Abstract
Endo-parasites that affect humans include Plasmodium, the causative agent of malaria, which remains one of the leading causes of death in human beings. Despite decades of research, vaccines to this and other endo-parasites remain elusive. This is in part due to the hyper-variability of the parasites surface proteins. Generally these surface proteins are encoded by a large family of genes, with only one being dominantly expressed at certain life stages. Another layer of complexity can be introduced through the alternative splicing of these surface proteins. The resulting isoforms may differ from each other with regard to cell localisation, substrate affinities and functions. They may even differ in structure to the extent that they are no longer recognised by the host’s immune system. In many cases this leads to changes in the N terminus of these proteins. The geographical localisation of endo-parasitic infections around the tropics and the highest incidences of HIV-1 infection in the same areas, adds a further layer of complexity as parasitic infections affect the host immune system resulting in higher HIV infection rates, faster disease progression, and an increase in the severity of infections and complications in HIV diagnosis. This review discusses some examples of parasite surface proteins that are alternatively spliced in trypanosomes, Plasmodium and the parasitic worm Schistosoma as well as what role alternate splicing may play in the interaction between HIV and these endo-parasites.
Keywords: Plasmodium, Schistosoma, Trypanosoma, HIV, Alternate splicing, Immune evasion
Review
Introduction
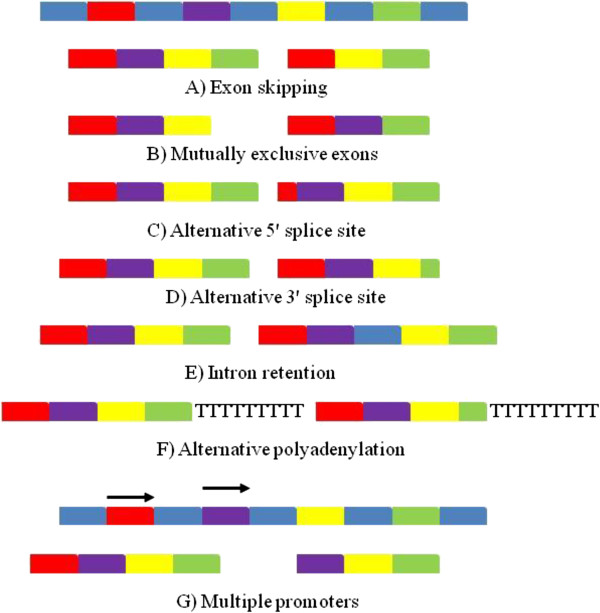
The expression of genes can be controlled at many levels, these include transcriptional regulation, post-transcriptional regulation, translational regulation, mRNA degradation, protein degradation and through the actions of inhibitory proteins. One of the most important and complex mechanisms of post-transcriptional control involves alternate selection of splice sites within the pre-mRNA, which allows multiple different protein products with different functions to be coded for by a single gene [1]. There are five basic forms of alternative splicing (Figure 1). These are exon skipping, mutually exclusive exons, alternative 5’ or 3’ splice sites and intron retention. Two other forms of splice variants occur but they are not normally defined as alternate splicing. These are multiple promoters and multiple polyadenylation sites [2]. Exon splicing involves the removal or retention of an exon. Mutually exclusive exons occur when only one of a set of exons is retained the other is spliced out. Alternative 3’ and 5’ splice sites result in the sequence being extended or truncated at the 3’ or 5’ end due the occurrence of an alternate splicing site at either end. Intron retention occurs when an intron is included and not spliced out of the transcript. Alternative polyadenylation sites can shorten the transcript from its 3’ end. Finally, multiple promoters can lead to transcription starting either upstream or downstream of the first promoter creating a different transcript [2]. RNA splicing is carried out by the spliceosome, which is a large complex of proteins that include five small nuclear RNAs (U1, U2, U4, U5 and U6) and over 150 proteins. The association of these proteins with the snRNAs forms the snRNP (RNA protein complex). Seven of the associated proteins (SM proteins) form a ring around a conserved motif on the snRNA. The pre-mRNA contains short conserved sequences at the 5’ splice site, 3’ splice site and the branch site which is located upstream of the 3’ splice site. U1 associates with the 5’ splice site and U2 with the branch site [3]. The tri snRNP complex U4, U5 and U6 then associate with the pre-mRNA, U1 and U4 are ejected from the complex and the now activated spliceosome performs the first step where the branch site of the intron carries out a nucleophillic attack of the 5’ splice site. The result is the formation of a lariat structure as the 5’ end of the intron is bound to the branch site. The second step is then carried out as the 5’ end of the exon attacks the 3’ splice site resulting in the 5’ and 3’ exons joining. The lariat is then ejected along with U2, U5 and U6 [3].
Figure 1.
Various forms of alternative splicing. The introns are marked in blue with the exons marked in red, purple, yellow and green as the sit in order in the gene. (A) Exon skipping occurs when an exon is excluded. (B) Mutually exclusive exons implies that one exon is included while another is excised (C and D). Alternative splice sites involve the 5’ and 3’ exons being shortened due to internal splice sites. (E) Intron retention occurs when an intron is retained and not excised. (F) Alternative polyadenylation sites can change the length of the 3’ untranslated region. (G) Alternate promoters can change transcription initiation sites.
In order to survive in the hostile environment presented by the host organism, parasites have evolved multiple mechanisms to evade the host’s immune system. Firstly, the parasites may hide in immune privileged tissue such as the central nervous system or fat tissue [4,5]. Secondly parasites can shield surface components to avoid recognition, which is commonly achieved by binding host molecules leading to the parasite being recognized as self by the host immune system [6-8]. Thirdly parasites may actively interfere with the hosts immune system by blocking signaling pathways required to activate different immune responses [9]. Finally they may change their antigenic surface molecules during infection. In order to do this many parasites have multiple surface variants within their genomes which are successively expressed [9]. Opportunistic infections by parasites in HIV patients are a common cause of mortality with most of these parasitic infections being due to protozoan and helminth parasites [10]. Of particular interest here are protozoan parasites of the genera Plasmodium, Trypanosoma and helminth parasites of the genus Schistosoma. The prevalence of these parasites varies geographically but collectively they are common in South America, Africa and South East Asia [10,11]. This coincides with the location of the highest prevalence of HIV infection [11,12].
This review will discuss the role that alternative splicing may play in the ability of a parasite to evade the immune system of the host. The discovery of alternate splicing giving rise to different isoforms of antigenic proteins seems to imply a further means of immune evasion by the parasite by avoiding immune detection. Some of these alternate splicing events give rise to isoforms with different functions allowing different interactions between the host and the parasite. Finally this review will examine the role played by alternative splicing in the interaction between HIV and parasites.
Alternate splicing in protozoan parasites
Trypanosomes
Trypanosoma is a genus of unicellular, flagellate, protozoan parasites, which require more than one host to complete their lifecycle (Figure 2). They are generally transmitted by blood feeding invertebrates and have a vertebrate host. In order to penetrate the host’s cells Trypanosoma use multiple methods including active penetration, active induction of receptor-mediated phagocytosis, and opsonin-mediated phagocytosis. Following invasion the parasite causes cells to rupture releasing trypomastigotes which then multiply. Trypanosomes belong to the class Kinetoplastida and all members of this class possess a DNA containing granule located within their single mitochondrion [13].
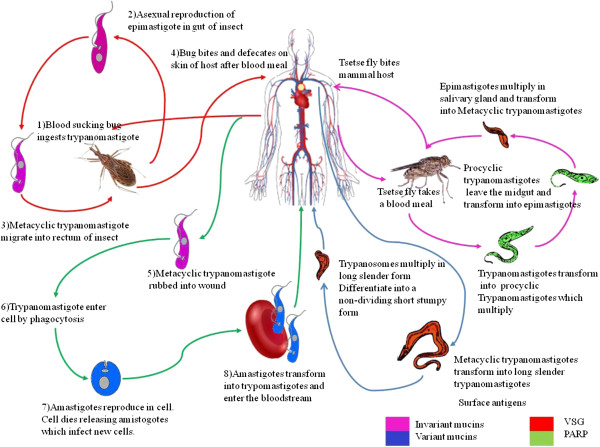
Figure 2.
The lifecycle of Trypanosoma cruzi and Trypanosoma brucei. The life cycle of T. cruzi is shown on the left, that of T. brucei on the right. The two parasites use different strategies to escape the immune system of the mammalian host. T. cruzi enters cells to get away from the immune system. T. brucei escapes from the immune response by expressing a new VSG. These surface antigens vary after a period of time allowing the parasite to continually evade the host immune system. The parasites are coloured according to the surface antigens they display. Antigenic variation most likely evolved to aid in the transmission between the arthropod and mammalian hosts. Both pathogens express highly variant surface molecules Variant surface glycoprotiens (VSG) and variant mucins. However, in the arthropod host the parasites express invariant mucins and procyclic acidic repertoire protein (PARP).
Trypanosoma brucei is a uniquely African trypanosome parasite that causes African sleeping sickness. Nearly 60 million people are at risk from this disease. T. brucei also causes damage to livestock populations. The parasite is spread by the tsetse fly Glossina moristans moristans. The genome of T. brucei is 11 mega bases in size and contains approximately 9000 predicted genes with 900 pseudogenes. Approximately 1700 of these genes are unique to T. brucei[14]. Studies indicate that on average there are 2.6 splice sites for every gene in T. brucei. The number of genes with internal splice sites was estimated to be around 488 [15]. Analysis of alternative splicing sites in the genome of T. brucei pointed to the RNA splicing patterns changing in the different life stages of the parasite. It was suggested that this could be due to differences between the different life stages with regards to: the initiation of transcription, the rate of mRNA degradation or the recognition of splice sites [15]. It can be concluded that alternative splicing is required for the development of the parasite in the different host organisms.
Trypanosoma cruzi is the infectious agent that causes Chagas disease. This chronic illness is characterized by cardiomyopathy and the degeneration of the smooth muscles in the digestive tract. The surface of the parasite is covered in mucintype O-glycosylated sialoglycoproteins. These mucin type O-glycosylated proteins are predicted to play a role in host cell attachment. They also help to protect the parasite from T cell associated killing. These molecules are also antigenic and were found to contain alternate translation start codons preceded by potential trans-splicing acceptor sites. This could give rise to two isoforms of protein although the expression of the smaller variant seems to be dominant [16].
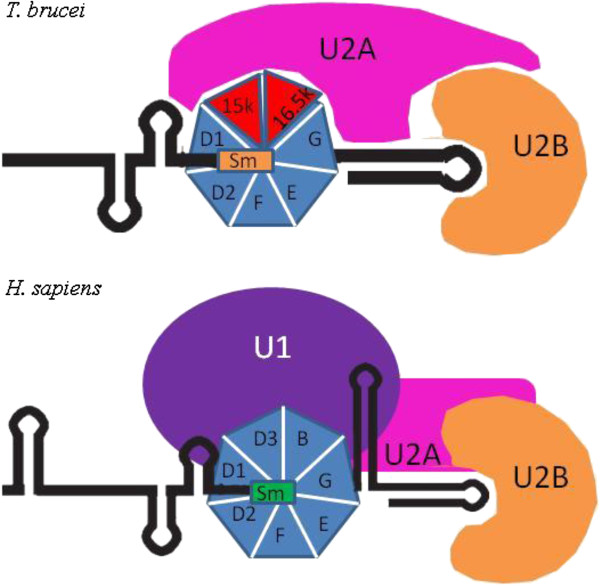
The spliceosome of trypanosomes differs from that of most eukaryotes due to a less stringent conserved motif on the snRNA’s SM site. Two Sm proteins (SmB and SmD3) are replaced by two proteins that are specific to U2 (SM15K and SM16.5K) (Figure 3). The unique Sm proteins seem to be an absolute requirement for the correct assembly of U2 SnRNP, associating with U2 and not U1 [13,17]. They may also be important for the correct positioning of mRNA as trypanosomes do not make use of sequence complementarity to define branch point sequences [18]. The differences between the structure of the human and trypanosome spliceosome are not limited to these Sm proteins, as the human stem loop RNA separating U2A and U1 is absent in trypanosomes [17]. Genes in trypanosomes are also arranged in long polycistronic transcription units. These genes are then transcribed by RNA polymerase II beginning at switch regions and proceeding bidirectionally. The resulting RNA undergoes a specialised form of splicing known as trans-splicing with a different mRNA transcript [13].
