Abstract
Many aspects of cellular physiology remain unstudied in somatic stem cells. For example, there are almost no data on protein synthesis in any somatic stem cell. We found that the amount of protein synthesized per hour in haematopoietic stem cells (HSCs) in vivo was lower than in most other haematopoietic cells, even if we controlled for differences in cell cycle status or forced HSCs to undergo self-renewing divisions. Reduced ribosome function in Rpl24Bst/+ mice further reduced protein synthesis in HSCs and impaired HSC function. Pten deletion increased protein synthesis in HSCs but also reduced HSC function. Rpl24Bst/+ cell-autonomously rescued the effects of Pten deletion in HSCs, blocking the increase in protein synthesis, restoring HSC function, and delaying leukaemogenesis. Pten deficiency thus depletes HSCs and promotes leukaemia partly by increasing protein synthesis. Either increased or decreased protein synthesis impairs HSC function.
Mutations in ribosomes and other gene products that affect protein synthesis are associated with human diseases marked by haematopoietic dysfunction1,2. Increased protein synthesis can promote the development and progression of certain cancers, including haematopoietic malignancies3–6. Ribosomal defects commonly impair HSC and erythroid progenitor function7–11. However, it is not clear whether these defects reflect a catastrophic reduction in protein synthesis below the level required for cellular homeostasis or whether HSCs require highly regulated protein synthesis.
Methods for measuring protein synthesis have depended upon the incorporation of radiolabeled amino acids, amino acid analogues12, or puromycin13–15 into nascent polypeptides in cultured cells. However, somatic stem cells profoundly change their properties in culture16 necessitating the analysis of protein synthesis in rare cells in vivo. A new fluorogenic assay using O-propargyl-puromycin (OP-Puro) has been developed to image protein synthesis in vivo17. OP-Puro, like puromycin, is taken up by cells in vivo, entering ribosome acceptor sites and incorporating into nascent polypeptides17. An azide-alkyne reaction can be used to fluorescently label OP-Puro to quantitate protein synthesis in individual cells17. We adapted this approach to quantify protein synthesis by haematopoietic cells using flow cytometry.
HSCs synthesize less protein per hour
We administered a single intraperitoneal injection of OP-Puro (50mg/kg body mass) then sacrificed mice one hour later and isolated bone marrow cells. We did not detect toxicity, signs of illness, changes in bone marrow cellularity, or changes in the frequencies of CD150+CD48−Lineage−Sca-1+c-kit+ (CD150+CD48−LSK) HSCs18, Annexin V+ bone marrow cells, Annexin V+ HSCs, or dividing HSCs (Extended Data Fig. 1a–e).
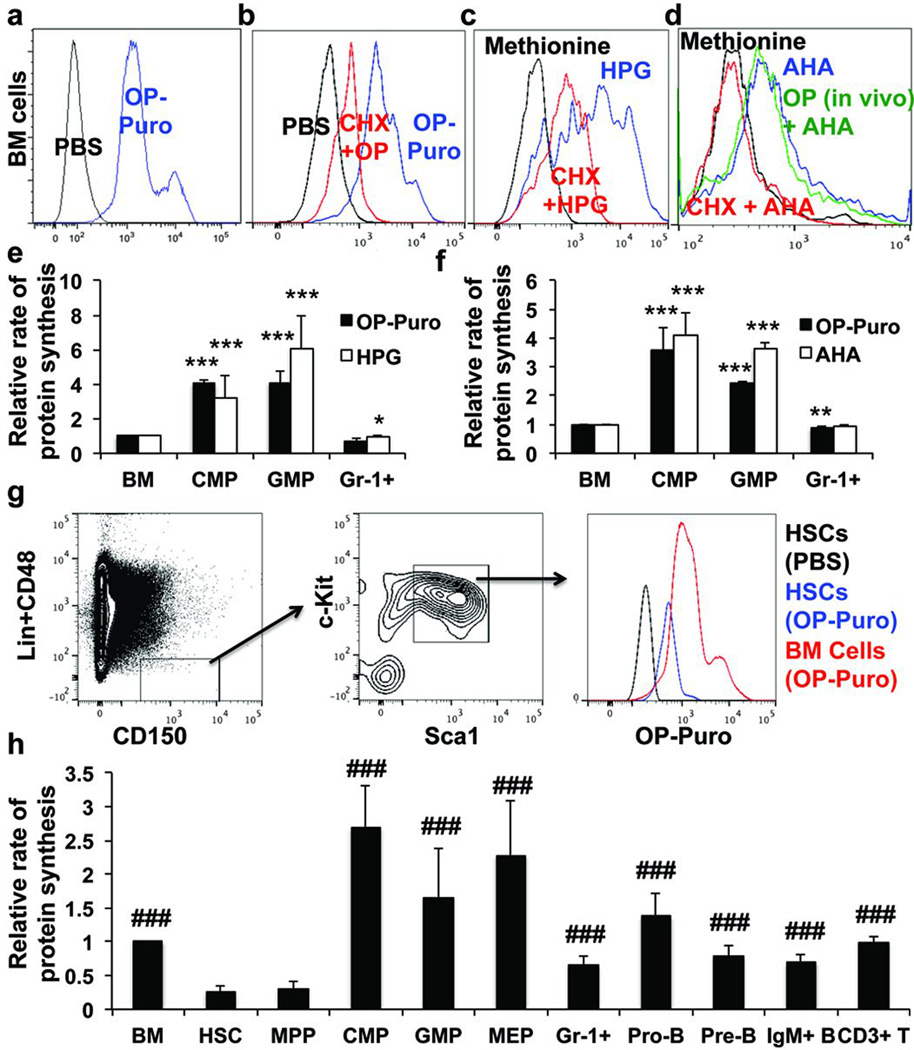
Bone marrow cells from OP-Puro treated mice exhibited a clear increase in fluorescence relative to untreated mice (Fig. 1a). The translation inhibitor, cycloheximide, profoundly blocked OP-Puro incorporation by bone marrow cells in culture (Fig. 1b). Incorporation of the methionine analogues, L-homopropargylglycine (HPG) and L-azidohomoalanine (AHA), into bone marrow cells, common myeloid progenitors (CMPs), granulocyte-macrophage progenitors (GMPs), and Gr-1+ myeloid cells correlated with OP-Puro incorporation in culture (Fig. 1c–f).
Figure 1. Quantifying protein synthesis in haematopoietic cells in vivo.
a, OP-Puro incorporation in bone marrow cells in vivo one hour after administration. b-d, OP-Puro (b), HPG (c), and AHA (d) incorporation in bone marrow cells in culture was inhibited by cycloheximide (CHX). d, Bone marrow cells from mice treated with OP-Puro in vivo exhibited normal AHA incorporation in culture, indicating OP-Puro did not block protein synthesis. e,f OP-Puro versus HPG (e; n=4 mice from 2 experiments) or AHA (f; n=3 mice from 3 experiments) incorporation by haematopoietic cells in culture. g, OP-Puro incorporation in CD150+CD48−LSK HSCs and unfractionated bone marrow cells one hour after administration in vivo. h, Protein synthesis in various haematopoietic stem and progenitor cell populations relative to unfractionated bone marrow cells (n=15 mice from 9 experiments). Extended Data Fig. 1j shows the data from Fig. 1h using a log2 scale. Data represent mean±s.d. Statistical significance was assessed using two-tailed Student’s t-tests (e-f) and differences relative to HSCs (h) were assessed using a repeated measures one way ANOVA followed by Dunnett’s test for multiple comparisons (*, p<0.05; **, p<0.01; ***, p<0.001 relative to bone marrow; ###, p<0.001 relative to HSCs).
HSCs incorporated less OP-Puro than most other bone marrow cells from the same mice (Fig. 1g). This suggested that HSCs synthesize less protein per hour than most other haematopoietic progenitors. CD150−CD48−LSK multipotent progenitors (MPPs)19 exhibited similar OP-Puro incorporation as HSCs (Fig. 1h); however, the mean rate of OP-Puro incorporation was significantly higher in unfractionated bone marrow cells, CMPs, GMPs, megakaryocyte-erythroid progenitors (MEPs), Gr-1+ myeloid cells, B220+IgM−CD43+ pro-B cells, B220+IgM−CD43− pre-B cells, B220+IgM+ B cells, CD3+ T cells, and CD71+Ter119+ erythroid progenitors (Fig. 1h). Extended Data Figures 1f–i show markers, gating strategies, and OP-Puro incorporation histograms for each cell population.
