 kDa | ||||||||
| 1 | 50.41 | 80 | gi 15963505 | 12 kDa salivary protein precursor | P. papatasi | 56 | ∼> 1e-50 | 7e-5 |
| 2 | 25 | 57.8 | gi 15963507 | 14 kDa salivary protein precursor | P. papatasi | 16 | ∼> 1e-25 | 7.3e-5 |
| 3 | 6 | 27.2 | gi 15963515 | 32 kDa salivary protein precursor | P. papatasi | 4 | ∼> 1e-6 | 7.54e-5 |
| 4 | 5.56 | 55.3 | gi 112497698 | 14.2 kDa salivary protein | P. duboscqi | 6 | ∼> 1e-5 | 7.66e-5 |
| 5 | 4 | 72.1 | gi 112497317 | 13.7 kDa salivary protein | P. duboscqi | 22 | ∼> 1e-4 | 7.85e-5 |
| 6 | 2 | 30.5 | gi 112497287 | 14.2 kDa salivary protein | P. duboscqi | 1 | 0.01 | 8.04e-5 |
| 14 kDa | ||||||||
| 1 | 33.3 | 86.1 | gi 335954166 | SP15 protein | P. papatasi | 50 | ∼> 1e-33 | 8.24e-5 |
| 2 | 11.18 | 86.1 | gi 335954098 | SP15 protein | P. papatasi | 44 | ∼> 1e-11 | 8.57e-5 |
| 3 | 6 | 28.2 | gi 15963507 | 14 kDa salivary protein precursor | P. papatasi | 3 | ∼> 1e-6 | 8.66e-5 |
| 4 | 4.05 | 90.9 | gi 112497496 | 14.5 kDa salivary protein | P. duboscqi | 39 | ∼> 1e-36 | 8.76e-5 |
| 5 | 4 | 10.6 | gi 15963515 | 32 kDa salivary protein precursor | P. papatasi | 2 | ∼> 1e-4 | 8.86e-5 |
| 6 | 2 | 39.4 | gi 112361953 | 14.4 kDa salivary protein | P. duboscqi | 3 | 0.01 | 9.05e-5 |
| 7 | 2 | 32.1 | gi 157361529 | 40S ribosomal protein S12-like protein | P. papatasi | 1 | 0.01 | 9.05e-5 |
| 8 | 0.26 | 86.1 | gi 335954106 | SP15 protein | P. papatasi | 40 | 0.55 | 9.55e-5 |
| 26 kDa | ||||||||
| 1 | 30.74 | 71.3 | gi 15963511 | 28 kDa salivary protein precursor | P. papatasi | 36 | ∼> 1e-30 | 8.78e-5 |
| 2 | 28.47 | 70 | gi 15963513 | 30 kDa salivary protein precursor | P. papatasi | 40 | ∼> 1e-28 | 8.85e-5 |
| 28 kDa | ||||||||
| 1 | 30.78 | 57.5 | gi 15963511 | 28 kDa salivary protein precursor | P. papatasi | 26 | ∼> 1e-30 | 4.69e-5 |
| 2 | 23.26 | 60.5 | gi 15963513 | 30 kDa salivary protein precursor | P. papatasi | 18 | ∼> 1e-23 | 4.72e-5 |
| 30 kDa | ||||||||
| 1 | 28.72 | 56.3 | gi 76589378 | 29 kDa salivary protein precursor | P. papatasi | 29 | ∼> 1e-28 | 9.06e-5 |
| 2 | 12 | 37.8 | gi 15963515 | 32 kDa salivary protein precursor | P. papatasi | 8 | ∼> 1e-12 | 9.25e-5 |
| 38 kDa | ||||||||
| 1 | 113.02 | 92 | gi 10443907 | Salivary apyrase | P. papatasi | 114 | ∼> 1e-113 | 4.94e-5 |
| 2 | 6.06 | 74.1 | gi 112496715 | 35.8 kDa salivary protein | P. duboscqi | 27 | ∼> 1e-6 | 5.21e-5 |
| 3 | 2.73 | 32 | gi 299829440 | 32.9 kDa salivary protein | P. sergenti | 7 | ∼> 1e-3 | 5.31e-5 |
| 40 kDa | ||||||||
| 1 | 72.21 | 80.8 | gi 15963517 | 42 kDa salivary protein precursor | P. papatasi | 70 | ∼> 1e-72 | 5.08e-5 |
| 2 | 9.68 | 51.1 | gi 112497236 | Yellow-related protein | P. duboscqi | 29 | ∼> 1e-9 | 5.15e-5 |
| 3 | 7.52 | 69.1 | gi 10443907 | Salivary apyrase | P. papatasi | 6 | ∼> 1e-72 | 5.24e-5 |
| 42 kDa | ||||||||
| 1 | 120.09 | 89.6 | gi 15963517 | 42 kDa salivary protein precursor | P. papatasi | 110 | ∼> 1e-120 | 1.04e-3 |
| 2 | 21.3 | 73 | gi 15963519 | 44 kDa salivary protein precursor | P. papatasi | 13 | ∼> 1e-21 | 1.09e-3 |
| 3 | 15.46 | 65.6 | gi 112497236 | Yellow-related protein | P. duboscqi | 46 | ∼> 1e-15 | 1.11e-3 |
| 4 | 4 | 55.2 | gi 299829410 | Yellow-related salivary protein | P. duboscqi | 19 | ∼> 1e-4 | 1.12e-3 |
| 44 kDa | ||||||||
| 1 | 91.06 | 85.8 | gi 15963517 | 42 kDa salivary protein precursor | P. papatasi | 100 | ∼> 1e-90 | 6.72e-7 |
| 2 | 81.47 | 80.3 | gi 15963519 | 44 kDa salivary protein precursor | P. papatasi | 97 | ∼> 1e-81 | 6.72e-7 |
| 3 | 15.45 | 61.8 | gi 112497236 | Yellow-related protein | P. duboscqi | 49 | ∼> 1e-15 | 6.75e-7 |
| 4 | 6.13 | 59.7 | gi 112497202 | Yellow-related protein | P. duboscqi | 41 | ∼> 1e-6 | 6.76e-7 |
| 46 kDa | ||||||||
| 1 | 130.07 | 98.3 | gi 15963519 | 44 kDa salivary protein precursor | P. papatasi | 188 | ∼> 1e-130 | 1.1e-5 |
| 2 | 43.93 | 77.2 | gi 15963517 | 42 kDa salivary protein precursor | P. papatasi | 41 | ∼> 1e-43 | 1.1e-5 |
| 3 | 8.89 | 74.7 | gi 112497202 | Yellow-related protein | P. duboscqi | 72 | ∼> 1e-8 | 1.11e-5 |
| 4 | 6.02 | 21.9 | gi 76446619 | Yellow-related salivary protein | P. perniciosus | 3 | ∼> 1e-6 | 1.12e-5 |
| 5 | 4.29 | 35.1 | gi 10443907 | Salivary apyrase | P. papatasi | 3 | ∼> 1e-4 | 1.12e-5 |
| 6 | 2.26 | 50.1 | gi 112497236 | Yellow-related protein | P. duboscqi | 26 | ∼> 1e-3 | 1.13e-5 |
| 7 | 2 | 38.7 | gi 52001011 | Yellow-related salivary protein | P. papatasi | 1 | 0.01 | 1.14e-5 |
Based on denatured protein bands excised for MS analysis (Figure 1B).
Percent representation of a protein based on unique peptide spectral assignments to that protein.
Negative log10 of the unused ProtScore, which is calculated using only peptides from spectra that have not been used by other proteins; a lower value indicates greater confidence of protein identification-based additive assignments of unique peptides.
Clustering of host antibody anti-salivary protein profiles.