Figure 3.
The spliceosome of Trypanosomes compared to that of H. sapiens. Unlike the spliceosome of most eukaryotes the SM protein complex of Trypanosomes contains two unique Sm proteins SM15K and SM16.5K. The Sm core associates with U2 and not U1 in Trypanosomes. The human stem loop RNA separating U2A and U1 is absent in Trypanosomes. The unique SM proteins are required for the correct assembly of U2A in Trypanosomes. The unique structure of the Trypanosome spliceosome seems to play a role in Trans-splicing, which is the dominant form of mRNA splicing in Trypanosomes.
Spliced leader trans-splicing
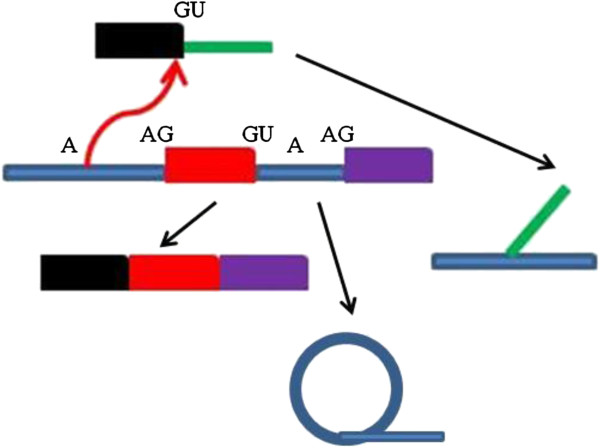
Another form of splicing known as trans-splicing, is used to generate a Short Leader sequence of RNA (the SL RNA), which performs the function of the U1 snRNP (Figure 4) [13]. As previously stated the Sm core associates with U2 and not U1 in trypanosomes and knockdown studies showed that U1 is not required for trans splicing activities [17,19]. With trans splicing being the most common form of splicing in trypanosomes, as most trypanosomal genes do not contain introns, with only two known examples in T. brucei[17]. Trypanosomes are able to successfully evade the host’s immune response by switching the expression of alleles coding for the Variant Surface Glycoprotein (VSG) [20]. In bloodstream forms of T. brucei 7-11% of all spliced transcripts code for VSG [15]. Variations in the surface antigen VSG are created through spliced leader trans-splicing [21]. This specific type of mRNA splicing leads to the replacement of the 5’ end of an mRNA transcript with a small nuclear mRNA referred to as an SL-RNA. The 3’ end of the trans-RNA contains a binding site for the Sm protein complex. SL trans splicing can provide a 5’ cap for protein coding RNAs when the RNA is transcribed by RNA polymerase 1 (Figure 4) [21]. It is also required to turn polycistronic RNA transcripts into individual monocistronic mRNA [22]. However, in terms of increasing the hyper variability of surface proteins of the parasite the most important role that can be played by SL trans-splicing is that of “trimming” or “sanitizing” the 5’ untranslated end of mRNA. By removing the 5’ end of the transcript it does not matter what sequence is present, and this would allow for less selective pressure on the 5’ end allowing for the evolution of the gene, by including new transcription start sites or to incorporate a wider variety of useful sequence components [22].
Figure 4.
Spliced Leader RNA trans-splicing. The SL RNA (black) attaches to the first exon (red) of an mRNA. The outron of the mRNA and the intron of the SL RNA attach and form a branch in the same way that in traditional splicing the intron forms a lariat.
The ability to vary the VSG structure and avoid recognition by the host immune system depends on the trans-splicing of mRNAs encoded by genes that have been rearranged and duplicated. There are over 1000 VSGs encoded in the genome of Trypanosomes [23]. This form of alternative splicing will have important consequences for the cellular location of the peptide as it changes the N terminal targeting sequence. It may also play a role in changing the stability and half-life of the protein.
Plasmodium
For thousands of years malaria has persistently claimed a large number of human lives. In 1880 a French army physician, Charles Louis Alphonse Laveran, identified the causative agent of malaria, in the form of gametocytes of the protozoan parasite Plasmodium falciparum. The genus Plasmodium is made up of unicellular parasites that infect a wide range of hosts. These parasites replicate asexually and target the host’s erythrocytes leading to a stiffening of the erythrocyte membrane. Plasmodium falciparum, as the leading cause of malaria infection (75% of malaria), is considered the most dangerous parasite. One of the features of malaria is the inability of the host’s immune system to prevent infection with the re-infection of hosts occurring even in endemic malaria areas. Malaria parasites are able to evade the immune response of the host, firstly by going through most of its lifecycle (Figure 5) without presenting any antigen on the surface of infected cells and secondly by increasing the hyper-variability of the surface proteins. This is achieved via the amplification of extensive repertoires of multicopy hyper-variable gene families that encode infected erythrocyte or merozoite surface proteins [24]. The genome of Plasmodium falciparum consists of roughly 5700 genes and at 19% has the lowest GC content of any genome sequenced thus far [25]. Sequencing of RNA transcripts or cDNA libraries pointed to the fact that the transcriptome contained variable length un-translated regions as well as prevalent alternate splicing of transcripts [26]. In addition to this over 50% of the genes in P. falciparum contain introns [27]. Using current sequencing technologies four types of alternate splicing strategies were identified in Plasmodium. These were exon skipping, intron retention or creation and 3’ or 5’ alternate splicing arising from coordinate changes to exons or alternate start or stop codons [26]. Splicing of genes in P. falciparum was found to have effects on the localisation of the protein within the cell as splicing results in the removal of a signaling peptide. Similarly the cell localization can be changed by affecting the solubility of the protein via the removal of a trans-membrane domain. Splicing has also been observed to drastically alter protein function due to the removal of an active domain. Premature stop codon can also contribute to protein regulation by resulting in the abolishment in the expression of the mature protein [28].
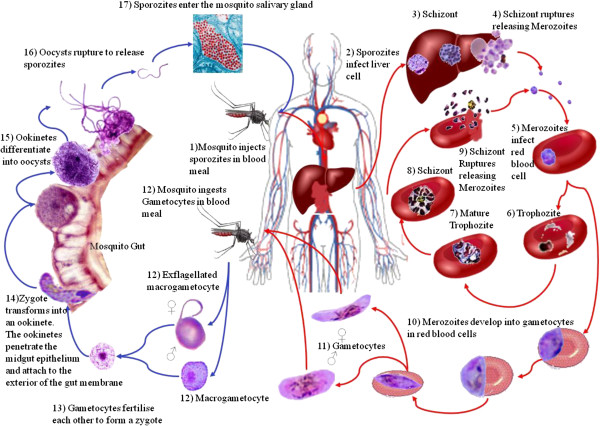
Figure 5.
Life cycle of Plasmodium falciparum. The human host harbouring sexually immature parasite is marked using red arrows. Here the parasite multiplies asexually in the liver (exoerythrocytic schizogony) and in red blood cells (erythrocytic schizogony). The sporozoites that invade the liver become trophozoites, divide and develop into a liver schizont. These contain thousands of nuclei. The resulting merozoites infect blood cells and turn into trophozoites. These develop into schizonts with 8–32 nuclei which are released when the erythrocytes burst and can re-infect new erythrocytes. Erythrocytes that contain other stages stick to the endothelial cells of the blood capillaries, escaping destruction in the spleen, allowing gametocytes to be produced. The cycle in the mosquito which harbours sexually mature parasites is shown in blue. Here the sexually differentiated gametocytes change into gametes in the mosquito's gut, where fertilisation of the macrogamete occurs. Large oocysts, containing up to 10,000 sporozoites, develop on the outer surface of the gut wall. When they burst the sporozoites enter the mosquito's hemolymph, and ultimately reach the salivary glands.
Splicing in plasmodium
A putative homolog of the SF2/ASF SR splicing protein was identified in Plasmodium. This component of the spliceosome affects alternative splice site selection by antagonizing other SR proteins as well as functioning to stimulate transcription by binding to ribosomes [28]. In addition to this, a functional SR protein PfSR1 as well as the SR-protein specific kinase that regulates it (PfSRPK1) have also been identified [29]. The splicing branch points, that give rise to the excised lariat structure of the intron, are also unusual in P. falciparum. The 5’ splice site is poorly conserved and is able to still function with various substitutions. Some introns also have numerous branch points indicating a further mechanism for alternate splicing [30].
Alignment of a cDNA library from P. falciparum revealed that there were 363 splicing events that demonstrated splicing of intron-exon junctions from the antisense strand. The majority of these antisense transcripts mapped to genes that contain introns, implying that they may play a role in intron splicing [31]. Antisense splicing may play a role in the control and regulation of transcription of sense RNA. This can be accomplished through steric hindrance as two polymerases cannot function on opposite DNA strands. An antisense RNA molecule may bind to and hide a splicing site on the sense mRNA leading to alternate splicing [31].
Surface antigens
Various antigenic proteins produced by Plasmodium species are known to have isoforms that are the result of alternate splicing. The blood antigen protein P41-3 of P. falciparum is a 375 amino acid protein encoded by a 2137 bp gene. It was the first parasite gene where alternative splicing was observed (Figure 6) [32]. Alternative splicing of the mRNA gives rise to three different transcripts of 2.4 kb, 2.1 kb, and 1.4 kb. The exact role of this protein is not known, however it appears to be a soluble protein that is exported from the parasite [32]. The P. falciparum surface antigen UB05, which is recognized by IgG antibodies from adults that are semi-immune to malaria infection [33], was found to be alternatively spliced through 3’ and 5’ alternate splicing [26,34]. Another suspected surface antigen, PF 70, is soluble in Triton X, suggesting that it is membrane bound. The protein is a novel 70 kDa protein that reacts with anti P. falciparum and anti P. yoelli serum [35]. The mRNA coding for PF 70 contains a 5’ alternative splice site, resulting in a protein that is 25 amino acids shorter than the protein translated from the full length transcript [34].
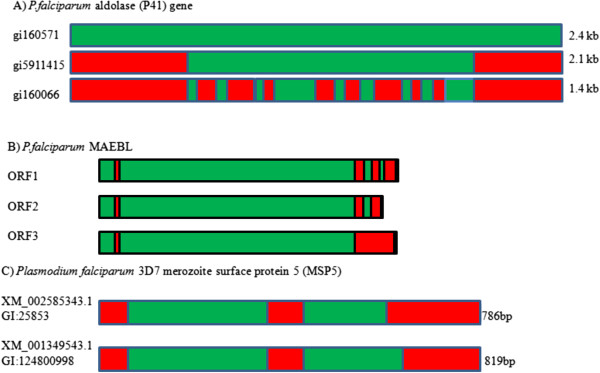
Figure 6.
