Abstract
Background
At present, malaria cases in Suriname occur predominantly in migrants and people living and/or working in areas with gold mining operations. A molecular survey was performed in Plasmodium falciparum isolates originating from persons from gold mining areas to assess the extent and role of mining areas as reservoirs of malaria resistance in Suriname.
Methods
The status of 14 putative resistance-associated single nucleotide polymorphisms in the pfdhfr, pfcrt, pfmdr1, and pfATP6 genes was assessed for 28 samples from gold miners diagnosed with P. falciparum malaria using polymerase chain reaction amplification and restriction fragment length polymorphism analysis, and the results were compared with earlier data from nonmining villagers.
Results
Isolates from miners showed a high degree of homogeneity, with a fixed pfdhfr Ile51/Asn108, pfmdr1 Phe184/Asp1042/Tyr1246, and pfcrt Thr76 mutant genotype, while an exclusively wild-type genotype was observed for pfmdr1 Asn86 and pfdhfr Ala16, Cys59, and Ile164, and for the pfATP6 positions Leu263/Ala623/Ser769. Small variations were observed for pfmdr1 S1034C. No statistically significant difference could be detected in allele frequencies between mining and nonmining villagers.
Conclusion
Despite the increased risk of malaria infection in individuals working/living in gold mining areas, we did not detect an increase in mutation frequency at the 14 analyzed single nucleotide polymorphisms. Therefore, mining areas in Suriname cannot yet be considered as reservoirs for malaria resistance.
Keywords: Plasmodium falciparum, gold mining, mutation frequency, Suriname
Background
Chloroquine has been used in Suriname as a treatment for malaria since 1958, and was first replaced with pyrimethamine/sulfadoxine and subsequently with quinine after reports of in vivo chloroquine-resistant Plasmodium falciparum in Suriname in 1973.
As part of a strengthened malaria program, artemisinin combination therapy was adopted in 2004, with artemether-lumefantrine (Coartem®; Novartis Pharma AG, Basel, Switzerland) as the first-line regimen for treatment of P. falciparum malaria, and mefloquine as prophylaxis for travelers to malaria endemic regions and as treatment for pregnant women. The different measures resulted in a large reduction in the number of cases, from 14,403 in 2003 to 1,477 in 2008.1 However, Suriname’s natural resources have attracted migrants from neighboring countries, mainly Brazil, to work as small-scale gold miners. An estimated 15,000 people are involved in mining activities, either as small-scale gold miners or as mining service providers, living in settlements mostly located in the eastern and central parts of the country. Malaria control and elimination efforts were challenged by a substantial influx of gold miners into these remote areas of the interior, where Anopheles darlingi is the principal vector. The illegal nature of the activities, alongside the increased migration of persons in and out of gold mining sites, a deficient or absent health care infrastructure, poor housing, and lack of compliance with national treatment policies have been known to increase the incidence of malaria and might promote emergence of drug resistance. The historical rapid spread of antimalarial drug resistance and slow development of new drugs to replace artemisinin dictate close monitoring of antimalarial drug resistance.
Many molecular markers have been associated with drug resistance. Chloroquine resistance has been linked to a specific point mutation in the P. falciparum chloroquine resistance transporter gene (pfcrt) gene,2 whereas mutations in the P. falciparum dihydrofolate reductase (pfdhfr) gene play a key role in conferring resistance to pyrimethamine.3 Likewise, molecular polymorphisms in the P. falciparum multidrug resistance 1 (pfmdr1) gene have been associated with resistance to mefloquine4 and artemisinin,5 while conflicting reports exist regarding the influence of certain mutations in the P. falciparum Ca2+-ATPase (pfATP6) coding gene on resistance to artemisinin.6,7
The availability of molecular data for P. falciparum isolates from gold mining populations can be an important tool for monitoring the emergence and dynamics of drug resistance. We determined the status of 14 resistance-associated mutations of four genes in P. falciparum isolates from patients living/working in active gold mining sites in Suriname. The collected molecular data in conjunction with our earlier results for nonmining villagers were used to assess the extent and role of mining areas as breeding pools for emerging resistant isolates in Suriname.
Materials and methods
Study site and study population
Suriname is situated north of Brazil, between Guyana and French Guiana. The country is inhabited by less than half a million people, with a small proportion (9.8%) of the population, mostly Amerindians and Maroons, living in the tropical rainforest in the interior.8 Since the 1990s, a migratory wave of Brazilians, working in the small-scale gold mining industry, accounts for an additional 15,000 persons in these regions.