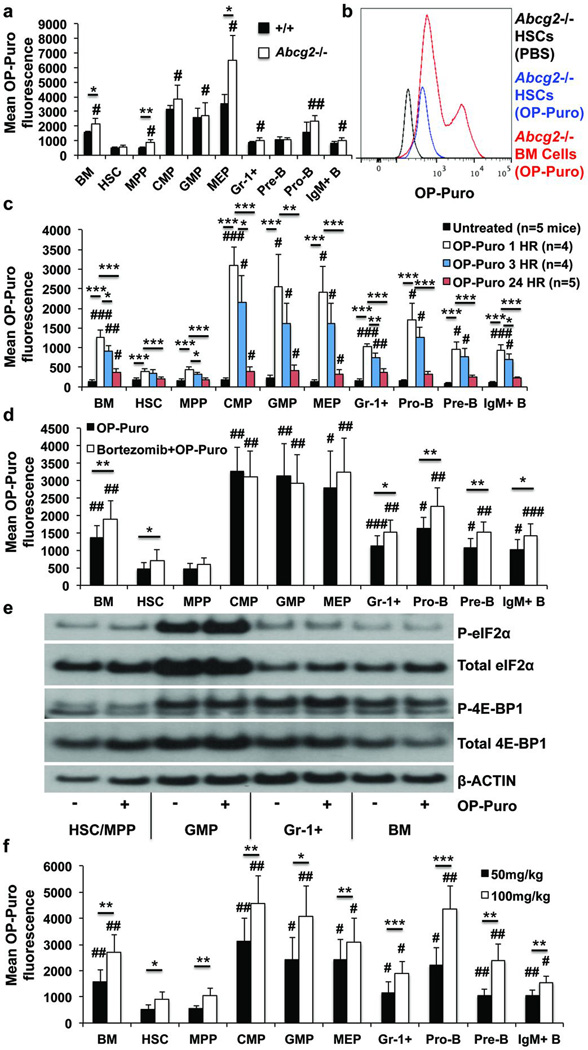
To test whether reduced OP-Puro incorporation into HSCs reflects OP-Puro efflux by the Abcg2/Bcrp1 transporter we administered OP-Puro to Abcg2-deficient mice, which lack efflux activity in HSCs20. Abcg2−/− HSCs continued to exhibit significantly lower mean rates of OP-Puro incorporation as compared to most other Abcg2−/− progenitors (Fig. 2a), similar to the lowest levels observed among bone marrow cells (Fig. 2b).
Figure 2. Lower rate of OP-Puro incorporation by HSCs does not reflect efflux or proteasomal degradation.
a,b, OP-Puro fluorescence in haematopoietic cells from Abcg2-deficient and control mice one hour after OP-Puro administration in vivo (n=4 mice from 3 experiments). c, OP-Puro fluorescence in haematopoietic cells one, three, or twenty-four hours after OP-Puro administration (n=5 experiments). d, OP-Puro fluorescence in haematopoietic cells two hours after bortezomib and one hour after OP-Puro administration in vivo (n=5 mice/treatment from 5 experiments). e, Western blots of 30,000 cells from each haematopoietic cell population from OP-Puro-treated or control mice. f, OP-Puro fluorescence in haematopoietic cells one hour after administering 50mg/kg or 100mg/kg OP-Puro (n=5 mice/dose from 5 experiments). All data represent mean±s.d. To assess the statistical significance of treatment effects within the same cells we performed two-tailed Student’s t-tests (*, p<0.05; **, p<0.01; ***, p<0.001). Differences between HSCs and other cell populations were assessed with a repeated measures one way ANOVA followed by Dunnett’s test for multiple comparisons (#, p<0.05; ##, p<0.01; ###, p<0.001).
Differences in OP-Puro incorporation did not reflect proteasomal degradation21. The maximum OP-Puro signal in haematopoietic cells was one hour after OP-Puro administration (data not shown). However, HSCs exhibited little decline in OP-Puro signal between 1 and 3 hours after administration and had significantly less OP-Puro incorporation than any restricted progenitor at both times (Fig. 2c). In contrast, the OP-Puro signal was profoundly reduced 24 hours after administration (Fig. 2c). This suggested that degradation of OP-Puro-containing polypeptides requires several hours. Consistent with this, incubation of bone marrow cells at 37°C for 30 minutes did not significantly reduce OP-Puro fluorescence in any cell population relative to an aliquot of the same cells kept on ice to arrest degradation (Extended Data Fig. 2a).
We pre-treated mice with bortezomib (1mg/kg i.v.) to inhibit proteasome activity22 one hour before OP-Puro administration. This significantly increased the OP-Puro signal in nearly all cell populations 24 hours after OP-Puro administration (Extended Data Fig. 2b). When we assessed OP-Puro incorporation one hour after administration, bortezomib pre-treatment only modestly increased OP-Puro fluorescence in certain cell populations (Fig. 2d). Even in the presence of bortezomib, HSCs had significantly less OP-Puro fluorescence than restricted progenitors. The lower OP-Puro incorporation by HSCs appears to primarily reflect reduced protein synthesis rather than accelerated proteosomal degradation.
OP-Puro also did not affect the levels of phosphorylated 4E-BP1 or eIF2α (Fig. 2e), key regulators of translation that can be influenced by proteotoxic stress23,24. HSC/MPPs had less p4E-BP1 and β-actin per cell than other hematopoietic cells but similar levels of total 4E-BP1. This raised the possibility that HSCs synthesize less protein as a consequence of increased 4E-BP1-mediated inhibition of translation (Fig. 2e).
Doubling the dose of OP-Puro to 100mg/kg significantly increased OP-Puro incorporation in all cell populations, but HSCs continued to have significantly lower levels than all other cell populations except MPPs (Fig. 2f). Thus, 50mg/kg OP-Puro did not significantly attenuate protein synthesis and uptake was not saturated in HSCs or most other cells.