Transcripts of alternatively spliced immune related genes in Plasmodium. (A) The blood antigen protein P41-3 is alternatively spliced to give rise to three different transcripts of 2.4 kb, 2.1 kb, and 1.4 kb. (B) The MAEBL trans-membrane receptor binds erythrocytes and is expressed as different isoforms depending on the developmental stage of the parasite. The maebl gene consists of two open reading frames ORF1 and ORF2. Some of the isoforms are soluble unlike the typical trans-membrane MAEBL and these soluble isoforms are the result of an ORF2 transcript. (C) Merozoite surface proteins found on the surface of the merozoite P. falciparum contain three MSPs: MSP2, MSP4 and MSP5. The 40 kDa MSP5 is alternatively spliced using an alternate stop codon.
Another class of surface antigens, Merozoite Surface Proteins (MSP) are proteins found on the surface of protozoan daughter cells. Interest has been expressed in the use of these proteins to create a malaria vaccine, as initial reports suggested that this protein is exposed on the surface of the merozoite and displays limited genetic diversity [36]. In P. falciparum chromosome 2 contains three MSPs: MSP2, MSP4 and MSP5. MSP5 is a 40 kDa glycosylphosphatidylinositol (GPI)-anchored protein with an epidermal growth factor domain [37]. However, recently it was discovered that MSP5 is alternatively spliced using an alternate stop codon (Figure 6) [26]. This may then result in increased variation despite a high sequence similarity, thus allowing the merozoite to avoid the immune system of the host and decreasing its usefulness as a vaccine target.
Malaria is associated with the adherence of infected erythrocytes to the endothelial lining of blood vessels [38]. The Merozoite Apical Erythrocyte-Binding Ligand (MAEBL) trans-membrane receptor was identified in the blood stage of P. falciparum. The protein binds erythrocytes and unlike many proteins from P. falciparum, which seem to be stage specific, MAEBL is expressed in both sporozoites and merozoites. However, the maebl gene consists of two open reading frames ORF1 and ORF2. This results in maebl being expressed as different isoforms depending on the developmental stage of the parasite (Figure 6) [39]. Some of the isoforms are soluble unlike the typical trans-membrane MAEBL and these soluble isoforms are the result of an ORF2 transcript. Transcripts of this soluble MAEBL are only found in the oocyst stage. It is unknown what the significance of these alternatively spliced isoforms are [39]. Another protein involved in erythrocyte binding is Cytoadherence linked asexual gene 9 (clag9). This is a trans-membrane protein found on the surface of infected erythrocytes that is required to bind to the endothelial cell receptors [40]. By measuring antibody response to synthetic clag9, it was determined that this protein is immunogenic [41]. Clag9 was found to be alternatively spliced through 3’ and 5’ alternative splicing [26]. The exact roles played by the isoforms of these various protein antigens remains unknown.
In addition to these Otto [26] concluded that there were an additional 32 conserved unidentified proteins in P. falciparum that could be expressed as different isoforms due to alternate splicing. Another member of the phylum Apicomplexa that is closely related to Plasmodium is Theileria annulata. Theileria are able to reversibly transform their host’s cells. These parasites are tick borne and cause lympho-proliferative diseases of cattle – tropical theileriosis and East Coast fever (ECF) [42]. The gene coding for an immunogenic protein, that was used to establish a diagnostic ELISA assay, was given the name clone 5. This gene is alternately spliced at its single intron. This splicing is a combination of intron retention and 3’ splice site alteration due to the introduction of another stop codon. Both mRNAs are translated into a corresponding long and short protein [43].
Hyper-variable gene families
Plasmodium species express four families of hyper-variable genes. These genes have many copies in the genome. The first of these families is used to prevent the parasite being sent through the spleen and destroyed. The P. falciparum erythrocyte membrane protein 1 (PfEMP1) family, are large proteins ranging between 250 and 300 kDa. They function as cytoadhesion proteins which results in infected erythrocytes sticking to the walls of blood vessels and not cycling through the spleen. These proteins are encoded by a multicopy gene family named var which consists of about 60 genes, of these only a single protein is expressed at a time leading to a wide variety of antigenic phenotypes and cyto-adherent abilities [24]. The Plasmodium vivax homologs of the var family, named the vir family, display varying numbers of exons as well as alternate splicing [44]. Var gene expression seems to be regulated through chromatin remodeling as var expression is silenced by histone deacetylases and enhanced by histone acetylation [45]. Two separate promoters control var gene expression. An upstream promoter creates coding mRNA, and an internal intron promoter which creates a non-coding RNA. The intron promoter seems to be bi-directional. This promoter gives rise to short antisense mRNA that appears to associate with the var genes on the chromosome. In this way the antisense RNA may regulate the transcription of var gene products [45].
The other three families all contain a trafficking motif known as pexel/TS, which allows the protein to be transported to the cytoplasm of the infected erythrocyte. They also share the same gene architecture consisting of a short first exon encoding a signal peptide followed by a longer exon. All three families code for trans-membrane proteins of varying size but similar structure. They consist of a two trans-membrane domains joined by a variable loop. This loop contains the hypervariable region which is exposed on the outside of the erythrocyte to antibody mediated immune selection [24]. The Pfmc-2TM family consists of 13 genes; the subtelomeric variable open reading frame family (stevor) consists of 30–40 genes [24] and the repetitive interspersed family (rif) consists of over 150 copies. This makes the Rif family the largest gene family in P. falciparum[46]. There is evidence that both the rif and stevor genes are alternatively spliced.
Stevor is suspected to play a role in antigenic variation [47,48]. It is expressed in the late stages of the asexual forms of the parasite, where it gathers in structures known as the Maurer’s cleft. This structure plays a role in the release of parasites from erythrocytes [48]. The expression of the protein at only the late stages seems to imply that it plays a role in the release of the parasite from erythrocytes or alternatively it acts to shield other cleft components from the immune system of the host [48]. Stevor is also expressed in the gametocytes and it is here where alternate splicing gives rise to a truncated version that lacks one of the trans-membrane regions [48], implying changes in the proteins function of localisation. These Stevor transcripts have a diverse structure resulting from the use of alternate splice sites giving rise to frame shifts and deletions [49]. Mature stage gametocyte infected erythrocytes need to re-enter circulation in order to infect blood feeding mosquitoes. This is achieved by increasing the severity of membrane deformation. Stevor has been shown to play a role in erythrocyte membrane deformation [50] where an increase in the rate of disassociation of Stevor from the membrane surface occurs prior to increased membrane deformation [51]. A switch between the transcription of various isoforms, especially those with fewer trans-membrane regions, may aid in this dissociation. The Rif family was found to be 3’ and 5’ alternately spliced [26]. The ORF of the genes are also spliced into short signaling peptides [46].
Alternative splicing of genes in the immune response of the mosquito host
The salivary gland of the female mosquito is the primary site of infection by Plasmodium parasites. An analysis of the transcriptome of the salivary gland before and after infection revealed changes in the levels and identities of the transcripts. In the case of the D7 odorant binding protein (OBP) there is a change in the identity of the transcripts due to alternate splicing [52]. There are instances in which odorant receptor molecules have been identified as being involved in innate immune signalling systems [53]. Additionally expression of OBPs may be in response to oxidative stress where they act as scavengers of reactive oxygen species [54] and this may be important to protect the mosquito from the negative effects of its own immune response to the parasite. Infection with dengue virus increased the expression of OBP in Aedes aegypti. This was shown to be a strategy by the virus to manipulate the behavior of the mosquito with an increase in the expression of OBPs leading to an increase in the frequency of the mosquito feeding, thereby increasing host infection [55].
Molecules with a more direct function in the immune system whose transcription is altered following infection with malaria parasites include serpins [56], peptidoglycan recognition proteins [57] and Down syndrome cell adhesion molecule (Figure 7) [58]. Serpins play an important role in the regulation of the immune response. The classic example is the role played by the serpin Nec in down-regulating the Toll pathway by inhibiting the serine protease Persephone [59]. As a result of alternate splicing the Serpin10 gene of Anopheles gambiae can give rise to four splice variants due to alternate splicing. The transcription of two of these isoforms is increased following infection with Plasmodium bergii while bacterial infection had a different effect on the levels of each isoforms mRNA [56]. Serpin10 is also known as plasmodium-related inhibitory serine protease inhibitor [60]. Alternative splicing also gives rise to three isoforms of Serpin4 from A. gambiae[61]. The functional role of these isoforms is unknown. However, the A. gambiae Serpin10 and Serpin4 show the highest sequence similarity with the Drosophila melanogaster Serpin42Da and Serpin28Dc respectively. Serpin28Dc is suspected to play a negative role in the regulation of melanization, the defense response and to be involved in the wound response [62].
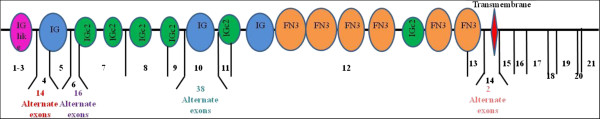
Figure 7.
Alternative splicing of the Anopheles gambiae Down syndrome cell adhesion molecule (DSCAM). This Ig domain protein has multiple alternative splice exons within its IG regions. The above image shows the different domains found within the protein and the exons encoding these domains. Exons 4, 6, 10 and 14 are alternately spliced with 14, 16, 38 and 2 alternate exons respectively. The different combinations of these exons can give rise to 31 920 splice forms. The protein plays a pathogen recognition role, with the large number of isoforms allowing it to recognize, bind and respond to a wide array of different pathogens.
An A. gambiae a homolog of the Peptidoglycan Recognition Protein long form C was found to encode for three different isoforms which carry alternative PGRP domains linked to a common backbone consisting of a trans-membrane region and signaling peptide [57]. In Drosophila this protein is required for the detection of bacterial pathogens and initiates an immune response via the Imd pathway [63]. The mosquito isoforms are, however, differentially transcribed depending on the nature of the infectious agent [57]. The mosquito PGRP-LC detects bacterial infection but also detects Plasmodium and this detection results in an anti-parasitic defense response. The isoform responsible for this detection and subsequent response is PGRP-LC3 [64].
The Down syndrome cell adhesion molecule (DSCAM) is an Ig domain containing protein that was identified in Drosophila as playing a role in neuronal guidance. This molecule contains multiple alternative splice exons within its IG regions (84) that can give rise to 31 920 splice forms (Figure 7) [58,65]. It was first suspected to be involved in the immune response of insects when it was found to be expressed in immune related cells within Drosophila and was later identified to be an essential recognition molecule in the insects immune response [65]. The hyper variability of the molecule allows it to recognize, bind and respond to a wide array of different pathogens with differently spliced forms being expressed in response to different pathogens [58,65]. Plasmodium infection results in the increase in variability of exon 4 and 6 in particular. The increase in the variability of exon 6 also seems indicative of infection by multiple strains of Plasmodium with different genotypes [58]. In this way the mosquito can respond to the hyper variability of the pathogen.
Schistosoma
The tropical disease schistosomiasis is caused by blood flukes of the genus Schistosoma. This disease affects 210 million people and is still a major source of morbidity, causing approximately 280, 000 deaths per year in sub-Saharan Africa. These worms require a vertebrate host and an intermediate gastropod host (Figure 8). Schistosomes are not hermaphroditic with male worms having a gynecophoral canal in which the female worm resides. The sequencing of the genome of Schistosoma mansoni revealed 11, 809 putative genes encoding 13,197 transcripts [66].