Artemether-lumefantrine has been used as first-line treatment for uncomplicated falciparum malaria in Suriname since 2004, while mefloquine is deployed for malaria during pregnancy and is recommended as prophylaxis for travelers before departure to malarious areas.
For this study, we used blood spotted 3 MM filters (Whatman, Clifton, NJ, USA) stored in zip lock bags at room temperature with desiccants from the National Malaria Gene Bank; these filters had been taken from patients with uncomplicated microscopically confirmed P. falciparum malaria working and/or living in different mining sites. From a pool of 47 blood spotted filters collected in the period January–August 2009, 28 filters were selected to represent the maximum number of different mining sites. Informed consent for this molecular malaria research was obtained on enrollment for the National Malaria Gene Bank, and the ethics committee of this institution approved the study protocols. Tableau version 8.0 software (Tableau, Seattle, WA, USA) was used for visualization of the data.
DNA extraction
Parasite DNA was extracted from the blood spotted filters using a slightly modified Chelex extraction method.9
Single nucleotide polymorphism analysis
SNPs in the pfdhfr gene
Mutation-specific polymerase chain reaction (PCR)-based identification was used to characterize five single nucleotide polymorphisms (SNPs, ie, A16V, N51I, C59R, S108N/T, and I164L) in the pfdhfr gene conferring pyrimethamine resistance.10
SNPs in the pfmdr1 gene
Identification of key mutations (N86Y, Y184F, S1034C, N1042D, D1246Y) in the pfmdr1 gene was performed using methods described earlier.11
SNP in the pfcrt gene
PCR in combination with restriction fragment length polymorphism was used to identify the mutation at codon 76 in the pfcrt gene;12 analysis with ApoI of the second round PCR allowed determination of the presence/absence of the mutation. Chloroquine-sensitive strains from Honduras provided by Dr J Alger (Universidad Nacional Autonoma de Honduras) were used as a positive control in the restriction analysis.
SNPs in the pfATP6 gene
Polymorphisms in the pfATP6 gene (A623E, L263E, S769N), supposedly associated with resistance to artemisinin and its derivatives, were assessed as described earlier.13 To monitor the lack of cross-contamination during DNA extraction and PCR, no template controls were included throughout the runs. Restriction efficacy in case of uncut PCR products was monitored by the inclusion of positive control samples. PCR performance was evaluated by inclusion of multiple positive controls, while random repeats were performed to monitor reproducibility.
Statistical analysis
Statistical analyses were performed using the χ2 (chi-squared) test.
Results
In this molecular survey, the frequency of a wide range of point mutations in four genes in P. falciparum, putatively associated with resistance to various antimalarial drugs, was determined in isolates from gold mining areas. The collected molecular data were compared with earlier results.
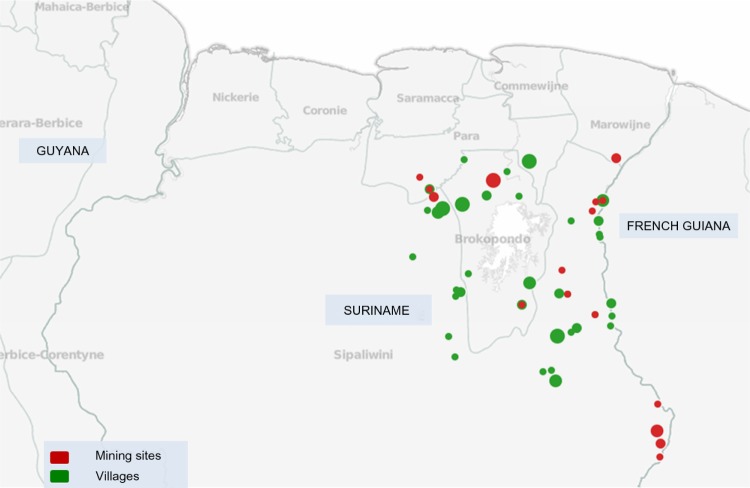
We tested 28 samples collected from P. falciparum-infected patients living/working in 17 different mining sites (Figure 1). The study population consisted of 61% males and 39% females and the median age was 36.5 (7–58) years.
Figure 1.
Map of sampling sites in Suriname. The locations of Plasmodium falciparum isolates collected in mining sites are represented as red circles, whereas locations in villages are shown as green circles. The size of each circle corresponds to the number of samples collected from that location. The geographical location (latitude/longitude) of four samples in the study could not be specified based on the patient travel history.