K-means clustering was performed on raw immunoblot score data, and therefore, the complete global antibody specificity profile for each individual donor was simultaneously considered. This approach helped to better characterize the contribution that regional and ecological characteristics played on host-acquired immunity to sand fly salivary proteins by distinguishing common patterns within the large complex dataset. K-means clustering was performed first with Egyptian and Jordanian donor immunoblot data using k = 6 (one cluster for each significant protein plus a null control) and again with k = 5 (the number of significant proteins). No distinguishing or consistently similar data patterns were observed using these numbers of clusters (data not shown). However, on using k = 4 clusters (Supplemental Figure 3), data patterns emerged that were both consistent among donors within each single cluster and significantly divergent between clusters by cumulative immunoblot intensity scores for the five immunogenic salivary proteins (P < 0.05, Student's t test). Similarly, in clustering immunoblot data from the US military donors, the use of k = 4 clusters generated cluster profiles with distinct patterns of specificity against the ∼38, 30, 26, 14, and 12 kDa salivary proteins (Supplemental Figure 4). Each cluster was also significantly divergent in this dataset by cumulative immunoblot intensity scores for this different set of five distinguishing salivary proteins (P < 0.05, Student's t test). Table 4 outlines the characteristic plasma antibody specificity patterns for each of the five immunogenic salivary protein sets.
Table 4.
Principle salivary proteins in K-means clusters (estimated MW)
| 12 kDa | 26 kDa | 30 kDa | 38 kDa | 44 kDa | |
|---|---|---|---|---|---|
| MENA clusters | |||||
| A | − | − | − | − | − |
| B | + | − | + | + | − |
| C | + | + | + | + | − |
| D | + | + | + | + | + |
| 12 kDa | 14 kDa | 26 kDa | 30 kDa | 38 kDa | |
| US military clusters | |||||
| E | − | − | − | − | − |
| F | − | − | − | − | + |
| G | − | + | + | + | + |
| H | + | + | − | + | − |
Cluster characteristics are defined as antibody recognition at the listed protein MW for > 50% of donors. Results details descriptions of clusters.
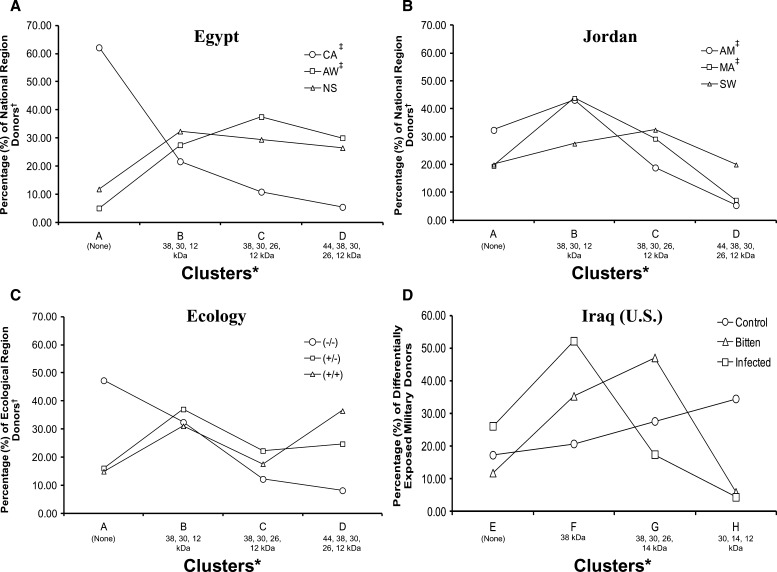
Clusters A–D pertain to Egyptian and Jordanian resident datasets. Cluster A is a null response cluster (populated predominantly by control region donors). Cluster B is a cluster with antibodies specific to ∼12, 30, and 38 kDa salivary proteins (minor difference between the numbers of donors from each region). Cluster C is a cluster with predominant specificity to ∼12, 30, 38 and 26 kDa proteins (again, no substantial differences in regional donor numbers). Cluster D is a cluster with specificity to ∼12, 26, 30, 38, and 44 kDa salivary proteins (substantially more donors residing in sand fly- and leishmaniasis-endemic regions compared with controls). Only the percentages of Egyptian CA and AW donors displayed a significant inverse Pearson correlation across clusters (P < 0.05, r = −0.94). However, the percentages of donors from both AW and NS trended positively across clusters, whereas the percentages of CA donors trended negatively (Figure 3A). For Jordanian donors, a significant positive correlation was found between AM and MA donors across clusters (P < 0.05, r = 0.96), and no correlations were observed between AM and SW (Figure 3B). No correlations existed across clusters between all donor immunoreactivity profiles combined by ecotope (Figure 3C).
Figure 3.
K-means clustering of total host plasma antibody specificity profiles to P. papatasi whole-salivary gland protein sonicates. Scores for human host plasma specificity to sand fly salivary proteins were separated by national and ecological region groups and then clustered by the k-means method across 1,000 iterations using Cluster v3.0, with k = 4 clusters and the Euclidean distance metric. (A) Total percentages of blood plasma donors from each Egyptian regional site assigned to each anti-SGS antibody specificity profile cluster by k-means clustering. (B) Total percentages of blood plasma donors from each Jordanian regional site assigned to each anti-SGS antibody specificity profile cluster by k-means clustering. (C) Total percentages of blood plasma donors from combined regional sites (grouped by defining ecological characteristics) (Table 1) assigned to each anti-SGS antibody specificity profile cluster by k-means clustering. (D) Total percentages of blood plasma donors from US military personnel differentially exposed to sand fly bites and Leishmania parasites assigned to each anti-SGS antibody specificity profile cluster by k-means clustering. *K-means cluster profile characteristics are outlined in Table 4. ‡A significant Pearson correlation was found between international donors from these sites.
Clusters E–H pertain to the US military donor dataset. Cluster E is a null response cluster (populated predominantly by control region donors). Cluster F is a cluster with antibodies specific almost exclusively to the ∼38 kDa salivary protein (a substantial difference in representation from each exposure group, where the greatest percentage of individuals was from the Leishmania-infected group). Cluster G is a cluster with predominant specificity to ∼14, 30, 26, and 38 kDa proteins (again, a substantial difference in representation from each exposure group, where the greatest percentage was from the bitten but not infected group). Cluster H is a cluster with specificity to ∼12, 14, and 30 kDa salivary proteins (a cluster populated largely of control donors). No statistically significant correlations existed between exposure groups across clusters E–H. The distribution of donors favored a single cluster, where one anti-salivary gland protein specificity profile was represented predominantly by donors of a single exposure type group (Figure 3D).
Anti-SGS antibody isotype assessments.
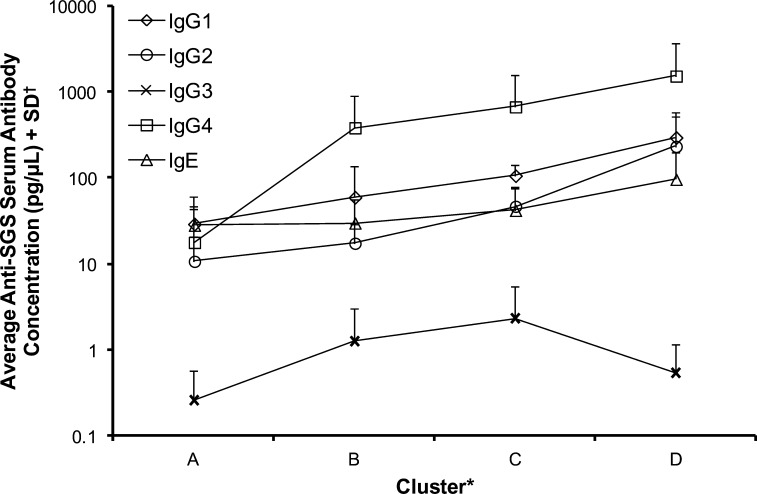
To further understand the acquired antibody-based immunity to sand fly salivary proteins, the six most representative donor plasmas from each of the MENA regional resident study sites (associated with each of the four k-means clusters) were assessed for specific anti-SGS Ig isotype classifications (Figure 4 and Supplemental Figure 5). Compared with concentrations for all other antibody isotypes, significantly greater cumulative concentrations of anti-SGS IgG4 were present in donor plasma (P < 0.05) (Figure 4). Notably, when data for all Ig isotypes was analyzed cumulatively, a significant, albeit weak, positive Pearson correlation was observed across the four clusters (P = 0.01, r = 0.23). When each isotype was analyzed separately, IgG1, −2, and −4 and IgE concentrations were positively correlated across the clusters (P = 0.01–0.05, r = 0.42–0.54). Only IgG3 did not correlate with clusters, likely as a result of such low relative concentrations. These correlations logically follow the defining characteristics of each cluster's immunoreactivity profile, wherein progressively, there is greater specificity to more salivary proteins in clusters A–D. Evidence exists for a correlation between anti-mosquito salivary IgG4 and IgE expression, which contributes to host type 1 hypersensitivity.40 Here, anti-P. papatasi SGS IgG4 and IgE concentrations did correlate modestly (P = 0.02, r = 0.5). Whether this correlation is actually related to the establishment of a true hypersensitivity among donors residing in our ecotopes of study remains unknown.