Even dividing HSCs make less protein
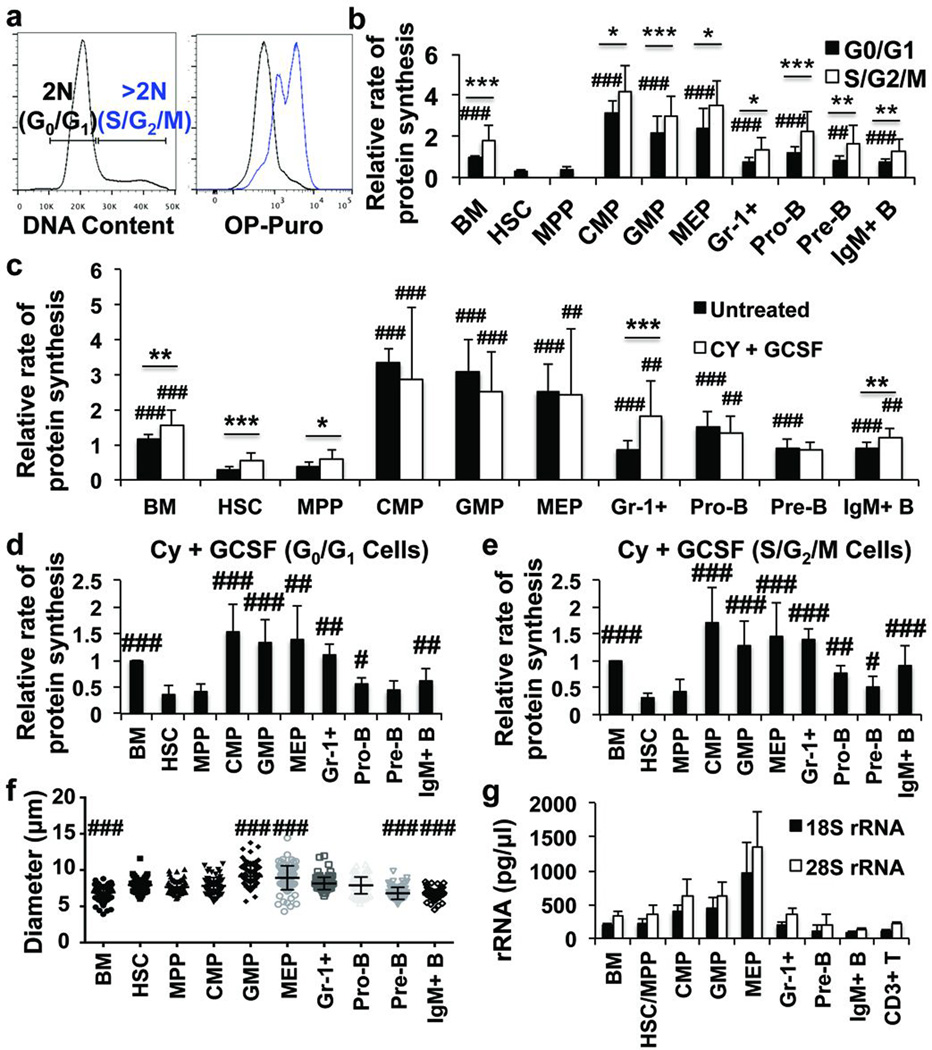
Haematopoietic progenitors in S/G2/M exhibited significantly higher rates of OP-Puro incorporation than cells in G0/G1 (Fig. 3a, b). Total protein was also higher in Gr-1+ cells and bone marrow cells in S/G2/M as compared to G0/G1 (Extended Data Fig. 3b). However, HSCs and MPPs exhibited less OP-Puro incorporation than restricted progenitors even when we compared only cells in G0/G1 (Fig. 3b). We treated mice with cyclophosphamide (Cy) followed by two daily injections of granulocyte colony stimulating factor (G-CSF) to induce self-renewing divisions by HSCs (Extended Data Fig. 3d)25. Cy/G-CSF also increased division by MPPs, Gr-1+ cells, and IgM+ B cells (Extended Data Fig. 3a, e). Each of these populations exhibited increased OP-Puro incorporation after Cy/G-CSF treatment (Fig. 3c). However, HSCs had significantly less protein synthesis as compared to most restricted progenitors irrespective of whether we compared S/G2/M cells (Fig. 3e) or G0/G1 cells (Fig. 3d, Extended Data Fig. 3f, g) from Cy/G-CSF-treated mice.
Figure 3. HSCs synthesize less protein than most haematopoietic progenitors, even when undergoing self-renewing divisions.
a, OP-Puro incorporation in vivo in bone marrow cells in G0/G1 versus S/G2/M. b, Protein synthesis in G0/G1 and S/G2/M cells from haematopoietic cell populations in vivo (n=10 mice from 6 experiments). We were unable to assess OP-Puro incorporation in S/G2/M HSCs and MPPs in these experiments because these cells are extraordinarily rare in normal bone marrow. c, Protein synthesis in haematopoietic cells after Cy/G-CSF treatment (n=10 mice from 6 experiments). d,e, Protein synthesis in G0/G1 (d) and S/G2/M (e) cells from Cy/G-CSF-treated mice (n=10 mice from 6 experiments). Extended Data Fig. 3f–g show the data from Fig. 3d–e side-by-side with data from untreated controls in Fig. 3b. f, Cell diameter (n>60 cells/population from 2 mice). g, 18S rRNA and 28S rRNA content in 15,000 cells from each stem/progenitor cell population (n=3 mice). All data represent mean±s.d. To assess the statistical significance of treatment effects within the same cells (b-c) we performed two-tailed Student’s t-tests (*, p<0.05; **, p<0.01; ***, p<0.001) and differences between HSCs and other cells (b-g) were assessed with a repeated measures one-way ANOVA followed by Dunnett’s test for multiple comparisons (#, p<0.05; ##, p<0.01; ###, p<0.001).
Differences in protein synthesis between HSCs and restricted progenitors were not fully explained by differences in cell diameter (Fig. 3f, Extended Data Fig 4b), ribosomal RNA (Fig. 3g, Extended Data Fig. 4c, d), or total RNA content (Extended Data Fig. 3c and 4e), which were similar among HSCs and lymphoid progenitors despite differences in protein synthesis.
Ribosomal mutant impairs HSC function
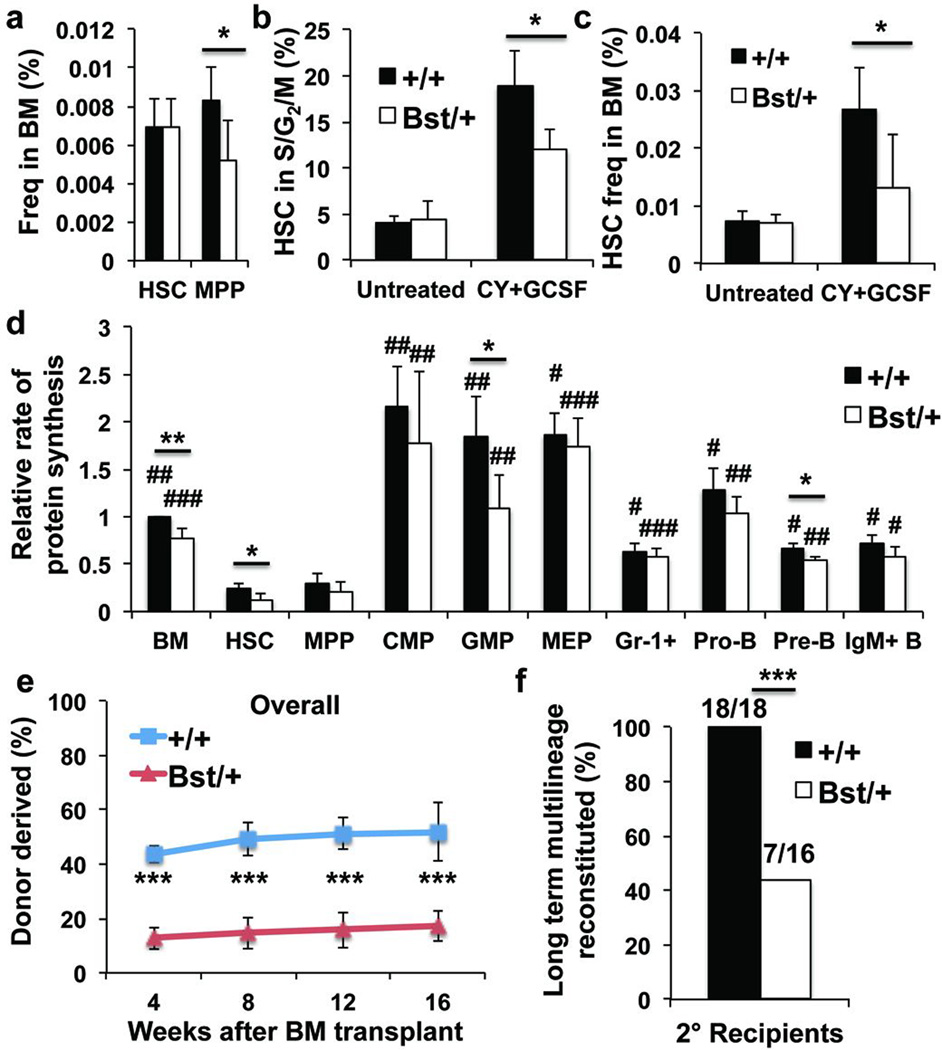
Rpl24Bst/+ mice have a hypomorphic mutation in the Rpl24 ribosome subunit, reducing protein synthesis in multiple cell types by 30% in culture3,4,26. Adult Rpl24Bst/+ mice are grossly normal but are 20% smaller than wild-type mice and have mild pigmentation and skeletal abnormalities26. These mice had normal bone marrow, spleen, and thymus cellularity, blood cell counts (Extended Data Fig. 5a, b), and HSC frequency (Fig. 4a). Frequencies of colony-forming progenitors (Extended Data Fig. 6b), restricted progenitors, and Annexin V+ HSCs and MPPs (Extended Data Fig. 5c–i) were also largely normal.