SNPs in the pfdhfr gene associated with pyrimethamine resistance
Assays to determine the status of the SNP sites in the pfdhfr gene were performed for the five amino acid positions (16, 51, 59, 108, and 164) associated with pyrimethamine resistance. The success rate for amplification of the PCRs for the positions 108 and 59 was 100% and 93%, respectively, while 96% of the samples could be amplified for the positions 16, 51, and 164. All successfully amplified samples displayed an identical pfdhfr profile; wild-type at position 16 (Ala16), 59 (Cys59), and 164 (Ile164) and mutated pfdhfr at codons 108 (Asn108) and 51 (Ile51). The Ser to Thr mutation at position 108 was not found in any of the tested isolates (Table 1).
Table 1.
Comparison of mutation frequencies of single nucleotide polymorphisms in the pfdhfr, pfmdr1, pfcrt, and pfATP6 genes associated with drug-resistance between samples collected from Plasmodium falciparum-infected patients working/living in mining areas and villagers (nonminers)
| Gene and positions | % mutation in miners | % mutation in villagers |
|---|---|---|
| pfdhfr | ||
| A16V | 0% (n=27) | 0% (n=30)a |
| N51I | 100% (n=27) | 100% (n=30)a |
| C59R | 0% (n=26) | 0% (n=30)a |
| S108N | 100% (n=28) | 100% (n=30)a |
| I164L | 0% (n=27) | 0% (n=30)a |
| pfmdr1 | ||
| N86Y | 0% (n=27) | 0% (n=40)b |
| Y184F | 100% (n=27) | 100% (n=40)b |
| S1034C | 96% (n=26) | 95% (n=40)b |
| N1042D | 100% (n=28) | 100% (n=40)b |
| D1246Y | 100% (n=28) | 100% (n=40)b |
| pfcrt | ||
| K76T | 100% (n=28) | 100% (n=40)b |
| pfATP6 | ||
| L263E | 0% (n=28) | 0% (n=24)c |
| A623E | 0% (n=27) | 0% (n=24)c |
| S769N | 0% (n=27) | 0% (n=24)c |
Notes:
Isolates from 2002 (unpublished data);
isolates from 2005–2006 (data from Adhin et al19);
isolates from 2002–2006 (data from Adhin et al13). The letters are abbreviations for different amino acids in the pfdhfr, pfmdr1, and pfcrt gene products. n represents the number of successfully amplified samples for the respective position in each study.
SNPs in the pfmdr1 gene associated with mefloquine resistance
In the pfmdr1 gene, encoding the glycoprotein Pgh-1, single nucleotide mutations at amino acid positions 86, 184, 1,034, 1,042, and 1,246 may modulate sensitivity to both mefloquine and artemisinin. The success rate for amplification for position 1,042 and 1,246 was 100%, while the PCRs for positions 1,034, 184, and 86 displayed failure rates of 7%, 4%, and 4%, respectively.
For the pfmdr1 gene position 1,034, 4% of the samples displayed the wild-type genotype (Ser1034) and 96% harbored the mutation (Cys1034). All investigated samples possessed the mutated single nucleotide polymorphisms at codons 184, 1,042, and 1,246 (Phe184, Asp1,042, and Tyr1,246), while exclusively wild-type was observed for position 86 (Asn86, Table 1).
SNPs in the pfcrt gene associated with chloroquine resistance
The success rate for PCR amplification of the pfcrt gene was 100%. The samples displayed a 100% prevalence of the pfcrt K76T mutation, which has been associated with chloroquine resistance.
SNPs in the pfATP6 gene putatively associated with artemisinin resistance
Mutations within the pfATP6 gene, encoding the Pf sarco-endoplasmic reticulum Ca2+-ATPase (SERCA), have been reported to decrease the susceptibility of P. falciparum to artemisinins in vitro.14
The amplification success rate for position 263 was 100% and 96% for the polymorphisms at position 623 and 769, respectively. All samples displayed the wild-type nucleotide at all three polymorphic sites (Leu263, Ala623, and Ser769, see Table 1).
No statistically significant difference could be observed for any of the 14 investigated SNPs between the isolates from the mining areas and the earlier data from nonmining villagers (Table 1).
Discussion
Suriname has been experiencing an influx of gold miners, and the malaria control program has been relentlessly challenged by these gold miners and activities associated with gold mining. The high degree of host mobility is a major factor affecting malaria prevalence in mining sites as well as in areas of low endemicity.