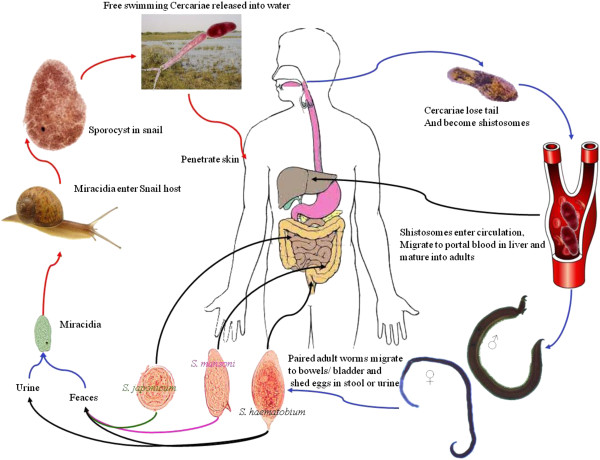
Figure 8.
Life cycle of Schistosomes. Unlike other Trematodes, schistosome adults have two sexes and are located in blood vessels. Eggs from human hosts hatch into free swimming miracidia which infect specific snail species. These develop into two generations of sporocysts and result in the production of cercariae which are released from the snail and swim freely in water where they encounter and penetrate the skin of the human host. The cercariae shed their tails to become schistosomules, which enter small veins and develop into adult worms in mesenteric veins. Their exact location varies with species. S. hematobium occurs in the bladder venous plexus and rectal veins. Eggs are deposited in the small veins of the portal and perivesical systems. In S. mansoni (pink arrows) and S. japonicum (green arrows) the eggs migrate to the lumen of the intestine and are excreted in the feces. In S. hematobium (black arrows) eggs migrate towards the lumen of the intestine or the bladder and uterus and are eliminated with feces or urine.
The first report of alternate splicing in S. mansoni concerned the use of sera from humans infected with S. mansoni to recognise an antigen from esophageal gland of S. mansoni. This antigen was given the name 10–3 and is made up of multiple alternatively spliced 27 bp exons and 81 bp repeats [67]. Alternative splicing also occurs due to different transcription start sites and changes in mRNA processing. This alternative splicing gives rise to isoforms that are differentially expressed in different life stages of the parasite (Figure 9). The 10–3 gene contains tandem repeats and all transcripts share the same 3’ sequence. However, the sequence 5’ of the tandem repeats is variable [68].
Figure 9.
Transcripts of alternatively spliced immune related genes in Schistosoma mansoni. (A) MEG 1 has 18 isoforms. These isoforms display very lifestage specific transcription. The exact role of MEG1 is unknown. (B) MEG 3 has four members with three, 10, three, and two isoforms four family members with 3, 1 and 4 isoforms each. MEG 3 is also known as GRAIL. (C) The 10–3 surface antigen also known as MEG 4 has different transcription start sites that give rise to two isoforms that are expressed at different stages in the parasites lifecycle. (D) SmVAL6 contains 38 exons, the four largest code for conserved protein motifs, while the smaller exons are alternately spliced to give rise to at least 5 isoforms. SmVAL6 probably arose due to a recombination event between a MEG gene and a SmVAL gene.
At least 45 genes were identified that contained a gene structure consisting of multiple very small exons (micro-exon structure) (Figure 9). Although micro-exon genes (MEG) have been identified in other organisms, S. mansoni is unusual in that the micro exons can make up to 75% of the coding sequence of the gene. Most of these genes contain a signal peptide at the 5’ end, while three have membrane anchors. Sequencing of transcripts of MEGs from families MEG-1, MEG-2 and MEG-3 showed that there were up to seven splice variants that arose due to exon skipping [66,67]. The 10–3 antigen described above was identified as a MEG and given the alternate name of MEG-4 [67]. The initial number of micro exon containing genes (45) that were identified in S. mansoni[66] was upon further analysis expanded by another 27 genes giving a total of 72 genes consisting of microexons. These genes can be divided into 18 families, which can be further divided into two groups, based on the number of members they contain [67]. Further transcriptome sequencing may have identified a further 11 micro-exon containing genes [69]. Protein expression studies show these genes code for small proteins with isoforms that have very similar size and charge that appear to be secreted. These proteins are expressed in epithelial and gland tissue. 16 MEG protein products have also been detected in secretions or detected in the glands of S. mansoni. The transcription of these genes increases during the mammalian stages of the life cycle where the parasite may interact with the host’s immune system, and appear to be associated with host invasion and persistence within the host [67,70]. Another feature of MEGs which allows for the neat segmentation of the coding region is the fact that the exons are symmetrical. This allows for the excision and without altering the reading frame [67].
Despite the fact that some of these proteins may be membrane bound and therefore exposed to the external environment of the host; it is unknown whether they are antigens. However, as they are detected in secretions and seem to be directly associated with a particular life stage, it is likely that these proteins are being directly exposed to the host and its immune system [67]. Therefore, the ability to vary their structure may provide protection against immune recognition by the host through an antigenic variation based immune evasion strategy similar to that seen in trypanosomes and Plasmodium. However unlike the antigenic variation genes within those parasites, these MEGs are expressed simultaneously and not sequentially [67]. A gender based alternative splicing of a transcription co-factor that interacts with the spliceosome was identified in S. mansoni. The gene SmCA150 codes for a protein that influences the alternative splicing of other gene transcripts. This may form the basis for other differences observed in RNA splicing observed between males and females [71].
The S. mansoni venom allergen like (SmVAL) protein family was given this name due to the similarity of the proteins with wasp venom allergen [72]. All these proteins contain a sperm coating protein domain. This domain is found in proteins that play a role in the immune response, testes development, as a venom toxin and host invasion, where parasitic nematode species secrete SCP/TAPS proteins into the host during infection [72]. In plants, proteins containing this domain are involved in the immune response, while in mammals they are known to be involved in lung development [73].
29 SmVAL genes have been identified and can be divided into two groups. The first group codes for proteins that contain a signal peptide and appear to be excreted. The second group codes for proteins that do not contain a signal peptide and should play an intracellular role [72]. Group 1 VALs such as 1, 2, 3, 4, 5, 7, 8, 9, 10, 12, 14, 15, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27 and 28) are associated with host invasion. SmVALs4 10 and 18 are associated with the invasion of the mammalian host. The other group 1 VALs appear to be involved with the invasion of the molluscan host. The role of most group two SmVALs such 6, 11, 13, 16 and 17 are unknown. However SmVAL6 is a group 2 VAL expressed during the mammalian life stages [73]. The presence of VALs in parasitic and non-parasitic helminthes indicates that they may play a role in life cycle changes [73].
Through the use of Rapid Amplification of cDNA Ends (RACE) mRNA isoforms were identified for multiple SmVALs. These included SmVAL1, SmVAL2, SmVAL4, SmVAL6, SmVAL7 and SmVAL11[72]. SmVAL6 was shown to contain micro-exons with alternative use of these exons giving rise to the different isoforms [72]. These isoforms were expressed at different lifestages of the parasite. For instance SmVAL1, SmVAL2 and SmVAL4 isoforms were isolated from mixed sex cercariae RNA while SmVAL6, SmVAL7 and SmVAL11 isoforms were isolated from mixed sex, 7-week adult RNA [72]. One of the members of this family SmVAL6 contains four exons of an average size and code for the conserved motifs in the protein family (Figure 9). However, this is followed by seven small exons, which is similar to the structure of MEGs. As many as 5 isoforms have been identified that occur due to alternate splicing of these smaller exons. SmVAL6 probably arose due to a recombination event between a MEG gene and a SmVAL gene. Additionally, both proteins are excreted and therefore will probably interact with the host’s immune system and generate an immune response [72]. The antigenic role played by different SmVALs was found to vary amongst individual family members. For example despite SmVAL4 and SmVAL26 both being secreted group 1 VALs, SmVal4 elicits an immune response in mice while smVAL26 does not, as assessed by a mouse model of airway inflammation. This difference in members of this family to elicit an immune response is most likely based on the localisation of the protein within the parasite. SmVAL4 and SmVAL26 are both group 1 SmVALs and are secreted. Transcription and expression data show that SmVAL4 is excreted during mammalian host skin invasion and is released during the cercariae stage, while SmVAL26 is excreted from the egg (Figure 8) [74].
The polymorphic mucins (SmPoMucs) are a protein family consisting of approximately ten members in S mansoni. The gene structure consists of between 0 and 20 tandem repeats of a 27 base pair exon near to the 5’ end of the genes. Alternative splicing as well as trans-splicing between transcripts generates hyper variability. Splicing also affects the proteins post-translational modification [75]. The mollusc host produces a highly variable immune receptor Fibrinogen Related Protein 3 (FREP3) that is required for host resistance to infection by S. mansoni[76,77]. These FREP3s interact and form complexes with the SmPoMucs [76]. Therefore the variation produced by alternate splicing plays a role in avoiding recognition by the invertebrate host’s immune system.
Parasites, host immunity and HIV
Like the immune response against most infectious agents, the immune response to parasites includes non-specific factors, such as the genetic factors of the host, and specific immune responses. Cellular immunity appears to be the most important immune response against parasites such as Plasmodium while antibody based responses are the most important when dealing with trypanosomes. Cytokine production by the different subsets of T cells is important in the immune response and patient pathology [78]. Different subsets of helper and cytotoxic T cells are activated in response to different parasite infections. The Th1 cells respond through the production of interferon (IFN)-interleukin (IL)-12 and results in the activation of monocytes and macrophages and CD8+ cytotoxic cells. The Th2 cells produce IL-4, IL-5, IL-13 and results in antibody production [79].
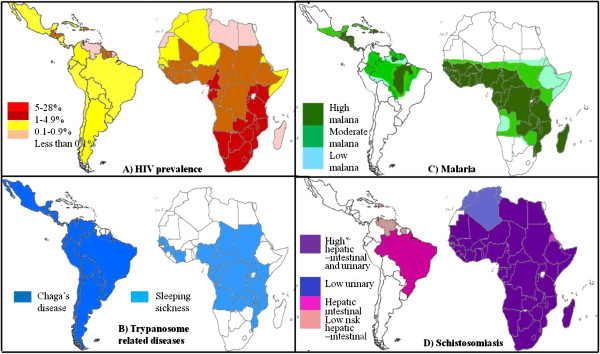
The ideal situation for these eukaryotic parasites is to exist in the host at a level where infection persists without resulting in the death of the host. This chronic infection will result in a high incidence of immunopathology [78]. Patients with an immunocompromised immune system resulting from immunosuppressant drugs, chemotherapy or HIV infection show impaired CD4+ cell mediated immune response and an exacerbation of disease associated pathology [79]. Due to the overlap in the geographical locations of the highest incidence of HIV and most of the protozoan parasite infections (Figure 10), the combined effect of parasitic and HIV infections has proved to be an important consideration for diagnosis, treatment and prognosis of these infections.
Figure 10.
The overlap in the geographical location of high HIV prevelance and parasitic diseases. The above maps of South America and Africa show the prevelance of HIV infection (A) and the occurrence of the parasitic infections by T. cruzi and T. brucei(B); malaria causing Plasmodium species (C) and Schistosoma parasites (D). These maps demonstrate that there is a geographical overlap amongst these infection. This implies that the increase in HIV prevelance observed in these areas may partly be due to the effect these paerasites have on host immunity.