Figure 4. Rpl24Bst/+ HSCs synthesize less protein and have less capacity to reconstitute irradiated mice.
a, Frequencies of HSCs and MPPs in Rpl24Bst/+ (n=7) versus littermate control (n=6) mice (n=5 experiments). b,c Frequency of HSCs in S/G2/M (b; n=4 untreated mice/genotype, n=3 (Rpl24Bst/+) or 4 (+/+) Cy/G-CSF treated mice/genotype in 3 experiments) and frequency of HSCs in the bone marrow (c; n=6 wild-type and 7 Rpl24Bst/+ untreated mice, n=7 wild-type and 5 Rpl24Bst/+ Cy/G-CSF treated mice in 4 experiments) after treatment with Cy/G-CSF. d, Protein synthesis in haematopoietic cells based on OP-Puro incorporation in vivo (n=4 mice/genotype in 4 experiments). e, Donor cell engraftment when 5×105 donor bone marrow cells were transplanted along with 5×105 recipient bone marrow cells into irradiated recipient mice (n=4 experiments with a total of 17 recipients for wild-type, and 20 for Rpl24Bst/+; myeloid, B, and T engraftment are in Extended Data Figure 5k). f, The number of long-term multilineage reconstituted secondary recipients (>0.5% donor myeloid and lymphoid cells for at least 16 weeks after transplantation) after secondary transplantation of 3×106 bone marrow cells from primary recipients in (e) (n=4 donors/genotype). All data represent mean±s.d. Statistical significance was assessed with two-tailed Student’s t-tests (a-e) and Fisher’s exact test (f) (*p<0.05, **p<0.01, ***p<0.001). Differences between HSCs and other cell populations (d) were assessed with a repeated measures one-way ANOVA followed by Dunnett’s test for multiple comparisons (#, p<0.05; ##, p<0.01; ###, p<0.001).
OP-Puro incorporation into unfractionated bone marrow cells, HSCs, GMPs and pre-B cells from Rpl24Bst/+ mice was significantly reduced relative to wild-type cells (Fig. 4d). OP-Puro incorporation into Rpl24Bst/+ HSCs was reduced by approximately 30% relative to control HSCs. Some cell populations appeared to depend more than others upon Rpl24 for protein synthesis. Rpl24 was highly differentially expressed among haematopoietic cells (Extended Data Fig. 5j).
Upon transplantation into irradiated mice, Rpl24Bst/+ bone marrow cells gave significantly lower levels of donor cell reconstitution in the myeloid, B, and T cell lineages relative to control donor cells (Fig. 4e and Extended Data Fig. 5k). We did not detect impaired homing of Rpl24Bst/+ LSK cells to the bone marrow (Extended Data Fig. 5l). Significantly impaired reconstitution by Rpl24Bst/+ cells was also evident in secondary recipient mice (Fig. 4f). After Cy/G-CSF treatment, significantly fewer Rpl24Bst/+ HSCs were in S/G2/M as compared to wild-type HSCs (Fig. 4b). HSC frequency in the bone marrow after Cy/G-CSF treatment was 2–3 fold higher in wild-type as compared to Rpl24Bst/+ mice (Fig. 4c). Colonies formed by individual Rpl24Bst/+ HSCs in methylcellulose contained significantly fewer cells than wild-type HSC colonies (Extended Data Fig. 6c). Rpl24Bst/+ HSCs are thus impaired in their proliferative potential.
Some ribosomal defects can induce p53 and its target p21cip1 (ref27). Comparing Rpl24Bst/+ versus wild-type cells, we did not detect any difference in p53 levels among Lineage− haematopoietic progenitors (Extended Data Fig. 5m) or in p21cip1 levels among LSK cells (Extended Data Fig. 5n). These data are consistent with prior studies28 indicating that p53 and p21cip1 are not induced in adult Rpl24Bst/+ haematopoietic cells. Moreover, loss of a single allele of p53 did not rescue the size of colonies formed by Rpl24Bst/+ HSCs in culture (Extended Data Fig. 6e) even though p53 heterozygosity largely rescues developmental phenotypes in Rpl24Bst/+ embryos28. HSC defects in Rpl24Bst/+ mice are therefore not caused by increased p53 function.
Pten deletion increases protein synthesis
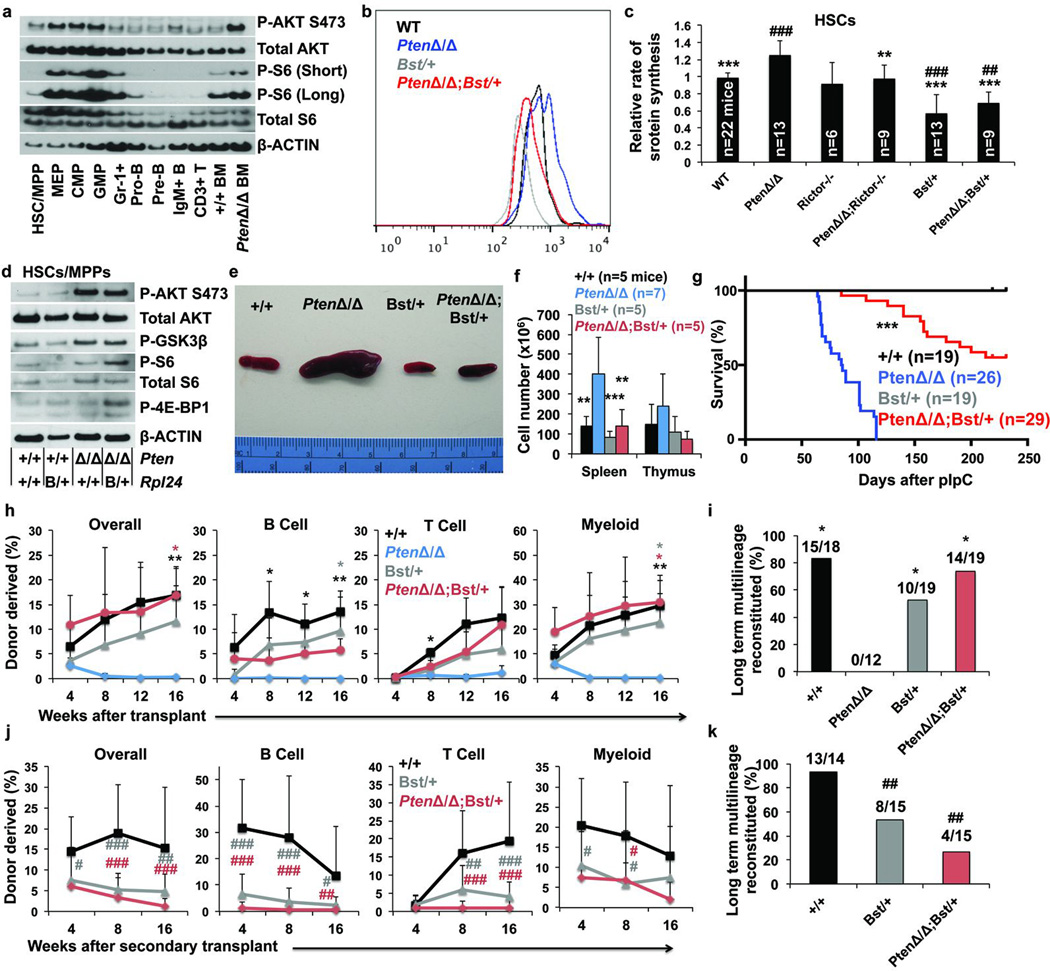
HSCs had pAkt and pS6 levels that were similar to lymphoid progenitors but lower than myeloid progenitors (Fig. 5a, Extended Data Fig. 4f). We conditionally deleted Pten from adult haematopoietic cells in Mx1-Cre; Ptenfl/fl mice. As previously shown29–33, Pten deletion strongly increased pAKT and pS6 levels in bone marrow cells (Fig. 5a) and in HSCs/MPPs (Fig. 5d). Consistent with this, we observed an approximately 30% increase in protein synthesis in Pten-deficient relative to control HSCs (p<0.01; Fig. 5b,c).