Figure 4.
Isotyping of anti-SGS human host plasma antibodies. For each k-means cluster, six plasmas most representative of that k-means cluster immunoreactivity profile and plasmas from individuals residing in each of the Middle Eastern regions were isotyped for total anti-SGS antibodies using custom indirect ELISA. Individual donor results for each antibody isotype may be found in Supplemental Figure 5. *Clusters are like those clusters described in Table 4. †Anti-SGS antibody concentrations were assessed by secondary antibody detection of a known concentration of purified Ig isotype standards, plated in dilution series as primary antigen, and fitted to the resultant standard curve; final values were averaged per cluster across all regional donors.
Cellular proliferation analysis.
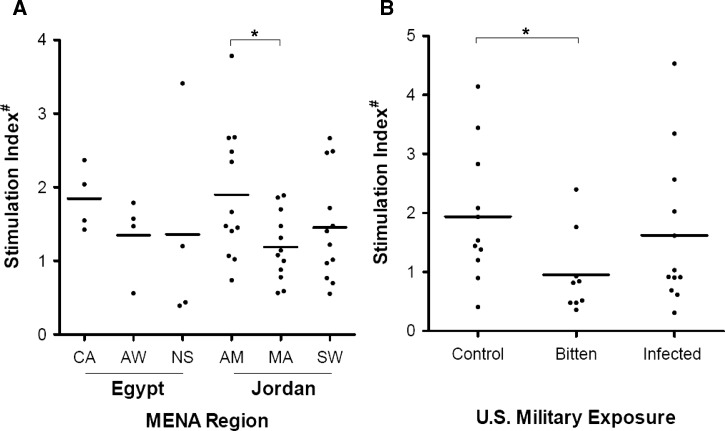
To assess donor cellular responsiveness to P. papatasi salivary proteins, peripheral blood mononuclear cells from a random subset of donors from each regional location and exposure group were assessed in a proliferation assay. Monocytic cells were adherence-selected from cryopreserved donor leukocytes and differentiated to dendritic cells by cytokine stimulation. Donor MDDCs were exposed to SGS 1 day before coculture with remaining autogenic leukocytes. Cellular proliferation between SGS pre-stimulated and unstimulated cocultures was measured by incorporation of tritiated thymidine (Figure 5). MDDCs of donors residing in sand fly-endemic regions elicited, on average, less proliferation after SGS exposure than those MDDCs from control region donors (Figure 5A). However, only the stimulation ratios compared between AM and MA site donors were significantly different (Student's t test, P < 0.05). Moreover, Iraq-deployed but uninfected US military donors exhibited less SGS-induced proliferation compared with control donors (Figure 5B).
Figure 5.
Mixed leukocyte proliferation in vitro of peripheral blood donors after stimulation of MDDCs with P. papatasi SGS. (A) Representative sampling of regional Egyptian and Jordanian resident donor cellular proliferations when exposed to SGS-stimulated autogeneic MDDCs. Proliferation was measured using coculture with tritiated thymidine for 24 hours and β-particle decay counts per minute. Values presented are ratios of stimulated over unstimulated assays normalized to a single positive control donor proliferation run concurrently. (B) Proliferation data for US military personnel differentially exposed to sand fly bites and Leishmania parasites. *Significant differences by Student's t test (P < 0.05). #Stimulation index is the ratio of β-counts (3H-thymidine uptake) between unstimulated and SGS pre-stimulated MDDCs and mixed leukocyte cocultures.
Cytokine secretion profiles.
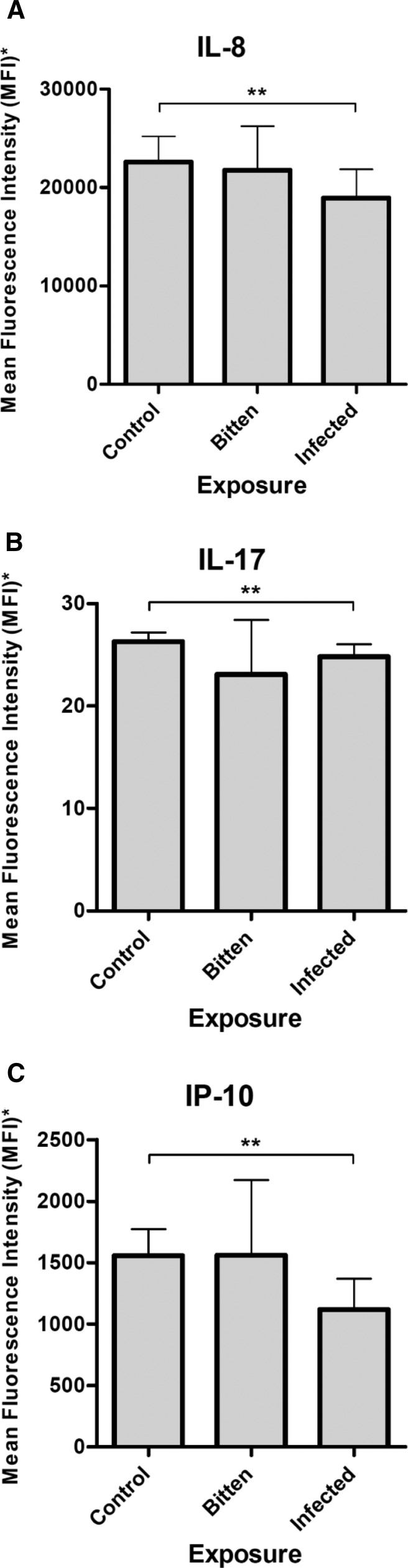
To better understand cellular sensitization of immunologically naïve individuals to P. papatasi salivary proteins, proliferation culture supernatants of Iraq-based military donors exposed to SGS were analyzed to quantify 21 secreted cytokine concentrations. Interleukin-8 (IL-8), IL-17, and interferon-γ–induced protein-10 (IP-10) of SGS-stimulated donor cultures were significantly greater in the supernatants of control exposure group donors than the supernatants of cells from the Leishmania-infected donors (P < 0.05, Student's t test) (Figure 6).
Figure 6.
Secreted cytokines/chemokines of SGS-stimulated MDDCs. Culture supernatant expression of (A) IL-8, (B) IL-17, and (C) IP-10 secreted by in vitro MDDCs from differential sand fly- and Leishmania-exposed US military personnel blood donors after stimulation with SGS. *Data are MFI + SD after normalization of SGS-stimulated MDDC supernatants to paired unstimulated supernatant values. **Significant differences by Student's t test (P < 0.05).