Infections with trypanosomes [80], malaria [81] and Schistosoma[82] are known to complicate the diagnosis of HIV infection using rapid diagnostic tests, due to the parasites inducing polyclonal B cell activation. This results in the production of a wide array of non-specific antibodies that can result in false positive diagnoses [80]. Chronic malaria infection also results in the increase in the levels of HIV RNA as well as a decrease in CD4+ cell number while HIV infection results in the increase in the severity of bouts of malaria [83]. Schistosoma infections accelerate HIV disease progression and transmission. This is largely due to physical damage in the urinary tract of the host as a result of the presence of the parasites eggs [12]. Schistosoma co-infection of Rhesus Macaques infected with Chronic Simian-Human Immunodeficiency Virus Clade C led to an increase in viral RNA expression and lower CD4+ counts [84]. S. mansoni infection leads to the increase in the number of cytokines cell surface receptors which can be used by HIV to infect a cell. The parasite also suppresses the Th1 immune response needed for the immune response to HIV [12,85]. For instance Schistosoma infection leads to increased CXCR4 expression levels on individual cells, but does not increase the number of cells that express this receptor. However, treatment of schistosomiasis leads to a decrease in the level of CCR5 and CXCR4 on CD4+ cells in both HIV infected and uninfected patients. Schistosomiasis was also found to lead to increased susceptibility to HIV infection, while treatment decreases the susceptibility to HIV infection and may slow disease progression [85]. Infection with T. brucei prior to HIV infection can however, decrease the viral infectivity, by inhibiting the ability of the virus from entering cells [86]. These interactions between parasites and HIV that alter infectivity and lowers the prognosis for AIDS appear to occur mainly at the level of the cell signaling molecules and cytokines. This may be due to the increase in the levels of cytokines such as (IFN)-γ, interleukin (IL)-12 and TNF, which can stimulate HIV replication [83], the inhibition of IL-2 synthesis [87] increase in CCR5 and CXCR4 synthesis [12]. Many of these cytokines, such as CD4, CD8 and CTLA4, are expressed as different isoforms, with the alternative splicing patterns for these genes changing in response to antigen levels (Figure 11) [88].
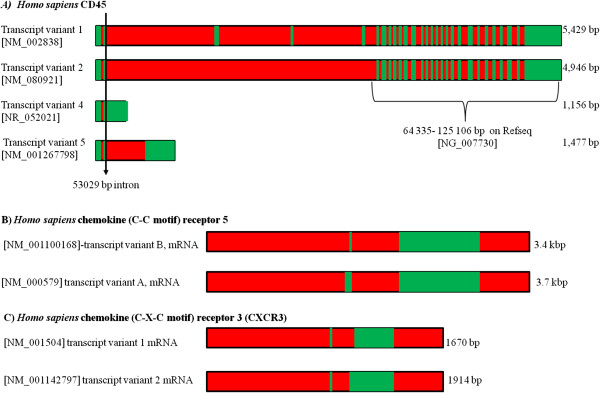
Figure 11.
Alternative splicing of cell surface receptors in Homo sapiens that play a role in HIV and parasite infections. (A) Sequence data was available to construct a diagram showing splicing for 4 CD45 isoforms. These isoforms arise due to the alternate splicing of introns 4–6. The resulting mRNAs differ drastically in size and sequence. The CD45 gene is 125 kbp in size with a large intron between exons 2 and 3. Isoforms 1 and 2 contain multiple exons dispersed across a large 60 kbp stretch of DNA. (B) Two CCR5 arise due to alternative splicing of a transcript derived from an alternative upstream promoter. The larger isoform negatively controls the expression of the smaller isoform. (C) Two CXCR3 isoforms have been identified which interact with the same chemokines but, this interaction results in opposing signaling based on the isoform present.
CD45
At least 5 different CD45 isoforms are expressed due to the alternate splicing of introns 4–6. These isoforms range in size from about 180–240 kDa (Figure 11) with the levels of the lower molecular weight isoforms increasing after the T cell has been exposed to an antigen. These T cells with higher levels of the low molecular weight forms also have a higher turnover rate [89,90]. These CD45 isoforms may also differ in their ability to promote IL-2 secretion, their ability to associate with CD4 and CD8 and their ability to promote B cell maturation. Evidence does exist for the homodimerisation of CD45 which may also regulate CD45 function further through the homodimerisation of different isoforms [90]. It has also been noted that the expression of these isoforms may be dependent on the type of cell or the state of cell activation and differentiation [91], while HIV infection results in reduced surface expression of all CD45 isoforms [92].
The T. evansi parasite that causes surra disease in animals possesses a membrane bound toxic protein that kills lymphocytes in a CD45 dependent manner. The presence of CD45 isoforms may explain why only some of the population of peripheral blood mononuclear cell (PBMCs) died after they were treated with T. evansi protein extracts [91]. Like HIV, trypanosome parasites are further able to suppress the immune system of their mammalian hosts via an increase in the apoptotic killing of T cells [87]. Infection with trypanosome parasites induces a strong CD8+ T cell response in mammalian hosts. These cells predominantly recognise the trans-sialdase epitope [93]. T. cruzi and T. brucei do not synthesize their own sialic acid for inclusion into their surface receptors. Instead they make use of the enzyme Trans-Sialidase found on the surface of the parasites, to transfer sialic residues from the sialic-glycosylates of the host. This enzyme is also shed into the bloodstream where it leads to the induction of apoptosis in immune related cells [94]. This is a result of the sialylation of surface receptors such as CD4, CD8 and CD45 [94,95]. The different isoforms of CD45 are the main targets of Trans Sialidase, which may result in changes to the functions of the different isoforms due to changes in glycosylation [94].
Chronic P. falciparum exposure results in atypical memory B cells (MBC) that express increased levels of CD45RA as well as negative regulators of T cell response Programmed Death 1 (PD-1) and lymphocyte-activation gene-3 (LAG-3) [96]. Atypical MBCs are also a characteristic of HIV infection. These MBCs are less responsive to infection and are thus known as exhausted CD8+ T cells [97]. Naïve T cells are known to express higher CD45-RA [98] and this combined with the expression of the negative regulators of T-cell activation, suggests another means for P. falciparum to evade the immune response of the host.
CCR5
The chemokine receptor CCR5 found on the surface of white blood cells plays a central role in HIV infection, where it acts as a co-receptor allowing the virus to enter target cells. Multiple isoforms of CCR5 arise due to alternative promoter regions as well as mRNA splicing (Figure 11). Of particular interest is two isoforms that arise due to the alternative splicing of a transcript derived from an upstream promoter [99]. Studies indicated that the longer isoform plays a role in negatively regulating the expression of the shorter isoform. Further isoforms that arise due to processing of transcripts from a downstream promoter may also play a role in expression by changing the stability of the mRNA [99]. HIV is too recent a human pathogen to have exerted any selective pressure on the various isoforms. However, pathogens that have infected humans for longer may have played an important role in exerting a selective pressure on the function and occurrence of these regulatory isoforms [100]. CCR5 isoforms that are unstable or down regulate the expression of active CCR5, result in an increased resistance to HIV and Plasmodium vivax infection [101]. CCR5 expression levels also influences the expression of IL-2 as well as the development of mature memory T-cells. These cells have higher levels of IL-2 and display an increased susceptibility to HIV infectivity [102]. Chronic T. cruzi infection, resulting in chronic Chagas disease can lead to cardiomyopathy due to the immune response damaging the tissues of the heart. This cardiomyopathy is accompanied by an increase in the expression of CCR5 on the surface of circulating lymphocytes and the use of a partial CCR5 antagonist protected mice from cardiomyopathy [103]. However, T. brucei infections can inhibit the entry of HIV into monocyte derived macrophages (MDM) partly due to a decrease in the expression of CCR5 [86]. Treatment of schistosomiasis leads to a decrease in the level of CCR5 and CXCR4 on CD4+ cells in both HIV infected and uninfected patients [85].
CXCR3
Expression of the chemokine CXCR3 is increased following Schistosoma infection. CXCR3 is primarily expressed on NK cells and activated T lymphocytes. Two isoforms of CXCR3, generated by alternative splicing (Figure 11), allow many of the chemokines known to interact with CXCR3 (CXCL-9, CXCL10 and CXCL11) to play opposing roles when it comes to cell proliferation. The original isoform named CXCR3-A results in an increase in cell proliferation while CXCR3-B increases apoptosis levels. CXCR3 expression and function is up-regulated following Schistosoma infection [104] while mice lacking active CXCR3 showed an increased pathology associated with Leishmania major infection [105].
Cerebral malaria is a form of encephalopathy that results from the introduction of inflammatory molecules into the Central Nervous System via a damaged blood brain barrier (BBB). Damage to the BBB is caused by the adherence of parasitised red blood cells to the endothelial layer of the barrier [106]. Studies using CXCR3 knockout mice have demonstrated that this damage is mediated by CD8+ T cells which are recruited by CXCR3, and CXCR3- mice fail to develop cerebral malaria [107,108]. The CXCR3 ligands CXCL9 and CXCL10 are also required for the development of cerebral malaria [108], but they are expressed in a tissue dependent manner with microglia having increased levels of CXCL9 and astrocytes having increased levels of CXCL10, while the levels of both were elevated in neurons [107,108]. A similar situation was observed following the intracranial infection of mice with Toxoplasma gondii. Here two populations of microglia formed in response to the infection, one containing the endotoxin response protein immunoresponsive gene 1 (IRG1) and CCR9; the other lacks IRG but expresses CXCR3. The IRG1+/CCR9+ microglia is neurotoxic and resistant to apoptosis while the IRG-/CXCR3+ microglia are neuro-supportive and sensitive to apoptosis [109]. The chronic activation of microglia in response to infection appears to be responsible for the neurodegeneration observed during AIDS. HIV targets microglia, which appear to be the only cells in the CNS which are productively infected with the virus, infected microglia then release neurotoxin mediators that result in the death of neurons and encephalopathy [110,111]. Low infection rates are observed in astrocytes and these may play the role of viral reservoirs [111]. The different roles played by CXCR3 in different infection and different stages of the infections may be governed by the various expression of one of the two isoforms. This would change the sensitivity of the cells to apoptotic stimuli and would allow the microglia to survive and act as a viral reservoir in HIV as well as allowing varied responses to different cytokines during the progression of cerebral malaria.
Conclusion
Despite decades of research there is currently no vaccine to any human parasite. One of the major reasons why vaccines seem to fail during phase II trials is the diversity of the parasite. Not only does the hypervariability of surface proteins make it difficult to target, but parasites can also show the ability to rapidly develop drug resistance due to rapid evolution. Immune evasion of parasites through antigenic variation is a complex process with a vast amount of literature devoted to it; however, the vast majority of studies and reviews examine antigenic variation through the expression of different members of a multigene family and not through the alternate splicing of mRNA. The control of RNA transcription is poorly understood within Trypanosoma and Plasmodium. With the genomic sequences of these parasites and a better understanding of the role played by Trans SL RNA splicing and transcriptional control through antisense RNA, we are gaining a better picture of gene regulation in these parasites. It is important to understand the role played by the expression of different isoforms of antigens in immune evasion as these isoforms may offer new targets for therapy. This would involve the preferential selection of one isoform over another. This could be beneficial by leading to a either a decrease in immune evasion or a decrease in parasite viability. As many isoforms are expressed in a life stage specific manner, the expression of unique sets of genes at different life stages of the parasite points to a useful strategy involving the targeting of one of these particular stages, interfering in the progression from one life stage into another. Alternatively the life stage expression of different isoforms could be used as a diagnostic tool.