In contrast with results from Brazil, where samples from gold mining sites displayed a higher mutation frequency in the pfdhfr gene than isolates from the city of Porto Velho, Brazil,15 the isolates from mining areas in Suriname exhibited a genotype for the pfdhfr gene identical to that of isolates obtained from nonmining villagers. Pyrimethamine/sulfadoxine treatment failure has been clinically observed in Suriname since 1983, accounting for the mutant alleles in the loci pfdhfr S108N and N51I, in all isolates. The single observed mutation at codon 108 (S108N) is reported to be enough to confer pyrimethamine resistance, and addition of the codon 51 mutation is associated with even higher levels of resistance.3 The common presence of pfdhfr mutations at codons 51 and 108 and lack of the mutation at codon 164 was also observed in samples from Bolivar State, Venezuela, collected mostly from gold miners.16
The relatively rare Thr108 mutation, possibly associated with cycloguanil-resistant isolates, which is sometimes found in conjunction with the presence of a second mutation at position 16,17 was not detected in any of the samples from Suriname. Although this Thr108 mutation has been observed in South America as a mixed parasite population, together with Asn108 in Porto Velho samples15 and scarcely in southwest Colombia,18 this result in Suriname could be expected due to the fixed presence of the Ala16 allele in Suriname. In both South American studies reporting Thr108, no mutation detection at position 16 had been performed.
For the pfmdr1 gene, the majority (96%) of the successfully amplified isolates from mining areas display the Asn86/Phe184/Cys1,034/Asp1,042/Tyr1,246 (NFCDY) haplotype, matching the allelic variation revealed in parasites, either isolated from nonmining villagers in Suriname19 or from Guyana20 and from mefloquine-sensitive isolates in Brazil.21 The minor (4%) haplotype showing Asn86/Phe184/Ser1,034/Asp1,042/Tyr1,246 (NFSDY), differing in only one locus, has also been observed in isolates from villagers (5%) and has been reported earlier in a higher incidence (37%) in Bolivar state, Venezuela,22 and in isolates from Colombia.20
According to the existing classification of pfmdr1 polymorphisms,23 both haplotypes could be characterized as category IV (Phe184 in combination with Ser/Cys1,034 and Asn/Asp1,042), which are reported to be less resistant to mefloquine, artesunate, and artemisinin than isolates in categories I (wild-type) and III (Phe184).23 The clinical success of mefloquine in Suriname is consistent with this characterization.
The prevalence of the pfcrt K76T mutation in mining samples does not differ from that in isolates originating from villages, as anticipated. All samples from Suriname show the K76T mutation, which has been strongly linked to chloroquine resistance,2 corresponding with the chloroquine resistance in our country reported since 1973 and analogous to the genotype observed in neighboring countries, ie, Brazil24 and Guyana.11
Studies in China25 and Africa26 reported a recovery of chloroquine efficacy in P. falciparum parasites and/or reduction in prevalence of the pfcrt K76T mutation 6–12 years after drug withdrawal. In this study, this tendency was not observed in parasites from Suriname, which might be explained by continuing drug pressure, since chloroquine is still the treatment of choice for Plasmodium vivax and Plasmodium malariae malaria.
For the pfATP6 gene, all mining samples showed the wild-type nucleotide at all three analyzed polymorphic sites (L263E, A623E, S769N), matching our earlier observations in the rest of the country.13 We could not detect a higher mutation frequency at these loci, not even for the S769N mutation, which was reported in isolates from neighboring French Guiana, despite frequent migration of miners between these regions.
In the currently surveyed mining environment with risk factors such as a high migration rate, deficient or absent health infrastructure, poor adherence to antimalarial medication, use of illegally acquired antimalarial drugs, all favorable for increased malaria infection, we did not detect an increase in mutation frequency at the 14 analyzed SNPs in mining isolates. This result resolves the use of samples from different time periods which would otherwise have been a confounding factor. In addition to the SNP analysis, no statistically significant difference could be detected in the pfmdr1 gene copy number in isolates collected from gold miners and nonmining villagers.27
Although the sample size is rather small, our results strongly indicate that, at least at present, mining areas in Suriname do not serve as breeding grounds for malaria resistance. This is in contrast with expectations, especially considering the outcry that current conditions in mining areas in neighboring French Guiana may spark artemisinin resistance.28 On the other hand, this might have been anticipated, given that mutational processes are not only governed by the threat of infection but also by a variety of other factors. These mediators, such as geographic conditions, intrahost dynamics, and level of human immunity are presumably equal for miners and villagers. In addition, the elapsed time is merely 20 years, which does not favor the occurrence of mutations, although changes in pfcrt76 have been detected in 6–12 years.25,26 Also, the direct association between molecular markers and artemisinin resistance is still disputed, and a recent study revealed that the artemisinin component of illegally acquired antimalarials in the mining areas in Suriname was effective.29 Furthermore, evolutionary history suggests that the de novo origin of resistance can be disregarded as a clinical concern, probably because resistance involves multiple mutations30 requiring a series of mutational events. Nevertheless, other indicators in Suriname, such as multiple pfmdr1 copy numbers and delayed day 3 parasite clearance after artemisinin combination therapy,31 warrant unabated monitoring of looming artemisinin resistance.