Discussion
We present here an examination of human host responses to P. papatasi salivary proteins. The overarching goal of this study was to understand the role of natural exposure to wild sand flies by characterizing host-acquired immunity dynamics with analyses of circulating antibody specificities and isotypes to whole-sand fly SGSs as well as the memory response of among leukocytes from those same donors. Together, these data serve to further our understanding of host responses to sand fly vector salivary proteins. Sites were selected in both Egypt and Jordan by presumed differential exposure of residents to wild P. papatasi populations. In the context of human antibody specificities against SGS proteins, a starker contrast between ecotope sites was observed among Egyptian sites compared with sites in Jordan (Figure 2A and B and Table 2). All collection sites in Egypt were located farther apart from one another compared with Jordanian collection sites (100+ miles versus ∼20 miles between sites, respectively), with far less intersite human traffic. Such proximity and easier travel throughout Jordan may be a factor contributing to SGS reactivity of donors residing in control regions. However, in addition to potential donor travel to endemic P. papatasi regions, the possibility of exposure to non-sand fly hematophagous insects, generating antibodies with specificity to conserved protein domains, should not be discounted.41
Our data for MENA residents compare well with the data in the work by Marzouki and others,12 which analyzed plasma antibody specificity of Tunisian donors to P. papatasi salivary proteins. Marzouki and others12 found that the majority of their donors exhibited positive reactivity to salivary proteins at ∼12, 15, 21, 28, 30, 36, and 44 kDa. Sand fly vectors in Tunisia are phylogenetically distinct from sand flies endemic to Egypt, Jordan, and Israel, which are part of a separate, albeit somewhat heterogeneous, population.42 The laboratory-reared P. papatasi used in the present study originated from Israel.
Glycoinositolphospholipids (GIPLs) on the surface of Trypanosoma spp. have been shown to alter the salivary profile of their insect vector,43 invoking the possibility that cross-strain variability among Leishmania spp. might affect vector saliva profiles and consequently, change human host antisalivary protein antibody reactivity. Failure of Egyptian and Jordanian donor plasma antibodies to recognize the salivary protein at ∼14 kDa (Figure 1), which was identified by Marzouki and others,12 may be because of the differential expression of this protein in distinct sand fly populations. However, US military personnel exposed to sand fly bites in Iraq displayed specificity to the protein at ∼14 kDa. The fact that this previously naïve group produced antibodies specific to this protein and endemic residents did not highlights the difference between chronic exposure to P. papatasi bites versus initial contact with new antigens in immunologically naive persons. Indeed, the study by Marzouki and others12 used blood samples collected from children between the ages of 6 and 12 years. The young age of donors in that study, likely lacking the prolonged history of sand fly bite exposure encountered by adult endemic residents in our current work, may account for the differential specificities observed against the ∼14 kDa salivary protein. The fact that the immunogenicity to this protein among US personnel was significantly less within the Leishmania-infected donor group compared with the sand fly bite only donors and controls suggests a possible role of the parasite in diminishing host antibody production against the ∼14 kDa protein or a possible impact on sand fly expression or post-translational modification of the protein before host contact.
The lack of expression of an ∼21 kDa protein (Figure 1B) by our Israeli colony of P. papatasi could be because of differential salivary gene expression between variant vector populations, especially given the seasonal and regional differences in expression of select salivary protein transcripts.15 Moreover, differences in post-translational modifications of salivary proteins, such as glycosylation or phosphorylation patterns,44 between the Tunisian and Israeli colonies may have led to differences in protein identification and even antibody specificities. Both studies used manual scoring of immunoblots, and designations of positive specificities at particular MWs may not be consistent between studies (i.e., what our observers agreed was specificity at 26 and 38 kDa may have been, for Marzouki and others12 observers, specificity at 28 and 36 kDa). It should also be noted that Marzouki and others12 identify their linearized 21 kDA salivary protein as PpSP42; we also identified this protein, corresponding to linearized salivary proteins in the ∼40–46 kDa range.
To assess the possible biological significance of immunogenic profile characteristics, P. papatasi salivary proteins were identified by MS analysis. Proteins determined to be ∼26 and 28 kDa by denaturing PAGE matched with approximately equivalent coverage and confidence to PpSP28 and PpSP30. The mature MWs for PpSP28 (gi∣15963511) and PpSP30 (gi∣15963513) are 27.2 and 27.8 kDa, respectively, and both proteins share 82.1% primary sequence identity by pairwise alignments, with multiple homologous domains. This information suggests that immunoblot scores for proteins in the 26–28 kDa range may be for one or both proteins by cross-immunoreactivity. PpSP28 and PpSP30 transcripts were differentially expressed among wild-caught P. papatasi across sites and time.15 Hence, exposure to sand fly bites at various seasons throughout a year may influence global immune responsiveness to P. papatasi. Although all blood samples from the MENA region in this study were collected during a 3-week window of late summer in 2007, circulating anti-salivary antibodies may be a result of prior exposures. The functional predictions for PpSP28 and PpSP30 implicate their role in binding thromboxane A,41 a host factor involved in vasoconstriction and prothromobosis, a process that would be advantageous to stifle the promotion of effective vector blood feeding.
Our results revealed that one majorly expressed salivary protein of ∼30 kDa is immunodominant to other salivary proteins among MENA donors (Figure 2 and Table 2). Another report implicated a ∼30 kDa salivary protein as strongly immunodominant,13 and Marzouki and others11 recently postulated that the strongest degree of antibody specificity among pre-exposed human and murine hosts was against a 32 kDa P. papatasi salivary protein (PpSP32, gi∣15963515). We analyzed an SGS protein band of approximately the same MW by MS (Figure 1B). The best match from among all published phlebotomine sand fly protein sequences33 was PpSP29 (gi∣76589378) (Table 2). PpSP32 emerged as a secondary match for the same protein gel band. PpSP29 and PpSP32 share little homology between primary sequences by pairwise alignment (data not shown). Most P. papatasi protein sequences in our local database were those sequences published from work with the Israeli colony, the same colony used throughout our study. Marzouki and others12 performed experiments using a Tunisian strain of P. papatasi but performed identifications using the same approximate database of protein sequences. It follows logically that our MS analysis yielded matches with higher degrees of confidence compared with the study by Marzouki and others,12 which used a population of differential phylogenetic origin42 from the common protein database. Abdeladhim and others41 recently performed an exhaustive analysis of the Tunisian strain P. papatasi salivary proteome through translated Basic Local Alignment Search Tool (BLAST) alignments of RNA transcripts to the same common protein sequence database. PpSP29 was found to display homology to insect allergen 5 (AG5), a protein expressed in most hematophagous insects. Immunoreactivity to PpSP29, given its membership in the pan-vector AG5 protein family, could be generated by exposure to other insect vectors because of conserved immunogenic epitopes. PpSP32 was predicted to be a potential mucin based on similarities of glycosylation sites and thus far, has been shown to be exclusively produced by sand flies.45 Exposure to mucin-like PpSP32 might stimulate T-cell proliferation and cytokine production, which has been known to occur in response to other true mucins.46 Given the agreed importance of proteins at the 30-kDa range, future analysis using recombinant PpSP29 and PpSP32 will be beneficial to ascertain which protein is the true immunodominant salivary antigen.
Conversely, for Iraq-based US military donors, the most immunodominant P. papatasi salivary protein was one of ∼38 kDa (Figure 2 and Table 2). Marzouki and others11,12 also noted that Tunisian donor antibodies displayed high specificity to this protein (referred to as an ∼36 kDa protein) but not the same degree as the ∼30/32 kDa protein(s). The ∼38 kDa protein matched best to a salivary apyrase (gi∣10443907). Apyrases hydrolyze both adenosine triphosphate (ATP) and adenosine diphosphate (ADP) to adenosine monophosphate (AMP), thus destroying an important physiologic stimulus of host platelet aggregation released from damaged tissues and blood cells. Apyrases have been found in most every hematophagous arthropod.47 The stark dichotomy between two donor groups (presumably chronically exposed regional residents and previously immunologically naive US military) in terms of reactivity to the ∼30 and 38 kDa proteins may suggest a population scale-acquired immune specificity for a protein that could be more specific to the Phelobotomus genus (∼30 kDa), versus one that is more encountered from insect bites occurring in the United States (∼38 kDa), or at least to which immunologically naive individuals more readily produce antibodies against after initial exposures.