The geographical co-localisation of parasite infections by members of the genera Plasmodium, Schistosoma and Trypanosoma with a high prevalence of HIV infection, suggests that infection with parasites contributes to HIV infection and disease progression. HIV and parasite infection both lead to a suppression of the cellular immune system. These pathogens interact with the cellular immune system at the level of cell surface receptors and cytokines. The alternate splicing of the transcripts for these genes leads to drastic changes in their regulation and function. This may alter the way that parasites and HIV interact with the host’s immune system and with each other at a molecular level. Once again the preferential expression of some isoforms over others may have therapeutic effects in the simultaneous treatment of these infections.
Abbreviations
clag9: Cytoadherence linked asexual gene 9; DSCAM: Down syndrome cell adhesionmolecule; FREP3: Fibrinogen related proteins; GPI: Glycosylphosphatidylinositol; LAG-3: lymphocyte-activation gene-3; MBC: Memory B cells; MSP: Merozoite surface proteins; MEG: Micro-exon gene; MDM: Monocyte derived macrophages; PfEMP1: P falciparum erythrocyte membrane protein1; PD-1: Programmed death 1; SL RNA: Short leader sequence of RNA; SmPoMucs: S. mansoni polymorphic mucins; SmVAL: S. mansoni venom allergen like; snRNAs: Small nuclear RNAs; stevor: Subtelomeric variable open reading frame family; VSG: Variant surface glycoprotein.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RH wrote the manuscript and ZD reviewed it critically. Both authors read and approved the final version of the manuscript.
Contributor Information
Rodney Hull, Email: hullr@unisa.ac.za.
Zodwa Dlamini, Email: dlamizl@unisa.ac.za.
References
- Marchetti G, Pinotti M, Lunghi B, Casari C, Bernardi F. Functional genetics. Thromb Res. 2012;129:336–340. doi: 10.1016/j.thromres.2011.10.028. [DOI] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhopale GM. Pathogenesis of toxoplasmosis. Comp Immunol Microbiol Infect Dis. 2003;26:213–222. doi: 10.1016/S0147-9571(02)00058-9. [DOI] [PubMed] [Google Scholar]
- Restrepo BI, Alvarez JI, Castańo JA, Arias LF, Restrepo M, Trujillo J, Colegial CH, Teale JM. Brain granulomas in neurocysticercosis patients are associated with a Th1 and Th2 profile. Infect Immun. 2001;69:4554–4560. doi: 10.1128/IAI.69.7.4554-4560.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill DR, Hewlett EL, Pearson RD. Lectin binding by Giardia lamblia. Infect Immun. 1981;34:733–738. doi: 10.1128/iai.34.3.733-738.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KM, Alderete JF. Acquisition of alpha 1-antitrypsin by a pathogenic strain of trichomonas vaginalis. Infect Immun. 1983;40:640–646. doi: 10.1128/iai.40.2.640-646.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom BR. Games parasites play: how parasites evade immune surveillance. Nature. 1979;279:21–26. doi: 10.1038/279021a0. [DOI] [PubMed] [Google Scholar]
- Schmid-Hempel P. Immune defence, parasite evasion strategies and their relevance for ‘macroscopic phenomena’ such as virulence. Philos T R Soc B. 2009;364:85–98. doi: 10.1098/rstb.2008.0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissapatorn V, Sawangjaroen N. Parasitic infections in HIV infected individuals: diagnostic & therapeutic challenges. Indian J Med Res. 2011;134:878–897. doi: 10.4103/0971-5916.92633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreani G, Lodge R, Richard D, Tremblay MJ. Mechanisms of interaction between protozoan parasites and HIV. Curr Opin HIV AIDS. 2012;7:276–282. doi: 10.1097/COH.0b013e32835211e9. [DOI] [PubMed] [Google Scholar]
- Mbabazi PS, Andan O, Fitzgerald DW, Chitsulo L, Engels D, Downs JA. Examining the relationship between urogenital Schistosomiasis and HIV infection. PLoS Neglect Trop D. 2011;5:e1396. doi: 10.1371/journal.pntd.0001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PreuBer C, Jae N, Bindereif A. mRNA splicing in trypanosomes. Int J Med Microbiol. 2012;302:221–224. doi: 10.1016/j.ijmm.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Böhme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ, Barron AJ, Bringaud F, Brooks K, Carrington M, Cherevach I, Chillingworth TJ, Churcher C, Clark LN, Corton CH, Cronin A. et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Nilsson D, Gunasekera K, Mani J, Osteras M, Farinelli L, Baerlocher L, Roditi I, Ochsenreiter T. Spliced leader trapping reveals widespread alternative splicing patterns in the highly dynamic transcriptome of Trypanosoma brucei. PLoS Pathog. 2010;6:e1001037. doi: 10.1371/journal.ppat.1001037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penha LL, Mendonça-Previato L, Previato JO, Scharfstein J, Heise N, Lima APCA. Cloning and characterization of the phosphoglucomutase of Trypanosoma cruzi and functional complementation of a Saccharomyces cerevisiae PGM null mutant. Glycobiology. 2005;15:1359–1367. doi: 10.1093/glycob/cwj023. [DOI] [PubMed] [Google Scholar]
- Günzl A. The Pre-mRNA splicing machinery of trypanosomes: complex or simplified? Eukaryot Cell. 2010;9:1159–1170. doi: 10.1128/EC.00113-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücke S, Jürchott K, Hung L-H, Bindereif A. MRNA splicing in Trypanosoma brucei: branch-point mapping reveals differences from the canonical U2 snRNA-mediated recognition. Mol Biochem Parasit. 2005;142:248–251. doi: 10.1016/j.molbiopara.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Tschudi C, Ullu E. Destruction of U2, U4, or U6 small nuclear RNA blocks trans splicing in trypanosome cells. Cell. 1990;61:459–466. doi: 10.1016/0092-8674(90)90527-L. [DOI] [PubMed] [Google Scholar]
- Horn D, McCulloch R. Molecular mechanisms underlying the control of antigenic variation in African trypanosomes. Curr Opin Microbiol. 2011;13:700–705. doi: 10.1016/j.mib.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover NA, Kaye MS, Cavalcanti ARO. Spliced leader trans-splicing. Curr Biol. 2006;16:R8–R9. doi: 10.1016/j.cub.2005.12.019. [DOI] [PubMed] [Google Scholar]
- Hastings KEM. SL trans-splicing: easy come or easy go? Trends Genet. 2005;21:240–247. doi: 10.1016/j.tig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Robinson NP, Burman N, Melville SE, Barry JD. Predominance of duplicative VSG gene conversion in antigenic variation in African trypanosomes. Mol Cell Biol. 1999;19:5839–5846. doi: 10.1128/mcb.19.9.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzikowski R, Templeton TJ, Deitsch K. Variant antigen gene expression in malaria. Cell Microbiol. 2006;8:1371–1381. doi: 10.1111/j.1462-5822.2006.00760.x. [DOI] [PubMed] [Google Scholar]
- Aravind L, Iyer LM, Wellems TE, Miller LH. Plasmodium biology: genomic gleanings. Cell. 2003;115:771–785. doi: 10.1016/S0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- Otto TD, Wilinski D, Assefa S, Keane TM, Sarry LR, Böhme U, Lemieux J, Barrell B, Pain A, Berriman M, Newbold C, Llinás M. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol Microbiol. 2010;76:12–24. doi: 10.1111/j.1365-2958.2009.07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM. et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iriko H, Jin L, Kaneko O, Takeo S, Han E-T, Tachibana M, Otsuki H, Torii M, Tsuboi T. A small-scale systematic analysis of alternative splicing in Plasmodium falciparum. Parasitol Int. 2009;58:196–199. doi: 10.1016/j.parint.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Eshar S, Allemand E, Sebag A, Glaser F, Muchardt C, Mandel-Gutfreund Y, Karni R, Dzikowski R. A novel Plasmodium falciparum SR protein is an alternative splicing factor required for the parasite’s proliferation in human erythrocytes. Nucleicl Acids Res. 2012;40:9903–9916. doi: 10.1093/nar/gks735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Tolzmann CA, Melcher M, Haas BJ, Gardner MJ, Smith JD, Feagin JE. Branch point identification and sequence requirements for intron splicing in Plasmodium falciparum. Eukaryot Cell. 2011;10:1422–1428. doi: 10.1128/EC.05193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Barragan MJ, Lemieux J, Quinones M, Williamson KC, Molina-Cruz A, Cui K, Barillas-Mury C, Zhao K, Su XZ. Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC Genomics. 2012;12:587. doi: 10.1186/1471-2164-12-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp B, Nau U, Hundt E, Küpper HA. Demonstration of alternative splicing of a pre-mRNA expressed in the blood stage form of Plasmodium falciparum. J Biol Chem. 1991;266:7148–7154. [PubMed] [Google Scholar]
- Kehdinga Titanji Vincent P, Amambua-Ngwa A, Anong Damian N, Mbandi Stanley K, Tangie E, Tening I, Yengo R. Isolation and expression of UB05, a Plasmodium falciparum antigen recognised by antibodies from semi-immune adults in a high transmission endemic area of the Cameroonian rainforest. Clin Chem Lab Med. 2009;47:1147. doi: 10.1515/CCLM.2009.255. ? [DOI] [PubMed] [Google Scholar]
- Sorber K, Dimon MT, DeRisi JL. RNA-Seq analysis of splicing in Plasmodium falciparum uncovers new splice junctions, alternative splicing and splicing of antisense transcripts. Nucleic Acids Res. 2011;39:3820–3835. doi: 10.1093/nar/gkq1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma HW, Ray P, Dhanda V, Das PK, Paliwal S, Sahoo N, Patra KP, Das LK, Singh B, Kironde FAS. A novel 70-kDa triton X-114-soluble antigen of Plasmodium falciparumThat contains interspecies-conserved epitopes. Exp Parasitol. 1996;83:322–334. doi: 10.1006/expr.1996.0080. [DOI] [PubMed] [Google Scholar]
- Goschnick MW, Black CG, Kedzierski L, Holder AA, Coppel RL. Merozoite surface protein 4/5 provides protection against lethal challenge with a heterologous malaria parasite strain. Infect Immun. 2004;72:5840–5849. doi: 10.1128/IAI.72.10.5840-5849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black CG, Wang L, Hibbs AR, Werner E, Coppel RL. Identification of the Plasmodium chabaudi homologue of merozoite surface proteins 4 and 5 of Plasmodium falciparum. Infect Immun. 1999;67:2075–2081. doi: 10.1128/iai.67.5.2075-2081.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnwell JW, Asch AS, Nachman RL, Yamaya M, Aikawa M, Ingravallo P. A human 88-kD membrane glycoprotein (CD36) functions in vitro as a receptor for a cytoadherence ligand on Plasmodium falciparum-infected erythrocytes. J Clin Invest. 1989;84:765–772. doi: 10.1172/JCI114234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Preiser P, Rénia L, Balu B, Barnwell J, Blair P, Jarra W, Voza T, Landau I, Adams JH. Conservation and developmental control of alternative splicing in maebl among malaria parasites. J Mol Biol. 2004;343:589–599. doi: 10.1016/j.jmb.2004.08.047. [DOI] [PubMed] [Google Scholar]