Figure 5. Rpl24Bst/+ blocks the increase in protein synthesis and restores HSC function after Pten deletion.
a, Western blots of 30,000 cells from each population. Long and short exposures for pS6 are shown. For total S6, a non-specific band is present below the specific band. Differences in β-Actin represent differences in β-Actin content per cell (1 representative blot from two experiments). b, Representative histograms of OP-Puro fluorescence in HSCs of each genotype. c, OP-Puro incorporation into HSCs of each genotype (n=15 experiments). d, Western blots of 30,000 HSCs/MPPs of the indicated genotypes (1 representative blot from two experiments). e, Representative spleens 2 weeks after pIpC administration to wild-type, Mx-1-Cre; Ptenfl/fl, Rpl24Bst/+, and Mx-1-Cre; Ptenfl/fl; Rpl24Bst/+ mice. f, Spleen and thymus cellularity (n=7 experiments). g, Time until mice had to be sacrificed due to illness after transplantation of 2×106 bone marrow cells of the indicated genotypes into irradiated recipient mice. h, i, 10 donor HSCs were transplanted along with 3×105 recipient bone marrow cells into irradiated recipients. Donor cell engraftment (h) and fraction of recipients that were long-term multilineage reconstituted (i; 3 experiments). j, k, Donor cell engraftment (j) and the fraction of secondary recipients that were long-term multilineage reconstituted (k) after transplantation of 3×106 bone marrow cells from primary recipients in (h) (n=4 donors/genotype). All data represent mean±s.d. Differences among genotypes (c, f, h) were assessed with a one way ANOVA followed by Dunnett’s test for multiple comparisons. Statistical significance was assessed by log-rank test (g), Chi-squared tests followed by Tukey's t-tests for pairwise comparisons (i,k), or a one way ANOVA followed by Tukey’s t-tests for multiple comparisons (j). Significance was expressed relative to wild-type (#, p<0.05; ##, p<0.01; ###, p<0.001) or Pten-deficient cells (*, p<0.05; **, p<0.01; ***, p<0.001).
HSCs are depleted after Pten deletion, even after transplantation into wild-type recipients that never develop leukaemia, by a mechanism that depends upon cell-autonomous mTORC1 and mTORC2 activation29–32. We examined OP-Puro incorporation into HSCs from Mx1-Cre; Rictorfl/fl mice, Mx1-Cre; Rictorfl/fl; Ptenfl/fl mice, and Mx1-Cre; Ptenfl/fl mice. Rictor deletion (which inactivates mTORC2) had no effect on the rate of protein synthesis in otherwise wild-type HSCs (Fig. 5c, Extended Data Fig. 6g), consistent with the normal reconstituting capacity of Rictor-deficient HSCs30. However, Rictor deficiency significantly reduced protein synthesis in Pten-deficient HSCs to normal levels (Fig. 5c, Extended Data Fig. 6g). The ability of Rictor deletion to rescue both protein synthesis and HSC function after Pten deletion30 suggested that Pten deletion depletes HSCs partly by cell-autonomously increasing protein synthesis. Rapamycin treatment, which also prevents HSC depletion after Pten deletion31,32, also blocked the increase in protein synthesis in Pten-deficient HSCs (data not shown).
HSCs from Mx1-Cre; Ptenfl/fl; Rpl24Bst/+ mice had significantly less protein synthesis than HSCs from Mx1-Cre; Ptenfl/fl mice (Fig. 5b,c). Although ribosomes can promote mTORC2 signaling34, Rpl24Bst/+ did not reduce levels of pAKT, pGSK3β, pS6, or p4E-BP1 in HSCs/MPPs (Fig. 5d), suggesting Rpl24Bst/+ did not reduce mTORC1 or mTORC2 signaling. In fact, Mx1-Cre; Ptenfl/fl; Rpl24Bst/+ HSCs/MPPs had slightly increased levels of pS6 and p4E-BP1 relative to Pten-deficient HSCs/MPPs, but this would be expected to further increase protein synthesis rather than reducing it. This suggested that Rpl24Bst/+ acted downstream of the PI3-kinase pathway to block the increase in protein synthesis in HSCs after Pten deletion, raising the question of whether Rpl24Bst/+ could also block leukaemogenesis or HSC depletion.
Rpl24Bst suppresses leukaemogenesis
Conditional deletion of Pten in haematopoietic cells leads to a myeloproliferative disorder (MPD) and T cell acute lymphoblastic leukaemia (T-ALL)31,33,35. Increased protein synthesis promotes some haematopoietic malignancies, including T cell leukaemias3,4,6,36. Two weeks after pIpC administration, Mx1-Cre; Ptenfl/fl mice exhibited significantly increased spleen and thymus cellularity (Fig. 5e, f, Extended Data Fig. 6h, i), consistent with the induction of MPD and T-ALL31,33,35. Mx1-Cre; Ptenfl/fl; Rpl24Bst/+ mice exhibited normal spleen and thymus cellularity (Fig. 5e, f, Extended Data Fig. 6h, i), suggesting that reduced ribosome function impaired the development of MPD and T-ALL after Pten deletion.
To compare MPD and T-ALL development we transplanted 2×106 bone marrow cells from each genetic background into irradiated mice then treated with pIpC four weeks later. Recipients of Mx1-Cre; Ptenfl/fl; Rpl24Bst/+ haematopoietic cells lived significantly longer than recipients of Mx1-Cre; Ptenfl/fl haematopoietic cells (Fig. 5g). We confirmed by PCR that the donor cells had completely excised Pten (data not shown). When ill recipients of cells of either genotype were sacrificed, they exhibited splenomegaly, thymomegaly and histological signs of MPD and T-ALL (Extended Data Fig. 6k). Rpl24Bst/+ therefore significantly (p<0.001; Fig. 5g) delayed, but did not entirely prevent, MPD and T-ALL after Pten deletion. It is unclear whether Rpl24Bst/+ impairs leukaemogenesis by acting within HSCs or within other haematopoietic cells.
Pten deletion mobilizes HSCs to the spleen29,30. Rpl24Bst/+ blocked this effect, restoring normal HSC numbers in spleens of Mx1-Cre; Ptenfl/fl; Rpl24Bst/+ mice (Extended Data Fig. 6j).
Rpl24Bst rescues Pten-deficient HSCs
We performed long-term reconstitution assays in which we transplanted 10 CD150+CD48−LSK HSCs from wild-type, Mx1-Cre; Ptenfl/fl, Rpl24Bst/+, or Mx1-Cre; Ptenfl/fl; Rpl24Bst/+ mice along with 300,000 wild-type bone marrow cells into irradiated wild-type mice. As expected, most (15 of 18) recipients of wild-type HSCs but no (0 of 12) recipients of Pten-deficient HSCs were long-term multilineage reconstituted by donor cells (Fig. 5h, i). Relative to control HSCs, Rpl24Bst/+ HSCs gave lower levels of donor cell reconstitution in all lineages (Fig. 5h) and a somewhat lower fraction of recipients were long-term multilineage reconstituted (10 of 19; Fig. 5i), though these differences were not statistically significant. Relative to Pten-deficient HSCs, Mx1-Cre; Ptenfl/fl; Rpl24Bst/+ compound mutant HSCs gave significantly higher levels of reconstitution and a significantly higher percentage of recipients were long-term multilineage reconstituted by donor cells (Fig. 5h, i). Reconstitution by Mx1-Cre; Ptenfl/fl; Rpl24Bst/+ HSCs was statistically indistinguishable from wild-type HSCs in all lineages except the B lineage (Pten deficiency impairs B lineage progenitors independent of its effects on HSCs33; Fig. 5h). We confirmed by PCR that the donor cells had completely excised Pten (data not shown). This demonstrates that reducing protein synthesis restores the ability of Pten-deficient HSCs to give long-term multilineage reconstitution.
Secondary transplantation of 3×106 bone marrow cells from long-term multilineage reconstituted primary recipients in Fig. 5h yielded significantly less donor cell reconstitution and long-term multilineage reconstitution among recipients of Rpl24Bst/+ as compared to control HSCs (Fig. 5j and 5k). Secondary recipients of Mx1-Cre; Ptenfl/fl; Rpl24Bst/+ compound mutant HSCs also exhibited significantly less donor cell reconstitution and long-term multilineage reconstitution as compared to control HSCs. Rpl24Bst/+ thus restored long-term multilineage reconstituting potential to Pten deficient HSCs but did not fully restore wild-type function. Pten deficiency thus impairs HSC function partly by increasing protein synthesis.