We used k-means clustering to identify patterns of plasma antibody immunoreactivity to SGS proteins across donors. Antibody specificity to the five salivary proteins that elicited significantly different immunoreactivity among MENA-endemic donors compared with control region data was the principle factor distinguishing clusters A–D (Table 4 and Supplemental Figure 3). Among Egyptian donors, CA and AW displayed a significant inverse Pearson correlation across clusters (P < 0.05, r = −0.94). No significant inverse correlation was observed between CA and NS, despite an inverse trend (Figure 3A). For Jordanian donors, a significant positive correlation was found between AM and MA donors across clusters (P < 0.05, r = 0.96), and no correlations were observed between AM and SW (Figure 3B). The shorter intersite distances and increased travel of residents in Jordan may have led to more homogeneous exposure conditions compared with the more distant and isolated Egyptian sites, accounting for the negative correlations in Jordan between control and exposure sites. Donors with strong immunoreactivity to the ∼12 and 38 kDa salivary proteins also displayed significant antibody specificity for the ∼30 kDa immunodominant protein. The concurrent presence of plasma antibodies to all proteins in these three MW ranges was the defining factor differentiating k-means clusters A from B (Table 4). By extension, immunoreactivity to all three salivary proteins (but not any one protein or pair of proteins) differentiated donors residing mostly in control regions from those donors residing in regions with endemic P. papatasi populations. Our data showed that immense antibody specificity to the ∼12 kDa protein (but not the ∼14 kDa protein) was present among many donors. Although the function of both proteins remains unknown, the differences in immunoreactivity could suggest divergent functions or expression patterns or differential antibody specificity caused by glycosylation. Given the conserved pan-vector expression of the ∼38 kDa salivary protein, the bloodfeeding function of proteins in the ∼30-kDa range, and the high degree of immunoreactivity to the ∼12 kDa protein, future investigations of sand fly salivary proteins should consider proteins in these three ranges in combination.
The same parameters used to cluster MENA immunoblot scores were used with US military personnel data. Although only two salivary proteins displayed immunogenicity significantly different between Leishmania-infected donors and unexposed controls (∼14 and 38 kDa), again, antibody specificity for five proteins was the defining characteristic for clusters E–H (Table 4 and Supplemental Figure 4). Like the MENA regional donor data, cumulative immunoblot scores for the five proteins (∼12, 14, 26, 30, and 38 kDa) were significantly different among donor members of each cluster (P < 0.05, Student's t test). The absence of specificity for proteins ∼44 kDa, the emergence of significantly differential specificity to the ∼14 kDa protein, and the apparent immunodominance of the ∼38 kDa protein (immunoreactivity to this protein alone defined the profile of cluster F) were key elements that defined the differences in profiles between US military donors and Egyptian or Jordanian donors. When assessing the exposure characteristics of donors in each of clusters E–H, the simple majority of Leishmania-infected donors made up the individuals grouped into cluster F, whereas cluster G was populated mostly by individuals who were bitten by sand flies but not infected (Figure 3D). Because cluster F is characterized by the singular specificity to the ∼38 kDa salivary protein, the average signal intensity of Leishmania-infected donors in that cluster to that protein was substantially greater than the average signal intensity of the bite exposure-only donors (2.2 versus 1.2, respectively). The same intracluster characteristics were true between donors of both groups in cluster G. Notably, the cluster with the greatest percentage of unexposed control donors was cluster H, characterized by antibody specificity for ∼12, 14, and 30 kDa salivary proteins. Specificity for the ∼30 kDa protein occurred in > 50% of all donors in this cluster H, but among unexposed control donors grouped into the cluster, the split was an even 50:50, making immunoreactivity to that protein a potentially conditional criteria for cluster definition (Supplemental Figure 2). The dominance of the ∼12 and 14 kDa protein specificity among control donors may suggest that it is these proteins that donors produce from circulating antibodies as a result of exposure to other hematophagous arthropods or that these proteins exhibit some post-translational modification to offer epitopes against which unexposed individuals have normal circulating antibodies.
Four majorly expressed proteins, ∼40, 42, 44, and 46 kDa, all matched with high coverage and confidence to either PpSP42 or PpSP44 (Table 3). Both proteins share 84.1% primary sequence homology and resemble yellow-related proteins, like those proteins found in L. longipalpis that function as antiinflammatory factors on dendritic cells.48 Because antibody specificities to both the ∼26 and 44 kDa proteins were differentiating characteristics between multiple k-means clusters among MENA donors (Table 4 and Supplemental Figure 3), the expression of such proteins in sand flies could occur in a conditional manner to elicit differential host responses. Despite that our data indicate the ∼26 and 44 kDa protein(s) to be less immunogenic than the other three major salivary proteins (Figure 2A and B and Table 2), the proposed functions and status in differentiating immunoreactivity clusters make the case for additional investigation of these proteins as combinatorial diagnostic markers of sand fly exposure.
Enzyme-linked immunosorbent assays (ELISAs) revealed that the dominant isotype and subclass of circulating anti-SGS antibodies is IgG4. Positive correlations were observed across k-means clusters for IgG1, -2, and -4 and IgE isotypes. We also observed a positive correlation between concentrations of IgG4 and IgE across clusters (P = 0.02, r = 0.5) (Figure 4). IgG4 and IgE are generalized as markers of Th2 immunity bias and allergenic type 1 hypersensitivity, respectively. The correlation between both antibody isotypes agrees with prior reports, where IgE production is dependent on Th2 responses and often associated with concurrent IgG4 production.49,50 Hence, because donors displayed circulating anti-SGS antibody profiles with progressive immunoreactivity for all five of the major salivary proteins (Table 2), a possibility emerged that allergic reactions to sand fly saliva could be part of the global host immunological response dictated by additive responses to those five proteins. These data further emphasize the notion that achieving acquired immunity to sand fly saliva may be the product of concurrent responses to multiple salivary proteins.
To broadly examine acquired cellular immunity, we monitored proliferation of donor leukocytes cocultured with autogeneic monocyte-derived dendritic cells pre-exposed to SGS. Using a random subset of MENA donor samples, we observed a trend to diminished cellular proliferation among donors from P. papatasi-endemic regions compared with those donors residing in control areas (Figure 5A), an observation similar to the observation by Rohousova and others.51 Despite the fact that the only proliferative responses between AM and MA were significantly different by Student's t test (P < 0.05), the trending patterns among both Jordanian and Egyptian donors would suggest that donors residing in regions with P. papatasi populations elicit a mitigated proliferative response on reexposure to sand fly saliva compared with unexposed donors. In similar fashion, MDDCs from Iraq-based military personnel donors bitten by sand flies but not infected with Leishmania induced significantly less cellular proliferation compared with unexposed donors (Figure 5B). A recent report52 showed that CD4+ T cells from sand fly-exposed humans stimulated with SGS expressed high levels of IFN-γ. When CD8+ T cells from the same donors were stimulated with SGS, high amounts of IL-10 were produced. When both cell types were cocultured and SGS was stimulated, only IL-10 was predominantly produced. However, when IL-10 was blocked during stimulation, cellular proliferation was substantially increased. The imbalance of Th1 and Th2 phenotypes at the lymphocyte point of response may also explain our observations in addition to potential reprogramming of antigen-presenting cells. Moreover, another report showed that P. papatasi saliva inhibited the function of murine dendritic cells in mice through increasing concentrations of free adenosine nucleosides and AMP.53 With the presence of the ∼38 kDa apyrase in our SGS, it may be possible that increased hydrolytic reactions by that protein to boost AMP levels in dendritic cell cultures could stymie the proliferative responses of subsequent mixed leukocyte cultures. No significant differences in stimulation indexes were observed when donors were grouped according to cluster membership (data not shown). Thus, although distinctive immunoreactivity profiles may allude to the biological activity induced by differential exposure characteristics, those same profiles do not seem to dictate overall MDDC licensing of immune responses on reexposure.