- Trenholme KR, Gardiner DL, Holt DC, Thomas EA, Cowman AF, Kemp DJ. clag9: A cytoadherence gene in Plasmodium falciparum essential for binding of parasitized erythrocytes to CD36. Proc Natl Acad Sci USA. 2000;97:4029–4033. doi: 10.1073/pnas.040561197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenholme KR, Boutlis CS, Kuns R, Lagog M, Bockarie MJ, Gatton ML, Kemp DJ, Good MF, Anstey NM, Gardiner DL. Antibdy reactions to linear epitopes of Plasmodium falciparum cytoadherence-linked asexual gene 9 in asymptomatic children and adults from Papua New Guinea. Am J Trop Med Hyg. 2005;72:708–713. [PMC free article] [PubMed] [Google Scholar]
- Pain A, Renauld H, Berriman M, Murphy L, Yeats CA, Weir W, Kerhornou A, Aslett M, Bishop R, Bouchier C, Cochet M, Coulson RM, Cronin A, de Villiers EP, Fraser A, Fosker N, Gardner M, Goble A, Griffiths-Jones S, Harris DE, Katzer F, Larke N, Lord A, Maser P, McKellar S, Mooney P, Morton F, Nene V, O'Neil S, Price C. et al. Genome of the host-cell transforming parasite Theileria annulata compared with T. Parva. Science. 2005;309:131–133. doi: 10.1126/science.1110418. [DOI] [PubMed] [Google Scholar]
- Bakheit MA, Ahmed JS, Seitzer U. Existence of splicing variants in homologues of Theileria lestoquardi clone-5 Gene’s transcripts in Theileria annulata and Theileria parva. Ann N Y Acad Sci. 2008;1149:212–213. doi: 10.1196/annals.1428.023. [DOI] [PubMed] [Google Scholar]
- Fernandez-Becerra C, Yamamoto MM, Vëncio RZN, Lacerda M, Rosanas-Urgell A, del Portillo HA. Plasmodium vivax and the importance of the subtelomeric multigene vir superfamily. Trends Parasitol. 2009;25:44–51. doi: 10.1016/j.pt.2008.09.012. [DOI] [PubMed] [Google Scholar]
- Epp C, Li F, Howitt CA, Chookajorn T, Deitsch KW. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA. 2009;15:116–127. doi: 10.1261/rna.1080109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyes SA, Rowe JA, Kriek N, Newbold CI. Rifins: A second family of clonally variant proteins expressed on the surface of red cells infected with Plasmodium falciparum. Proc Natl Acad Sci U S A. 1999;96:9333–9338. doi: 10.1073/pnas.96.16.9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niang M, Yan Yam X, Preiser PR. The Plasmodium falciparum STEVOR multigene family mediates antigenic variation of the infected erythrocyte. PLoS Pathog. 2009;5:e1000307. doi: 10.1371/journal.ppat.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe JE, Surentheran T, Preiser PR. STEVORa multifunctional protein? Mol Biochem Parasit. 2004;134:11–15. doi: 10.1016/j.molbiopara.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Sutherland CJ. Stevor transcripts from Plasmodium falciparum gametocytes encode truncated polypeptides. Mol Biochem Parasit. 2001;113:331–335. doi: 10.1016/S0166-6851(01)00225-0. [DOI] [PubMed] [Google Scholar]
- Sanyal S, Egée S, Bouyer G, Perrot S, Safeukui I, Bischoff E, Buffet P, Deitsch KW, Mercereau-Puijalon O, David PH, Templeton TJ, Lavazec C. Plasmodium falciparum STEVOR proteins impact erythrocyte mechanical properties. Blood. 2012;119:e1–e8. doi: 10.1182/blood-2011-08-370734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibúrcio M, Niang M, Deplaine G, Perrot S, Bischoff E, Ndour PA, Silvestrini F, Khattab A, Milon G, David PH, Hardeman M, Vernick KD, Sauerwein RW, Preiser PR, Mercereau-Puijalon O, Buffet P, Alano P, Lavazec C. A switch in infected erythrocyte deformability at the maturation and blood circulation of Plasmodium falciparum transmission stages. Blood. 2012;119:e172–e180. doi: 10.1182/blood-2012-03-414557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Sharma A, Mourya DT, Kamaraju R, Patole MS, Shouche YS. Salivary gland transcriptome analysis during Plasmodium infection in malaria vector Anopheles stephensi. Int J Infect Dis. 2009;13:636–646. doi: 10.1016/j.ijid.2008.07.027. [DOI] [PubMed] [Google Scholar]
- Levy F, Bulet P, Ehret-Sabatier L. Proteomic analysis of the systemic immune response of Drosophila. Mol Cell Proteomics. 2004;3:156–166. doi: 10.1074/mcp.M300114-MCP200. [DOI] [PubMed] [Google Scholar]
- Grolli S, Merli E, Conti V, Scaltriti E, Ramoni R. Odorant binding protein has the biochemical properties of a scavenger for 4-hydroxy-2-nonenal in mammalian nasal mucosa. FEBS J. 2006;273:5131–5142. doi: 10.1111/j.1742-4658.2006.05510.x. [DOI] [PubMed] [Google Scholar]
- Sim S, Ramirez JL, Dimopoulos G. Dengue virus infection of the Aedes aegypti salivary gland and chemosensory apparatus induces genes that modulate infection and blood-feeding behavior. PLoS Pathog. 2012;8:e1002631. doi: 10.1371/journal.ppat.1002631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielli A, Kafatos FC, Loukeris TG. Cloning and characterization of four Anopheles gambiae serpin isoforms, differentially induced in the midgut by Plasmodium berghei invasion. J Biol Chem. 2003;278:4184–4193. doi: 10.1074/jbc.M208187200. [DOI] [PubMed] [Google Scholar]
- Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa NT, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Moita LF, Müller HM, Osta MA, Paskewitz SM, Reichhart JM, Rzhetsky A, Troxler L, Vernick KD, Vlachou D, Volz J. et al. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- Smith PH, Mwangi JM, Afrane YA, Yan G, Obbard DJ, Ranford-Cartwright LC, Little TJ. Alternative splicing of the Anopheles gambiae Dscam gene in diverse Plasmodium falciparum infections. Malaria J. 2011;10:156. doi: 10.1186/1475-2875-10-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson AS, Belorgey D, Lilley KS, Lomas DA, Gubb D, Dafforn TR. Characterization of the necrotic protein that regulates the toll-mediated immune response in Drosophila. J Biol Chem. 2003;278:6175–6180. doi: 10.1074/jbc.M209277200. [DOI] [PubMed] [Google Scholar]
- The UniProt C. Reorganizing the protein space at the universal protein resource (UniProt) Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwanchaichinda C, Kanost MR. The serpin gene family in Anopheles gambiae. Gene. 2009;442:47–54. doi: 10.1016/j.gene.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold SJ, Leyland PC, Seal RL, Goodman JL, Thurmond J, Strelets VB, Wilson RJ. the FlyBase c. FlyBase: improvements to the bibliography. Nucleic Acids Res. 2013;41:D751–D757. doi: 10.1093/nar/gks1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe K-M, Lee H, Anderson KV. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc Natl Acad Sci U S A. 2005;102:1122–1126. doi: 10.1073/pnas.0404952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister S, Agianian B, Turlure F, Relógio A, Morlais I, Kafatos FC, Christophides GK. Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathog. 2009;5:e1000542. doi: 10.1371/journal.ppat.1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Taylor HE, Dimopoulos G. AgDscam, a hypervariable immunoglobulin domain-containing receptor of the anopheles gambiae innate immune system. PLoS Biol. 2006;4:e229. doi: 10.1371/journal.pbio.0040229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, Cerqueira GC, Mashiyama ST, Al-Lazikani B, Andrade LF, Ashton PD, Aslett MA, Bartholomeu DC, Blandin G, Caffrey CR, Coghlan A, Coulson R, Day TA, Delcher A, DeMarco R, Djikeng A, Eyre T, Gamble JA, Ghedin E, Gu Y, Hertz-Fowler C, Hirai H, Hirai Y, Houston R, Ivens A, Johnston DA. et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarco R, Mathieson W, Manuel SJ, Dillon GP, Curwen RS, Ashton PD, Ivens AC, Berriman M, Verjovski-Almeida S, Wilson RA. Protein variation in blood-dwelling schistosome worms generated by differential splicing of micro-exon gene transcripts. Genome Res. 2010;20:1112–1121. doi: 10.1101/gr.100099.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RE, Davis AH, Carroll SM, Rajkovic A, Rottman FM. Tandemly repeated exons encode 81-base repeats in multiple, developmentally regulated Schistosoma mansoni transcripts. Mol Cell Biol. 1988;8:4745–4755. doi: 10.1128/mcb.8.11.4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida GT, Amaral MS, Beckedorff FCF, Kitajima JOP, DeMarco R, Verjovski-Almeida S. Exploring the Schistosoma mansoni adult male transcriptome using RNA-seq. Exp Parasitol. 2010;132:22–31. doi: 10.1016/j.exppara.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Parker-Manuel SJ, Ivens AC, Dillon GP, Wilson RA. Gene expression patterns in larval Schistosoma mansoni associated with infection of the mammalian host. PLoS Negl Trop D. 2011;5:e1274. doi: 10.1371/journal.pntd.0001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMarco R, Oliveira KC, Venancio TM, Verjovski-Almeida S. Gender biased differential alternative splicing patterns of the transcriptional cofactor CA150 gene in Schistosoma mansoni. Mol Biochem Parasit. 2006;150:123–131. doi: 10.1016/j.molbiopara.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Chalmers IW, McArdle AJ, Coulson RM, Wagner MA, Schmid R, Hirai H, Hoffmann KF. Developmentally regulated expression, alternative splicing and distinct sub-groupings in members of the Schistosoma mansoni venom allergen-like (SmVAL) gene family. BMC Genomics. 2008;9:89. doi: 10.1186/1471-2164-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers IW, Hoffmann KF. Platyhelminth venom allergen-like (VAL) proteins: revealing structural diversity, class-specific features and biological associations across the phylum. Parasitology. 2012;139:1231–1245. doi: 10.1017/S0031182012000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias LP, Rodrigues D, Cunna V, Rofatto HK, Faquim-Mauro EL, Leite LCC. Schistosoma mansoni venom allergen like proteins present differential allergic responses in a murine model of airway inflammation. PLoS Negl Trop D. 2012;6:e1510. doi: 10.1371/journal.pntd.0001510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger E, Grunau C, Pierce RJ, Hirai H, Gourbal B, Galinier R, Emans RM, Cesari IM, Cosseau C, Mitta G. Controlled chaos of polymorphic mucins in a metazoan parasite (Schistosoma mansoni) interacting with its invertebrate host (Biomphalaria glabrata) PLoS Negl Trop D. 2008;2:e330. doi: 10.1371/journal.pntd.0000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mone Y, Gourbal B, Duval D, Du Pasquier L, Kieffer-Jaquinod S, Mitta G. A large repertoire of parasite epitopes matched by a large repertoire of host immune receptors in an invertebrate host/parasite model. PLoS Negl Trop D. 2010;4:e813. doi: 10.1371/journal.pntd.0000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanington PC, Forys MA, Dragoo JW, Zhang S-M, Adema CM, Loker ES. Role for a somatically diversified lectin in resistance of an invertebrate to parasite infection. Proc Natl Acad Sci U S A. 2010;107:21087–21092. doi: 10.1073/pnas.1011242107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed JR. Protozoa: Pathogenesis and Defenses. 4. Galveston: University of Texas Medical Branch; 1996. [PubMed] [Google Scholar]