To test MPP function we competitively transplanted 100 donor CD150−CD48−LSK MPPs19 from Mx-1-Cre; Ptenfl/fl, Rpl24Bst/+, or control mice into irradiated recipients. There was a trend toward lower reconstitution by Rpl24Bst/+ MPPs and Pten-deficient MPPs relative to control MPPs but the differences were not statistically significant (Extended Data Fig. 7a, b). Neither Rpl24Bst/+ nor Pten deletion significantly affected the percentage of bone marrow cells that formed colonies or the cellularity of those colonies (Extended Data Fig. 6b, d). Thus, we have not detected clear effects of Rpl24Bst/+ or Pten deletion on the proliferative potential of MPPs or restricted progenitors, suggesting they are not as sensitive as HSCs to changes in protein synthesis, though it remains possible they are also impaired by changes in protein synthesis.
DISCUSSION
When we added HSCs to culture, OP-Puro incorporation increased dramatically (data not shown). This raises the possibility that the failure to sustainably maintain HSCs under any known culture conditions37 may reflect a limitation imposed by increased protein synthesis. Consistent with this, rapamycin promotes HSC maintenance in culture38, though it remains to be determined whether rapamycin attenuates the increase in protein synthesis in cultured HSCs.
Low rates of protein synthesis may be essential for maintaining metabolic homeostasis in HSCs and potentially in other kinds of somatic stem cells. Changes in protein synthesis may cause undesirable changes in the quality and/or content of the proteome, such as due to misfolding. There may also be changes in the translation of certain subsets of transcripts (potentially including key HSC regulators) when protein synthesis increases, similar to what occurs in cancer cells5.
Embryonic stem (ES) cells have high proteasome activity and proteasome activity increases both ES cell maintenance and C. elegans lifespan39,40. Rapamycin treatment, and mutations that reduce mTOR signaling, also increase lifespan (see citations in ref41) and would be predicted to reduce protein synthesis. In the context of these results, our observations raise the possibility that reduced protein synthesis and/or increased proteasome activity is required to maintain certain long-lived somatic cells in addition to increasing organismal lifespan.
METHODS
Mice
Rpl24Bst/+ 26, Ptenfl/fl 44, Mx1-Cre45, Rictorfl/fl 30, Abcg2 46 and p53+/− (ref 47) mutant mice have been previously described. These mice were all backcrossed for at least eight generations onto a C57BL background, with the exception of Abcg2−/− mice, which were on an FVB.129 N7 background (Taconic). C57BL/Ka-Thy-1.1 (CD45.2) and C57BL/Ka-Thy-1.2 (CD45.1) mice were used in transplantation experiments. Both male and female mice were used in all studies. Expression of Mx1-Cre was induced by three or four intraperitoneal injections of 10μg pIpC (GE Healthcare) administered every other day beginning around 6 weeks of age. For cyclophosphamide and G-CSF experiments, 4mg of cyclophosphamide (Baxter) was administered by intraperitoneal injection on day 0, and 5μg of G-CSF (Neupogen; Amgen) was administered by subcutaneous injection on days 1 and 2. Mice were analyzed on day 3. All mice were housed in the Unit for Laboratory Animal Medicine at the University of Michigan (UM), where breeding for these studies was initiated, or in the Animal Resource Center at the University of Texas (UT) Southwestern Medical Center, where these studies were performed. All protocols were approved by the UM Committee on the Use and Care of Animals and by the UT Southwestern Institutional Animal Care and Use Committee.
Measuring protein synthesis
For in vitro analysis 103-104 bone marrow or sorted cells were plated in 100 μl of methionine free Dulbecco’s Modified Eagle’s Medium (Sigma) supplemented with 200 μM L-cysteine (Sigma), 50 μM 2-mercaptoethanol (Sigma), 1mM L-glutamine (Gibco) and 0.1% bovine serum albumin (BSA; Sigma). For analysis of HPG and AHA incorporation, cells were pre-cultured for 45 minutes to deplete endogenous methionine. For OP-Puro, the medium was supplemented with 1mM L-methionine (Sigma). HPG (Life Technologies; 1mM final concentration), AHA (Life Technologies; 1mM final concentration) or OP-Puro (Medchem Source; 50 μM final concentration) were added to the culture medium for 1 hour (HPG and OP-Puro) or 2.5 hours (AHA), then cells were removed from wells and washed twice in Ca2+ and Mg2+ free phosphate buffered saline (PBS). Cells were fixed in 0.5ml of 1% paraformaldehyde (Affymetrix) in PBS for 15 minutes on ice. Cells were washed in PBS, then permeabilized in 200 μl PBS supplemented with 3% fetal bovine serum (Sigma) and 0.1% saponin (Sigma) for 5 minutes at room temperature. The azide-alkyne cycloaddition was performed using the Click-iT Cell Reaction Buffer Kit (Life Technologies) and azide conjugated to Alexa Fluor 488 or Alexa Fluor 555 (Life Technologies) at 5μM final concentration. After the 30 minute reaction, the cells were washed twice in PBS supplemented with 3% fetal bovine serum and 0.1% saponin, then resuspended in PBS supplemented with 4’,6-diamidino-2-phenylindole (DAPI; 4 μg/ml final concentration) and analyzed by flow cytometry. To inhibit OP-Puro, HPG or AHA incorporation, cycloheximide (Sigma) was added 30 minutes prior to OP-Puro or HPG at a final concentration of 100 μg/ml. All cultures were incubated at 37°C in 6.5% CO2 and constant humidity.
For in vivo analysis, OP-Puro (50mg/kg body mass; pH 6.4–6.6 in PBS) was injected intraperitoneally. One hour later mice were euthanized, unless indicated otherwise. Bone marrow was harvested, and 3×106 cells were stained with combinations of antibodies against cell surface markers as described below. After washing, the cells were fixed, permeabilized, and the azide-alkyne cycloaddition was performed as described above. “Relative rates of protein synthesis” were calculated by normalizing OP-Puro signals to whole bone marrow after subtracting autofluorescence background. “Mean OP-Puro fluorescence” reflected absolute fluorescence values for each cell population from multiple independent experiments.
To assess the effect of proteasome activity on OP-Puro incorporation mice were administered an intravenous injection of bortezomib (Cell Signaling; 1mg/kg body mass) 1 hour before OP-Puro administration. OP-Puro incorporation was assessed as described above 1 hour later unless indicated otherwise.
Flow cytometry and cell isolation
Bone marrow cells were isolated by flushing the long bones (femurs and tibias) or by crushing the long bones, vertebrae, and pelvic bones with a mortar and pestle in Ca2+ and Mg2+ free Hank’s buffered salt solution (HBSS; Gibco) supplemented with 2% heat-inactivated bovine serum (Gibco). Spleens and thymuses were prepared by crushing tissues between frosted slides. All cells were filtered through a 40 μm cell strainer to obtain single cell suspensions. Cell number and viability were assessed by a Vi-CELL cell viability analyzer (Beckman Coulter) or by counting with a hemocytometer.
For flow cytometric analysis and isolation of specific haematopoietic progenitors, cells were incubated with combinations of antibodies to the following cell-surface markers, conjugated to FITC, PE, PerCP-Cy5.5, Cy5, APC, PE-Cy7, eFluor 660, Alexa Fluor 700, APC-eFluor 780, or biotin: CD2 (RM2-5), CD3ε (17A2), CD4 (GK1.5), CD5 (53-7.3), CD8α (53-6.7), CD11b (M1/70), CD16/32 (FcΥRII/III; 93), CD24 (M1/69), CD25 (PC61.5), CD34 (RAM34), CD43 (1B11), CD44 (IM7), CD45.1 (A20), CD45.2 (104), CD45R (B220; RA3-6B2), CD48 (HM48-1), CD71 (C2), CD117 (cKit; 2B8), CD127 (IL7Rα; A7R34), CD138 (281-2), CD150 (TC15-12F12.2), Ter119 (TER-119), Sca1 (D7, E13-161.7), Gr-1 (RB6-8C5), and IgM (II/41). For isolation of HSCs and MPPs, Lineage markers included CD3, CD5, CD8, B220, Gr-1, and Ter119. For isolation of CMPs, GMPs, and MEPs, these Lineage markers were supplemented with additional antibodies against CD4 and CD11b.