Culture supernatants from SGS-stimulated US military personnel donors were used to assess the production of secreted cytokines and chemokines. The significant difference in total cellular proliferation between control and sand fly bite-exposed donors suggested the likelihood of differential cell signaling profiles to drive or inhibit that response. Leishmania-infected donors produced significantly less secreted IL-8, IL-17, and IP-10 compared with unexposed control donors (P < 0.05, Student's t test) (Figure 6). IL-17 is a major proinflammatory signaling molecule in the context of cutaneous leishmaniasis infections shown to promote cutaneous disease progression and severity in the BALB/c mouse model.54 Bone marrow-derived dendritic cells pre-exposed to salivary gland extracts from the South American Leishmania sand fly vector L. longipalpis exhibited diminished IL-17 expression when secondarily challenged to inflammation.48 Moreover, peripheral blood mononuclear cells from human donors who self-healed from cutaneous disease expressed less IL-17 compared with individuals presenting with active disease after in vitro rechallenge with soluble Leishmania antigens.55 IL-8 and IP-10 (CXCL10) are potent chemotactic agents responsible for recruitment of neutrophils, monocytes, and other immune cell types and have been shown to be responsible for the abrogation of the visceral forms of leishmaniasis.56 Pre-exposure of murine cells to L. intermedia salivary sonicates resulted in decreased IP-10 expression during subsequent secondary exposures.57 The combination of both sand fly salivary protein and Leishmania parasite exposure may have generated immunological memory sufficient to significantly decrease the relative expression of IL-17, IL-8, and IP-10 after challenge with P. papatasi saliva alone.
Taken together, it seems that, as investigations go forward to identify vaccine candidates from the saliva of sand flies, a combinatorial approach will likely be necessary. Indeed, stronger immune responses are evident in hosts pre-exposed to homogenous laboratory colony strains of P. papatasi than heterogeneous wild populations,58,59 with protein polymorphisms implicated as a possible explanation for the phenomena; this finding further emphasizes the need for development of a clear universal baseline profile for host-acquired immunity to vector salivary components.
Supplementary Material
ACKNOWLEDGMENTS
We would like to express our gratitude to the Multi National Force and Observes (MFO) North Camp and the Preventive Medicine Service, 1st US Army Support Battalion, El-Arish, North Sinai, Egypt. In addition, the authors thank the Egyptian Ministry of Health for their support in this project. We are also grateful to Drs. Shaden Kamhawi and Jesus Valenzuela and their staff for providing us with salivary glands to begin our project and advising our initial work.
Disclaimer: The views expressed in this article are the views of the authors and do not necessarily reflect the official policy or position of the Department of Army or Navy, the Department of Defense, or the US Government. The study protocol was approved by the Naval Medical Research Unit No. 3 Institutional Review Board in compliance with all applicable federal regulations governing the protection of human subjects (Institutional Review Board #193, Department of Defense #NAMRU3.2006.0011). One author is a military service member, and some authors are employees of the US Government. This work was prepared as part of our official duties. Title 17 U.S.C. §105 provides that “Copyright protection under this title is not available for any work of the United States Government.” Title 17 U.S.C. §101 defines a US Government work as a work prepared by a military service member or employee of the US Government as part of that person's official duties.
Footnotes
Financial support: This work was supported by Defense Advanced Research Projects Agency Contract W911NF0410380 (to M.A.D.). Major funding was provided by the US Defense Advanced Research Projects Agency (BAA 04-12).
Authors' addresses: Nicholas S. Geraci, Rami M. Mukbel, Michael T. Kemp, Mariha N. Wadsworth, Gwen M. Stayback, Matthew M. Champion, Megan A. Bernard, and Mary Ann McDowell, Eck Institute for Global Health, University of Notre Dame, Notre Dame, IN, E-mails: ngeraci1@nd.edu, rmmukbel@just.edu.jo, mikemp@umich.edu, mwadswor@nd.edu, staybac@nd.edu, Matthew.M.Champion.8@nd.edu, mbernar2@nd.edu, and mcdowell.11@nd.edu. Emil Lesho, Walter Reed Army Institute of Research, Silver Spring, MD, and Walter Reed Army Medical Center, Washington, DC, E-mail: emil.p.lesho.mil@mail.mil. Mahmoud Abo-Shehada, Department of Basic Veterinary Sciences, Jordan University of Science and Technology, Irbid, Jordan, E-mail: mshehada@e-parasite.com. Iliano V. Coutinho-Abreu and Marcelo Ramalho-Ortigão, Department of Entomology, Kansas State University, Manhattan, KS, E-mails: iliano@ksu.edu and mortigao@ksu.edu. Hanafi A. Hanafi, Emadeldin Y. Fawaz, Shabaan S. El-Hossary, and David F. Hoel, Research Sciences Directorate, US Naval Medical Research Unit No. 3 (NAMRU-3), Cairo, Egypt, E-mails: yahanafi@yahoo.com, emadel-din.yehia.eg@namru3.med.navy.mil, shabaan.elhossary.ctr.eg@med.navy.mil, and david.hoel@med.navy.mil. Glenn Wortmann, Walter Reed Army Medical Center, Washington, DC, E-mail: glenn.w.wortmann@medstar.net.
References
- 1.Alvar J, Velez ID, Bern C, Herrero M, Desjeux P, Cano J, Jannin J, den Boer M. Team WHOLC Leishmaniasis worldwide and global estimates of its incidence. PLoS One. 2012;7:e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Konecny P, Stark DJ. An Australian case of New World cutaneous leishmaniasis. Med J Aust. 2007;186:315–317. doi: 10.5694/j.1326-5377.2007.tb00910.x. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization Leishmaniasis: Burden of Disease. 2012. http://www.who.int/leishmaniasis/burden/en/ Available at. Accessed August 15, 2012.
- 4.de Oliveira CI, Nascimento IP, Barral A, Soto M, Barral-Netto M. Challenges and perspectives in vaccination against leishmaniasis. Parasitol Int. 2009;58:319–324. doi: 10.1016/j.parint.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 5.Kobets T, Grekov I, Lipoldova M. Leishmaniasis: prevention, parasite detection and treatment. Curr Med Chem. 2012;19:1443–1474. doi: 10.2174/092986712799828300. [DOI] [PubMed] [Google Scholar]
- 6.Aquino DM, Caldas AJ, Miranda JC, Silva AA, Barral-Netto M, Barral A. Epidemiological study of the association between anti-Lutzomyia longipalpis saliva antibodies and development of delayed-type hypersensitivity to Leishmania antigen. Am J Trop Med Hyg. 2010;83:825–827. doi: 10.4269/ajtmh.2010.10-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barral A, Honda E, Caldas A, Costa J, Vinhas V, Rowton ED, Valenzuela JG, Charlab R, Barral-Netto M, Ribeiro JM. Human immune response to sand fly salivary gland antigens: a useful epidemiological marker? Am J Trop Med Hyg. 2000;62:740–745. doi: 10.4269/ajtmh.2000.62.740. [DOI] [PubMed] [Google Scholar]