- Pfaff AW, Candolfi E. Immune responses to protozoan parasites and its relevance to diagnosis in immunocompromised patients. Eur J Protistol. 2003;39:428–434. doi: 10.1078/0932-4739-00016. [DOI] [Google Scholar]
- Lejon V, Ngoyi DM, Ilunga M, Beelaert G, Maes I, Büscher P, Fransen K. Low specificities of HIV diagnostic tests caused by Trypanosoma brucei gambiense sleeping sickness. J Clin Microbiol. 2010;48:2836–2839. doi: 10.1128/JCM.00456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasasira AF, Dorsey G, Kamya MR, Havlir D, Kiggundu M, Rosenthal PJ, Charlebois ED. False-positive results of enzyme immunoassays for human immunodeficiency virus in patients with uncomplicated malaria. J Clin Microbiol. 2006;44:3021–3024. doi: 10.1128/JCM.02207-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett DB, Baisely KJ, McNerney R, Hambleton I, Chirwa T, Ross DA, Changalucha J, Watson-Jones D, Helmby H, Dunne DW, Mabey D, Hayes RJ. Association of Schistosomiasis with false-positive HIV test results in an African adolescent population. J Clinl Microbiol. 2010;48:1570–1577. doi: 10.1128/JCM.02264-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlov M, Vaida F, Finney OC, Smith DM, Talley AK, Wang R, Kappe SH, Deng Q, Schooley RT, Duffy PE. P. falciparum enhances HIV replication in an experimental malaria challenge system. PLoS One. 2012;7:26. doi: 10.1371/journal.pone.0039000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayash-Rashkovsky M, Chenine AL, Steele LN, Lee SJ, Song R, Ong H, Rasmussen RA, Hofmann-Lehmann R, Else JG, Augostini P, McClure HM, Secor WE, Ruprecht RM. Coinfection with Schistosoma mansoni reactivates viremia in rhesus macaques with chronic simian-human immunodeficiency virus clade C infection. Infect Immun. 2007;75:1751–1756. doi: 10.1128/IAI.01703-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secor WE, Shah A, Mwinzi PMN, Ndenga BA, Watta CO, Karanja DMS. Increased density of human immunodeficiency virus type 1 coreceptors CCR5 and CXCR4 on the surfaces of CD4+ T cells and monocytes of patients with Schistosoma mansoni infection. Infect Immun. 2003;71:6668–6671. doi: 10.1128/IAI.71.11.6668-6671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreani G, Celentano AM, Solana ME, Cazorla SI, Malchiodi EL, Martinez Peralta LA, Dolcini GL. Inhibition of HIV-1 replication in human monocyte-derived macrophages by parasite Trypanosoma cruzi. PLoS ONE. 2009;4:e8246. doi: 10.1371/journal.pone.0008246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcaide P, Fresno M. The Trypanosoma cruzi membrane mucin AgC10 inhibits T cell activation and IL-2 transcription through L-selectin. Int Immunol. 2004;16:1365–1375. doi: 10.1093/intimm/dxh138. [DOI] [PubMed] [Google Scholar]
- Ip JY, Tong A, Pan Q, Topp JD, Blencowe BJ, Lynch KW. Global analysis of alternative splicing during T-cell activation. RNA. 2007;13:563–572. doi: 10.1261/rna.457207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara K, Okumura M, Shiono H, Inoue M, Kadota Y, Miyoshi S, Matsuda H. A study on CD45 isoform expression during T-cell development and selection events in the human thymus. Human Immunol. 2002;63:394–404. doi: 10.1016/S0198-8859(02)00379-8. [DOI] [PubMed] [Google Scholar]
- McNeill L, Cassady RL, Sarkardei S, Cooper JC, Morgan G, Alexander DR. CD45 isoforms in T cell signalling and development. Immunol Lett. 2004;92:125–134. doi: 10.1016/j.imlet.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Antoine-Moussiaux N, Cornet A, Cornet F, Glineur S, Dermine M, Desmecht D. A Non-cytosolic protein of Trypanosoma evansi induces CD45-dependent lymphocyte death. PLoS ONE. 2009;4:e5728. doi: 10.1371/journal.pone.0005728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahalingam M, Pozniak A, McManus TJ, Senaldi G, Vergani D, Peakman M. Abnormalities of CD45 isoform expression in HIV infection. Clin Immunol Immunop. 1996;81:210–214. doi: 10.1006/clin.1996.0178. [DOI] [PubMed] [Google Scholar]
- Martin DL, Weatherly DB, Laucella SA, Cabinian MA, Crim MT, Sullivan S, Heiges M, Craven SH, Rosenberg CS, Collins MH, Sette A, Postan M, Tarleton RL. CD8+ T-cell responses to trypanosoma cruzi Are highly focused on strain-variant trans sialidase epitopes. PLoS Pathog. 2006;2:e77. doi: 10.1371/journal.ppat.0020077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muiá RP, Yu H, Prescher JA, Hellman U, Chen X, Bertozzi CR, Campetella O. Identification of glycoproteins targeted by Trypanosoma cruzi trans-sialidase, a virulence factor that disturbs lymphocyte glycosylation. Glycobiology. 2010;20:833–842. doi: 10.1093/glycob/cwq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucci J, Risso MG, Leguizamón MS, Frasch ACC, Campetella O. The trans-sialidase from Trypanosoma cruzi triggers apoptosis by target cell sialylation. Cell Microbiol. 2006;8:1086–1095. doi: 10.1111/j.1462-5822.2006.00689.x. [DOI] [PubMed] [Google Scholar]
- Illingworth J, Butler NS, Roetynck S, Mwacharo J, Pierce SK, Bejon P, Crompton PD, Marsh K, Ndungu FM. Chronic exposure to Plasmodium falciparum is associated with phenotypic evidence of B and T cell exhaustion. J Immunol. 2013;190:1038–1047. doi: 10.4049/jimmunol.1202438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir S, Fauci AS. B cells in HIV infection and disease. Nat Rev Immunol. 2009;9:235–245. doi: 10.1038/nri2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall SR, Heffernan BM, Thompson NT, Rowan WC. CD4+CD45RA+ and CD4+CD45RO+ T cells differ in their TCR-associated signaling responses. Eur J Immunol. 1999;29:2098–2106. doi: 10.1002/(SICI)1521-4141(199907)29:07<2098::AID-IMMU2098>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Mummidi S, Ahuja SS, McDaniel BL, Ahuja SK. The human CC chemokine receptor 5 (CCR5) gene. Multiple transcripts with 5′-end heterogeneity, dual promoter usage, and evidence for polymorphisms within the regulatory regions and noncoding exons. J Biol Chem. 1997;272:30662–30671. doi: 10.1074/jbc.272.49.30662. [DOI] [PubMed] [Google Scholar]
- Bamshad MJ, Mummidi S, Gonzalez E, Ahuja SS, Dunn DM, Watkins WS, Wooding S, Stone AC, Jorde LB, Weiss RB, Ahuja SK. A strong signature of balancing selection in the 5′ cis-regulatory region of CCR5. Proc Natl Acad Sci U S A. 2002;99:10539–10544. doi: 10.1073/pnas.162046399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pease JE, Murphy PM. Microbial corruption of the chemokine system: an expanding paradigm. Semin Immunol. 1998;10:169–178. doi: 10.1006/smim.1998.0129. [DOI] [PubMed] [Google Scholar]
- Camargo JF, Quinones MP, Mummidi S, Srinivas S, Gaitan AA, Begum K, Jimenez F, VanCompernolle S, Unutmaz D, Ahuja SS, Ahuja SK. CCR5 Expression levels influence NFAT translocation, IL-2 production, and subsequent signaling events during T lymphocyte activation. J Immunol. 2009;182:171–182. doi: 10.4049/jimmunol.182.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros GA, Silvério JC, Marino APMP, Roffê E, Vieira V, Kroll-Palhares K, Carvalho CE, Silva AA, Teixeira MM, Lannes-Vieira J. Treatment of chronically Trypanosoma cruzi-infected mice with a CCR1/CCR5 antagonist (Met-RANTES) results in amelioration of cardiac tissue damage. Microbes Infect. 2009;11:264–273. doi: 10.1016/j.micinf.2008.11.012. [DOI] [PubMed] [Google Scholar]
- Lasagni L, Francalanci M, Annunziato F, Lazzeri E, Giannini S, Cosmi L, Sagrinati C, Mazzinghi B, Orlando C, Maggi E, Marra F, Romagnani S, Serio M, Romagnani P. An alternatively spliced variant of CXCR3 mediates the inhibition of endothelial cell growth induced by IP-10, Mig, and I-TAC, and acts as functional receptor for platelet factor 4. J Exp Med. 2003;197:1537–1549. doi: 10.1084/jem.20021897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas LE, Barbi J, Lu B, Fujiwara Y, Gerard C, Sanders VM, Satoskar AR. CXCR3−/− Mice mount an efficient Th1 response but fail to control Leishmania major infection. Eur J Immunol. 2005;35:515–523. doi: 10.1002/eji.200425422. [DOI] [PubMed] [Google Scholar]
- Shikani HJ, Freeman BD, Lisanti MP, Weiss LM, Tanowitz HB, Desruisseaux MS. Cerebral malaria: we have come a long way. Am J Pathol. 2012;181:1484–1492. doi: 10.1016/j.ajpath.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miu J, Mitchell AJ, Müller M, Carter SL, Manders PM, McQuillan JA, Saunders BM, Ball HJ, Lu B, Campbell IL, Hunt NH. Chemokine gene expression during fatal murine cerebral malaria and protection due to CXCR3 deficiency. J Immunol. 2008;180:1217–1230. doi: 10.4049/jimmunol.180.2.1217. [DOI] [PubMed] [Google Scholar]
- Campanella GSV, Tager AM, El Khoury JK, Thomas SY, Abrazinski TA, Manice LA, Colvin RA, Luster AD. Chemokine receptor CXCR3 and its ligands CXCL9 and CXCL10 are required for the development of murine cerebral malaria. Proc Natl Acad Sci U S A. 2008;105:4814–4819. doi: 10.1073/pnas.0801544105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Gang Z, Yuling H, Luokun X, Jie X, Hao L, Li W, Chunsong H, Junyan L, Mingshen J, Youxin J, Feili G, Boquan J, Jinquan T. Different neurotropic pathogens elicit neurotoxic CCR9- or neurosupportive CXCR3-expressing microglia. J Immunol. 2006;177:3644–3656. doi: 10.4049/jimmunol.177.6.3644. [DOI] [PubMed] [Google Scholar]
- Streit WJ. Microglia and Alzheimer’s disease pathogenesis. J Neurosci Res. 2004;77:1–8. doi: 10.1002/jnr.20093. [DOI] [PubMed] [Google Scholar]
- Rock RB, Gekker G, Hu S, Sheng WS, Cheeran M, Lokensgard JR, Peterson PK. Role of microglia in central nervous system infections. Clin Microbiol Rev. 2004;17:942–964. doi: 10.1128/CMR.17.4.942-964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]