Biotinylated antibodies were visualized by incubation with PE-Cy7 or APC-Alexa Fluor 750 conjugated streptavidin. All reagents were acquired from BD Biosciences, eBiosciences, or BioLegend. All incubations were for approximately 30 minutes on ice. HSCs, MPPs, CD34+CD16/32lowCD127−Lineage−Sca-1−ckit+ CMPs, CD34+CD16/32highCD127−Lineage−Sca-1−ckit+ GMPs, and CD34−CD16/32−/lowCD127−Lineage−Sca-1−ckit+ MEPs were sometimes pre-enriched by selecting c-kit+ cells using paramagnetic microbeads and an autoMACS magnetic separator (Miltenyi Biotec).
Non-viable cells were excluded from sorts and analyses using DAPI. Apoptotic cells were identified using APC Annexin V (BD Biosciences). Data acquisition and cell sorting were performed on a FACSAria flow cytometer (BD Biosciences). All sorted fractions were double sorted to ensure high purity. Data were analyzed by FACSDiva (BD Biosciences) or FlowJo (Tree Star) software.
Long-term competitive repopulation assay
Adult recipient mice (CD45.1) were administered a minimum lethal dose of radiation using an XRAD 320 X-ray irradiator (Precision X-Ray Inc.) to deliver two doses of 540 rad (1,080 rad in total) at least 3 hours apart. Cells were injected into the retro-orbital venous sinus of anesthetized recipients. For competitive bone marrow transplants 5×105 donor and 5×105 recipient cells were transplanted. For HSC transplants 10 donor CD150+CD48−Lin−Sca1+c-kit+ HSCs and 3×105 recipient bone marrow cells were transplanted. Blood was obtained from the tail veins of recipient mice every 4 weeks for at least 16 weeks after transplantation. For MPP transplants 100 donor CD150−CD48−Lin−Sca1+c-kit+ MPPs and 3×105 recipient bone marrow cells were transplanted. Blood was obtained from the tail veins of recipient mice 3, 5 and 7 weeks after transplantation. Red blood cells were lysed with ammonium chloride potassium buffer. The remaining cells were stained with antibodies against CD45.2, CD45.1, CD45R (B220), CD11b, CD3, and Gr-1 to assess donor cell engraftment. Mice that died or developed obvious haematopoietic neoplasms were omitted from the analyses. For secondary transplants, 3×106 bone marrow cells harvested from primary recipients were transplanted non-competitively into irradiated recipient mice. Primary recipients used for secondary transplantation had long-term multilineage reconstitution by donor cells and levels of donor cell reconstitution that were typical (closest to mean values) for the treatments from which they originated. For survival studies in Figure 5, irradiated mice were transplanted with 2×106 bone marrow cells of each genotype and pIpC was administered 4 weeks later to delete Pten. Three to four donors per genotype were used with 5–10 recipients per donor. Mice were sacrificed when they appeared moribund.
Western blotting
Equal numbers of cells from each stem/progenitor population (unless indicated otherwise) were sorted into, or resuspended in, trichloracetic acid (TCA, Sigma). The final concentration was adjusted to 10% TCA. Extracts were incubated on ice for at least 15 minutes and centrifuged at 16,100 × g at 4°C for 15 minutes. Precipitates were washed in acetone twice and dried. The pellets were solubilized in 9M urea, 2% Triton X-100, and 1% DTT. LDS loading buffer (Life Technologies) was added and the pellet was heated at 70°C for 10 minutes. Samples were separated on Bis-Trispolyacrylamide gels (Life Technologies) and transferred to PVDF membrane (Millipore). Western blotting was performed according to the protocol from Cell Signaling Technologies and blots were developed with the SuperSignal West Femto chemiluminescence kit (Thermo Scientific). Blots were stripped with 1% SDS, 25 mM glycine (pH 2) prior to reprobing. The following primary antibodies were used for western blots (obtained from Cell Signaling Technologies unless indicated otherwise): phos-Akt (Ser473; D9E), AKT (C67E7), phos-S6 (SER240/244; polyclonal), S6 (5G10), phos-4E-BP1 (T37/46; 236B4), 4E-BP1 (polyclonal), phos-eIF2α (SER51; polyclonal), eIF2α (D7D3), phos-GSK3β (Ser9; D85E12), β-Actin (AC-74; Sigma), p53 (cm5p; Leica), p21cip1 (F-5; Santa Cruz), and Rpl24 (polyclonal; Abcam). For p21cip1, the membranes were treated with the SuperSignal Western Blot Enhancer (Thermo Scientific). Band intensity was quantified with ImageJ software.
Methylcellulose cultures
200 live bone marrow cells or single CD150+CD48−LSK cells were sorted per well of a 96 well plate containing methylcellulose culture medium (M3434, Stemcell Technologies) and incubated at 37°C in 6.5% CO2 and constant humidity. Colony formation was assessed 14 days after plating, except for Extended Data Fig. 6e where colonies were assessed 15 days after plating. Colony size was assessed by microscopically picking individual colonies with a pipette, washing the cells in PBS, and counting live cells by Trypan blue exclusion on a hemocytometer.
Cell size
The average diameter of sorted cells was measured by sorting cells into flat bottom 96 well plates and analyzing micrographs with ImageJ software.
RNA content
15,000 cells of each haematopoietic cell population were sorted into RLT Plus buffer (Qiagen) supplemented with 2-mercaptoethanol. RNA from each sample was extracted with the RNeasy micro plus kit (Qiagen) into 14 μl of water. 2 μl of each sample was analysed in duplicate for 18S rRNA, 28S rRNA, and total RNA concentration using an Agilent RNA 6000 Pico kit and a Bioanalyzer (Agilent) at the UT Southwestern Genomics & Microarray Core.
Protein content
50,000 cells of each population were sorted into PBS. G0/G1 cells were distinguished from S/G2/M cells based on DNA content using Hoechst 33342 (Sigma) staining of live cells. Cells were spun down and resuspended in 20 μl of RIPA buffer (Pierce) supplemented with complete protease inhibitor (Roche) and rotated at 4°C for 30 minutes. Lysates were spun down at 16,100 × g at 4°C for 15 minutes. The protein content in the supernatant was assessed with the BCA assay (Pierce).
Statistical Methods
In all cases, multiple independent experiments were performed on different days to verify the reproducibility of experimental findings. Group data are always represented by mean±standard deviation. Numbers of experiments noted in figure legends reflect independent experiments performed on different days.
To test statistical significance between two samples, two-tailed Student’s t-tests were used. When multiple samples were compared, statistical significance was assessed using a one way ANOVA or a repeated measures one way ANOVA (when comparing multiple time points or populations from the same mouse) followed by Dunnett’s test for multiple comparisons. When multiple samples were each compared to one another, statistical significance was assessed using a one way ANOVA followed by Tukey’s t-tests for multiple comparisons. Statistical significance comparing overall numbers of long term multilineage reconstituted mice was assessed either by a Fisher’s exact test (Fig. 4f) or by Chi-squared tests followed by Tukey’s t-tests for pairwise comparisons (Fig. 5i and 5k). Statistical significance with respect to differences in survival (Fig. 5g) was calculated using a log-rank test. The specific type of test used for each figure panel is described in the figure legends.
For normalized protein synthesis rates and normalized mRNA expression, means were calculated and statistical tests were performed using log10 transformed data and then means were back transformed to prevent data skewing.
No randomization or blinding was used in any experiments. The only mice excluded from any experiment were those that developed leukaemia after transplantation in Figure 5h,i. Since the purpose of this experiment was to compare the reconstituting capacity of normal HSCs, the presence of leukaemia in a minority of recipient mice was a confounding factor that had the potential to inappropriately skew the results; therefore, the experiment was initiated with the intention of excluding data from any mouse that died during the experiment. We excluded 0–4 mice per treatment.