- 8.Clements MF, Gidwani K, Kumar R, Hostomska J, Dinesh DS, Kumar V, Das P, Muller I, Hamilton G, Volfova V, Boelaert M, Das M, Rijal S, Picado A, Volf P, Sundar S, Davies CR, Rogers ME. Measurement of recent exposure to Phlebotomus argentipes, the vector of Indian visceral leishmaniasis, by using human antibody responses to sand fly saliva. Am J Trop Med Hyg. 2010;82:801–807. doi: 10.4269/ajtmh.2010.09-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gidwani K, Picado A, Rijal S, Singh SP, Roy L, Volfova V, Andersen EW, Uranw S, Ostyn B, Sudarshan M, Chakravarty J, Volf P, Sundar S, Boelaert M, Rogers ME. Serological markers of sand fly exposure to evaluate insecticidal nets against visceral leishmaniasis in India and Nepal: a cluster-randomized trial. PLoS Negl Trop Dis. 2011;5:e1296. doi: 10.1371/journal.pntd.0001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomes RB, Brodskyn C, de Oliveira CI, Costa J, Miranda JC, Caldas A, Valenzuela JG, Barral-Netto M, Barral A. Seroconversion against Lutzomyia longipalpis saliva concurrent with the development of anti-Leishmania chagasi delayed-type hypersensitivity. J Infect Dis. 2002;186:1530–1534. doi: 10.1086/344733. [DOI] [PubMed] [Google Scholar]
- 11.Marzouki S, Abdeladhim M, Abdessalem CB, Oliveira F, Ferjani B, Gilmore D, Louzir H, Valenzuela JG, Ben Ahmed M. Salivary antigen SP32 is the immunodominant target of the antibody response to phlebotomus papatasi bites in humans. PLoS Negl Trop Dis. 2012;6:e1911. doi: 10.1371/journal.pntd.0001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marzouki S, Ben Ahmed M, Boussoffara T, Abdeladhim M, Ben Aleya-Bouafif N, Namane A, Hamida NB, Ben Salah A, Louzir H. Characterization of the antibody response to the saliva of Phlebotomus papatasi in people living in endemic areas of cutaneous leishmaniasis. Am J Trop Med Hyg. 2011;84:653–661. doi: 10.4269/ajtmh.2011.10-0598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rohousova I, Ozensoy S, Ozbel Y, Volf P. Detection of species-specific antibody response of humans and mice bitten by sand flies. Parasitology. 2005;130:493–499. doi: 10.1017/s003118200400681x. [DOI] [PubMed] [Google Scholar]
- 14.de Moura TR, Oliveira F, Novais FO, Miranda JC, Clarencio J, Follador I, Carvalho EM, Valenzuela JG, Barral-Netto M, Barral A, Brodskyn C, de Oliveira CI. Enhanced Leishmania braziliensis infection following pre-exposure to sandfly saliva. PLoS Negl Trop Dis. 2007;1:e84. doi: 10.1371/journal.pntd.0000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coutinho-Abreu IV, Mukbel R, Hanafi HA, Fawaz EY, El-Hossary SS, Wadsworth M, Stayback G, Pitts DA, Abo-Shehada M, Hoel DF, Kamhawi S, Ramalho-Ortigao M, McDowell MA. Expression plasticity of Phlebotomus papatasi salivary gland genes in distinct ecotopes through the sand fly season. BMC Ecol. 2011;11:24. doi: 10.1186/1472-6785-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janini R, Saliba E, Kamhawi S. Species composition of sand flies and population dynamics of Phlebotomus papatasi (Diptera: Psychodidae) in the southern Jordan Valley, an endemic focus of cutaneous leishmaniasis. J Med Entomol. 1995;32:822–826. doi: 10.1093/jmedent/32.6.822. [DOI] [PubMed] [Google Scholar]
- 17.Janini R, Saliba E, Khoury S, Oumeish O, Adwan S, Kamhawi S. Incrimination of Phlebotomus papatasi as vector of Leishmania major in the southern Jordan Valley. Med Vet Entomol. 1995;9:420–422. doi: 10.1111/j.1365-2915.1995.tb00016.x. [DOI] [PubMed] [Google Scholar]
- 18.Kamhawi S, Arbagi A, Adwan S, Rida M. Environmental manipulation in the control of a zoonotic cutaneous leishmaniasis focus. Arch Inst Pasteur Tunis. 1993;70:383–390. [PubMed] [Google Scholar]
- 19.Saliba EK, Pratlong F, Dedet JP, Saleh N, Khoury SA, Oumeish OY, Batayneh O, Al-Oran R. Identification of Leishmania strains from Jordan. Ann Trop Med Parasitol. 2004;98:677–683. doi: 10.1179/000349804X3144. [DOI] [PubMed] [Google Scholar]
- 20.Schlein Y, Jacobson RL. Sugar meals and longevity of the sandfly Phlebotomus papatasi in an arid focus of Leishmania major in the Jordan Valley. Med Vet Entomol. 1999;13:65–71. doi: 10.1046/j.1365-2915.1999.00138.x. [DOI] [PubMed] [Google Scholar]
- 21.Schlein Y, Warburg A, Schnur LF, Gunders AE. Leishmaniasis in the Jordan Valley II. Sandflies and transmission in the central endemic area. Trans R Soc Trop Med Hyg. 1982;76:582–586. doi: 10.1016/0035-9203(82)90215-2. [DOI] [PubMed] [Google Scholar]
- 22.Yuval B. Populations of Phlebotomus papatasi (Diptera: Psychodidae) and the risk of Leishmania major transmission in three Jordan Valley habitats. J Med Entomol. 1991;28:492–495. doi: 10.1093/jmedent/28.4.492. [DOI] [PubMed] [Google Scholar]
- 23.Yuval B, Warburg A, Schlein Y. Leishmaniasis in the Jordan Valley. V. Dispersal characteristics of the sandfly Phlebotomus papatasi. Med Vet Entomol. 1988;2:391–395. doi: 10.1111/j.1365-2915.1988.tb00213.x. [DOI] [PubMed] [Google Scholar]
- 24.el Said SM, Kenawy MA, el Sawaf BM, Beier JC, el Sawy FM. Seasonal abundance and distribution of Phlebotomus papatasi (Diptera: Psychodidae) inside houses in Aswan Governorate, Egypt. J Egypt Soc Parasitol. 1985;15:371–380. [PubMed] [Google Scholar]
- 25.Fryauff DJ, Modi GB, Mansour NS, Kreutzer RD, Soliman S, Youssef FG. Epidemiology of cutaneous leishmaniasis at a focus monitored by the multinational force and observers in the northeastern Sinai Desert of Egypt. Am J Trop Med Hyg. 1993;49:598–607. doi: 10.4269/ajtmh.1993.49.598. [DOI] [PubMed] [Google Scholar]
- 26.Hanafi HA, el Sawaf BM, Fryauff DJ, Beavers GM, Tetreault GE. Susceptibility to Leishmania major of different populations of Phlebotomus papatasi (Diptera: Psychodidae) from endemic and non-endemic regions of Egypt. Ann Trop Med Parasitol. 1998;92:57–64. doi: 10.1080/00034989860175. [DOI] [PubMed] [Google Scholar]
- 27.Hogsette JA, Hanafi HA, Bernier UR, Kline DL, Fawaz EY, Furman BD, Hoel DF. Discovery of diurnal resting sites of phlebotomine sand flies in a village in southern Egypt. J Am Mosq Control Assoc. 2008;24:601–603. doi: 10.2987/08-5789.1. [DOI] [PubMed] [Google Scholar]
- 28.Morsy TA, Aboul Ela RG, Sarwat MA, Arafa MA, el Gozamy BM. Some aspects of Phlebotomus papatasi (Scopoli) in greater Cairo, Egypt. J Egypt Soc Parasitol. 1993;23:399–416. [PubMed] [Google Scholar]
- 29.Morsy TA, Shoukry A, Schnur LF, Sulitzeanu A. Gerbillus pyramidum is a host of Leishmania major in the Sinai Peninsula. Ann Trop Med Parasitol. 1987;81:741–742. doi: 10.1080/00034983.1987.11812180. [DOI] [PubMed] [Google Scholar]
- 30.Prates DB, Santos LD, Miranda JC, Souza AP, Palma MS, Barral-Netto M, Barral A. Changes in amounts of total salivary gland proteins of Lutzomyia longipallpis (Diptera: Psychodidae) according to age and diet. J Med Entomol. 2008;45:409–413. doi: 10.1603/0022-2585(2008)45[409:ciaots]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 31.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 32.Champion MM, Williams EA, Kennedy GM, DiGiuseppe Champion PA. Direct detection of bacterial protein secretion using whole colony proteomics. Mol Cell Proteomics. 2012;11:596–604. doi: 10.1074/mcp.M112.017533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Information NCfB Phlebotomine Sandfly Proteins. 2012. http://www.ncbi.nlm.nih.gov/protein/?term=txid29031[Organism:noexp] Available at. Accessed December 4, 2012.
- 34.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 35.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang XQ, Hardison RC, Miller W. A space-efficient algorithm for local similarities. Comput Appl Biosci. 1990;6:373–381. doi: 10.1093/bioinformatics/6.4.373. [DOI] [PubMed] [Google Scholar]
- 37.Belkaid Y, Kamhawi S, Modi G, Valenzuela J, Noben-Trauth N, Rowton E, Ribeiro J, Sacks DL. Development of a natural model of cutaneous leishmaniasis: powerful effects of vector saliva and saliva preexposure on the long-term outcome of Leishmania major infection in the mouse ear dermis. J Exp Med. 1998;188:1941–1953. doi: 10.1084/jem.188.10.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 39.Saldanha AJ. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 40.Reunala T, Brummer-Korvenkontio H, Palosuo T. Are we really allergic to mosquito bites? Ann Med. 1994;26:301–306. doi: 10.3109/07853899409147906. [DOI] [PubMed] [Google Scholar]
- 41.Abdeladhim M, Jochim RC, Ben Ahmed M, Zhioua E, Chelbi I, Cherni S, Louzir H, Ribeiro JM, Valenzuela JG. Updating the salivary gland transcriptome of Phlebotomus papatasi (Tunisian strain): the search for sand fly-secreted immunogenic proteins for humans. PLoS One. 2012;7:e47347. doi: 10.1371/journal.pone.0047347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamarsheh O, Presber W, Yaghoobi-Ershadi MR, Amro A, Al-Jawabreh A, Sawalha S, Al-Lahem A, Das ML, Guernaoui S, Seridi N, Dhiman RC, Hashiguchi Y, Ghrab J, Hassan M, Schonian G. Population structure and geographical subdivision of the Leishmania major vector Phlebotomus papatasi as revealed by microsatellite variation. Med Vet Entomol. 2009;23:69–77. doi: 10.1111/j.1365-2915.2008.00784.x. [DOI] [PubMed] [Google Scholar]
- 43.Gazos-Lopes F, Mesquita RD, Silva-Cardoso L, Senna R, Silveira AB, Jablonka W, Cudischevitch CO, Carneiro AB, Machado EA, Lima LG, Monteiro RQ, Nussenzveig RH, Folly E, Romeiro A, Vanbeselaere J, Mendonca-Previato L, Previato JO, Valenzuela JG, Ribeiro JM, Atella GC, Silva-Neto MA. Glycoinositolphospholipids from Trypanosomatids subvert nitric oxide production in Rhodnius prolixus salivary glands. PLoS One. 2012;7:e47285. doi: 10.1371/journal.pone.0047285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin-Martin I, Molina R, Jimenez M. Identifying salivary antigens of Phlebotomus argentipes by a 2DE approach. Acta Trop. 2013;126:229–239. doi: 10.1016/j.actatropica.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 45.Hostomska J, Volfova V, Mu J, Garfield M, Rohousova I, Volf P, Valenzuela JG, Jochim RC. Analysis of salivary transcripts and antigens of the sand fly Phlebotomus arabicus. BMC Genomics. 2009;10:282. doi: 10.1186/1471-2164-10-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kane LP. T cell Ig and mucin domain proteins and immunity. J Immunol. 2010;184:2743–2749. doi: 10.4049/jimmunol.0902937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Francischetti IM. Platelet aggregation inhibitors from hematophagous animals. Toxicon. 2010;56:1130–1144. doi: 10.1016/j.toxicon.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grespan R, Lemos HP, Carregaro V, Verri WA, Jr, Souto FO, de Oliveira CJ, Teixeira C, Ribeiro JM, Valenzuela JG, Cunha FQ. The protein LJM 111 from Lutzomyia longipalpis salivary gland extract (SGE) accounts for the SGE-inhibitory effects upon inflammatory parameters in experimental arthritis model. Int Immunopharmacol. 2012;12:603–610. doi: 10.1016/j.intimp.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng Z, Rasic N, Liu Y, Simons FE. Mosquito saliva-specific IgE and IgG antibodies in 1059 blood donors. J Allergy Clin Immunol. 2002;110:816–817. doi: 10.1067/mai.2002.128736. [DOI] [PubMed] [Google Scholar]
- 50.Shan EZ, Taniguchi Y, Shimizu M, Ando K, Chinzei Y, Suto C, Ohtaki T, Ohtaki N. Immunoglobulins specific to mosquito salivary gland proteins in the sera of persons with common or hypersensitive reactions to mosquito bites. J Dermatol. 1995;22:411–418. doi: 10.1111/j.1346-8138.1995.tb03415.x. [DOI] [PubMed] [Google Scholar]
- 51.Rohousova I, Hostomska J, Vlkova M, Kobets T, Lipoldova M, Volf P. The protective effect against Leishmania infection conferred by sand fly bites is limited to short-term exposure. Int J Parasitol. 2011;41:481–485. doi: 10.1016/j.ijpara.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 52.Abdeladhim M, Ben Ahmed M, Marzouki S, Belhadj Hmida N, Boussoffara T, Belhaj Hamida N, Ben Salah A, Louzir H. Human cellular immune response to the saliva of Phlebotomus papatasi is mediated by IL-10-producing CD8+ T cells and Th1-polarized CD4+ lymphocytes. PLoS Negl Trop Dis. 2011;5:e1345. doi: 10.1371/journal.pntd.0001345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carregaro V, Sa-Nunes A, Cunha TM, Grespan R, Oliveira CJ, Lima-Junior DS, Costa DL, Verri WA, Jr, Milanezi CM, Pham VM, Brand DD, Valenzuela JG, Silva JS, Ribeiro JM, Cunha FQ. Nucleosides from Phlebotomus papatasi salivary gland ameliorate murine collagen-induced arthritis by impairing dendritic cell functions. J Immunol. 2011;187:4347–4359. doi: 10.4049/jimmunol.1003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lopez Kostka S, Dinges S, Griewank K, Iwakura Y, Udey MC, von Stebut E. IL-17 promotes progression of cutaneous leishmaniasis in susceptible mice. J Immunol. 2009;182:3039–3046. doi: 10.4049/jimmunol.0713598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Souza MA, Castro MC, Oliveira AP, Almeida AF, Reis LC, Silva CJ, Brito ME, Pereira VR. American tegumentary leishmaniasis: cytokines and nitric oxide in active disease and after clinical cure, with or without chemotherapy. Scand J Immunol. 2012;76:175–180. doi: 10.1111/j.1365-3083.2012.02717.x. [DOI] [PubMed] [Google Scholar]
- 56.Gupta G, Bhattacharjee S, Bhattacharyya S, Bhattacharya P, Adhikari A, Mukherjee A, Bhattacharyya Majumdar S, Majumdar S. CXC chemokine-mediated protection against visceral leishmaniasis: involvement of the proinflammatory response. J Infect Dis. 2009;200:1300–1310. doi: 10.1086/605895. [DOI] [PubMed] [Google Scholar]
- 57.de Moura TR, Oliveira F, Rodrigues GC, Carneiro MW, Fukutani KF, Novais FO, Miranda JC, Barral-Netto M, Brodskyn C, Barral A, de Oliveira CI. Immunity to Lutzomyia intermedia saliva modulates the inflammatory environment induced by Leishmania braziliensis. PLoS Negl Trop Dis. 2010;4:e712. doi: 10.1371/journal.pntd.0000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ben Hadj Ahmed S, Chelbi I, Kaabi B, Cherni S, Derbali M, Zhioua E. Differences in the salivary effects of wild-caught versus colonized Phlebotomus papatasi (Diptera: Psychodidae) on the development of zoonotic cutaneous leishmaniasis in BALB/c mice. J Med Entomol. 2010;47:74–79. doi: 10.1603/033.047.0110. [DOI] [PubMed] [Google Scholar]
- 59.Ben Hadj Ahmed S, Kaabi B, Chelbi I, Cherni S, Derbali M, Laouini D, Zhioua E. Colonization of Phlebotomus papatasi changes the effect of pre-immunization with saliva from lack of protection towards protection against experimental challenge with Leishmania major and saliva. Parasit Vectors. 2011;4:126. doi: 10.1186/1756-3305-4-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.