In the case of measurements in which variation among experiments tends to be low (e.g. HSC frequency) we generally examined 3–6 mice. In the case of measurements in which variation among experiments tends to be higher (e.g. reconstitution assays) we examined larger numbers of mice (>10). In the case of assays that assess protein synthesis, there was no historical data on which to base sample sizes. In that case we performed multiple independent experiments with multiple biological replicates to ensure the reproducibility of our findings.
Supplementary Material
ACKNOWLEDGEMENTS
SJM is a Howard Hughes Medical Institute Investigator, the Mary McDermott Cook Chair in Pediatric Genetics, and the director of the Hamon Laboratory for Stem Cells and Cancer. This work was supported by the Cancer Prevention and Research Institute of Texas and the National Institute on Aging (R37 AG024945). R.A.J.S. was supported by fellowships from the Leukemia & Lymphoma Society (5541-11) and the Canadian Institutes of Health Research (MFE-106993). J.A.M. was supported by the UT Southwestern K12 Pediatrics Training Grant (K12-HD068369). We thank A. Pineda, K. Cowan, E. Daniel, M. Acar, H. Oguro, J. Peyer, K. Rajagopalan, and E. Piskounova for technical support and advice, N. Loof and the Moody Foundation Flow Cytometry Facility, L. Hynan and J. Reisch for advice regarding statistics, J. Shelton for histology, and R. Coolon, S. Manning, M. Gross, and K. Correll for mouse colony management.
Footnotes
AUTHOR CONTRIBUTIONS
R.A.J.S. conceived the project. A.S. developed the OP-Puro reagent and provided advice in the early stages of the project. R.A.J.S. performed all of the experiments with the exception of the western blots in Figure 2e, 5a,d, and Extended Data Fig. 5j which were performed by J.A.M. R.A.J.S., J.A.M. and S.J.M. designed the experiments and interpreted results. R.A.J.S. and S.J.M. wrote the manuscript.
The authors declare no competing financial interests.
REFERENCES
- 1.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakamoto KM, Shimamura A, Davies SM. Congenital disorders of ribosome biogenesis and bone marrow failure. Biology of Blood and Marrow Transplantation. 2010;16:S12–S17. doi: 10.1016/j.bbmt.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsieh AC, et al. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barna M, et al. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456:971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nature Reviews Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 7.Jaako P, et al. Mice with ribosomal protein S19 deficiency develop bone marrow failure and symptoms like patients with Diamond-Blackfan anemia. Blood. 2011;118:6087–6096. doi: 10.1182/blood-2011-08-371963. [DOI] [PubMed] [Google Scholar]
- 8.Wong CC, Traynor D, Basse N, Kay RR, Warren AJ. Defective ribosome assembly in Shwachman-Diamond syndrome. Blood. 2011;118:4305–4312. doi: 10.1182/blood-2011-06-353938. [DOI] [PubMed] [Google Scholar]
- 9.Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112:5228–5237. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- 10.Sen S, et al. The ribosome-related protein, SBDS, is critical for normal erythropoiesis. Blood. 2011;118:6407–6417. doi: 10.1182/blood-2011-02-335190. [DOI] [PubMed] [Google Scholar]
- 11.Payne EM, et al. L-Leucine improves the anemia and developmental defects associated with Diamond-Blackfan anemia and del(5q) MDS by activating the mTOR pathway. Blood. 2012;120:2214–2224. doi: 10.1182/blood-2011-10-382986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beatty KE, et al. Fluorescence visualization of newly synthesized proteins in mammalian cells. Angew Chem Int Ed Engl. 2006;45:7364–7367. doi: 10.1002/anie.200602114. [DOI] [PubMed] [Google Scholar]
- 13.Nathans D. Puromycin Inhibition of Protein Synthesis: Incorporation of Puromycin into Peptide Chains. Proc. Natl. Acad. Sci. USA. 1964;51:585–592. doi: 10.1073/pnas.51.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nature Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- 15.Starck SR, Green HM, Alberola-Ila J, Roberts RW. A general approach to detect protein expression in vivo using fluorescent puromycin conjugates. Chemistry & Biology. 2004;11:999–1008. doi: 10.1016/j.chembiol.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 16.Joseph NM, Morrison SJ. Toward and understanding of the physiological function of mammalian stem cells. Developmental Cell. 2005 doi: 10.1016/j.devcel.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Xu Y, Stoleru D, Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc. Natl. Acad. Sci. USA. 2012;109:413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiel MJ, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 19.Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou S, et al. Bcrp1 gene expression is required for normal numbers of side population stem cells in mice, and confers relative protection to mitoxantrone in hematopoietic cells in vivo. Proc. Natl. Acad. Sci. USA. 2002;99:12339–12344. doi: 10.1073/pnas.192276999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ciechanover A, Finley D, Varshavsky A. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell. 1984;37:57–66. doi: 10.1016/0092-8674(84)90300-3. [DOI] [PubMed] [Google Scholar]
- 22.Luker GD, Pica CM, Song J, Luker KE, Piwnica-Worms D. Imaging 26S proteasome activity and inhibition in living mice. Nature Medicine. 2003;9:969–973. doi: 10.1038/nm894. [DOI] [PubMed] [Google Scholar]
- 23.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes & Development. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 24.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 25.Morrison SJ, Wright D, Weissman IL. Cyclophosphamide/granulocyte colony-stimulating factor induces hematopoietic stem cells to proliferate prior to mobilization. Proc. Natl. Acad. Sci. USA. 1997;94:1908–1913. doi: 10.1073/pnas.94.5.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver ER, Saunders TL, Tarle SA, Glaser T. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development. 2004;131:3907–3920. doi: 10.1242/dev.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fumagalli S, Thomas G. The role of p53 in ribosomopathies. Seminars in Hematology. 2011;48:97–105. doi: 10.1053/j.seminhematol.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Barkic M, et al. The p53 tumor suppressor causes congenital malformations in Rpl24-deficient mice and promotes their survival. Molecular and Cellular Biology. 2009;29:2489–2504. doi: 10.1128/MCB.01588-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JY, et al. mTOR activation induces tumor suppressors that inhibit leukemogenesis and deplete hematopoietic stem cells after Pten deletion. Cell Stem Cell. 2010;7:593–605. doi: 10.1016/j.stem.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magee JA, et al. Temporal changes in PTEN and mTORC2 regulation of hematopoietic stem cell self-renewal and leukemia suppression. Cell Stem Cell. 2012;11:415–428. doi: 10.1016/j.stem.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yilmaz OH, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 32.Kalaitzidis D, et al. mTOR Complex 1 Plays Critical Roles in Hematopoiesis and Pten-Loss-Evoked Leukemogenesis. Cell Stem Cell. 2012;11:429–439. doi: 10.1016/j.stem.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, et al. PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- 34.Zinzalla V, Stracka D, Oppliger W, Hall MN. Activation of mTORC2 by association with the ribosome. Cell. 2011;144:757–768. doi: 10.1016/j.cell.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 35.Guo W, et al. Multi-genetic events collaboratively contribute to Pten-null leukaemia stem-cell formation. Nature. 2008;453:529–533. doi: 10.1038/nature06933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruggero D. Translational control in cancer etiology. Cold Spring Harbor Perspectives in Biology. 2013;5 doi: 10.1101/cshperspect.a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dahlberg A, Delaney C, Bernstein ID. Ex vivo expansion of human hematopoietic stem and progenitor cells. Blood. 2011;117:6083–6090. doi: 10.1182/blood-2011-01-283606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang J, Nguyen-McCarty M, Hexner EO, Danet-Desnoyers G, Klein PS. Maintenance of hematopoietic stem cells through regulation of Wnt and mTOR pathways. Nature Medicine. 2012;18:1778–1785. doi: 10.1038/nm.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vilchez D, et al. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature. 2012;489:304–308. doi: 10.1038/nature11468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vilchez D, et al. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature. 2012;489:263–268. doi: 10.1038/nature11315. [DOI] [PubMed] [Google Scholar]
- 41.Signer RA, Morrison SJ. Mechanisms that regulate stem cell aging and life span. Cell Stem Cell. 2013;12:152–165. doi: 10.1016/j.stem.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.