Acknowledgments
This work was supported by the Pan American Health Organization’s Amazon Network for the Surveillance of Antimalarial Drug Resistance in cooperation with the US Agency for International Development’s Amazon Malaria Initiative SU/LOA/0900001.001. We express our gratitude to J Kartowidjojo, M Grünberg, and J Faerber for laboratory assistance, to J Alger for providing control samples, and to R Ardjosentono and H Cairo for support with preparation of the map.
Footnotes
Author contributions
MRA conceived the study, participated in its design and coordination, and drafted the manuscript. MLB participated in the laboratory analysis and in preparation of the manuscript. SV participated in the design of the study and in revision of the manuscript. All authors read and approved the final draft.
Disclosure
The authors report that they have no competing interests in this work. The views expressed herein are the views of the authors and do not necessarily reflect the opinions of the Pan American Health Organization, the US Agency for International Development, or the Amazon Network for the Surveillance of Antimalarial Drug Resistance.
References
- 1.Pan American Health Organization Report on the Situation of Malaria in the Americas. 2008. [Accessed February 25, 2014]. Available from: http://www.paho.org/hq/index.php?option=com_content&view=article&id=5175:report-on-the-situation-of-malaria-in-the-americas-2008&catid=1841:hsd0107j--malaria-publications&Itemid=2258&lang=en.
- 2.Fidock DA, Nomura T, Talley AK, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6(4):861. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci U S A. 1988;85(23):9114–9118. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403(6772):906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 5.Duraisingh MT, Roper C, Walliker D, Warhurst DC. Increased sensitivity to the antimalarials mefloquine and artemisinin is conferred by mutations in the pfmdr1 gene of Plasmodium falciparum. Mol Microbiol. 2000;36(4):955–961. doi: 10.1046/j.1365-2958.2000.01914.x. [DOI] [PubMed] [Google Scholar]
- 6.Imwong M, Dondorp AM, Nosten F, et al. Exploring the contribution of candidate genes to artemisinin resistance in Plasmodium falciparum. Antimicrob Agents Chemother. 2010;54(7):2886–2892. doi: 10.1128/AAC.00032-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pillai DR, Lau R, Khairnar K, et al. Artemether resistance in vitro is linked to mutations in PfATP6 that also interact with mutations in PfMDR1 in travellers returning with Plasmodium falciparum infections. Malar J. 2012;11(1):131. doi: 10.1186/1475-2875-11-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Algemeen Bureau voor de Statistiek . Zevende algemene volks- en woningtelling in Suriname: landelijke resultaten volume I, demografische en sociale karakteristieken. Suriname in Cijfers. (No 213-2005/02) Statistiek; 2005. 7th general population census in Suriname: country results I, demographic and social characteristics. Dutch. [Google Scholar]
- 9.Fischer A, Lejczak C, Lambert C, et al. Simple DNA extraction method for dried blood spots and comparison of two PCR assays for diagnosis of vertical human immunodeficiency virus type 1 transmission in Rwanda. J Clin Microbiol. 2004;42(1):16–20. doi: 10.1128/JCM.42.1.16-20.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plowe CV, Cortese JF, Djimde A, et al. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J Infect Dis. 1997;176(6):1590–1596. doi: 10.1086/514159. [DOI] [PubMed] [Google Scholar]
- 11.Plummer WB, Pereira LMP, Carrington CV. Pfcrt and pfmdr1 alleles associated with chloroquine resistance in Plasmodium falciparum from Guyana, South America. Mem Inst Oswaldo Cruz. 2004;99(4):389–392. doi: 10.1590/s0074-02762004000400008. [DOI] [PubMed] [Google Scholar]
- 12.Djimdé A, Doumbo OK, Cortese JF, et al. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001;344(4):257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 13.Adhin MR, Labadie-Bracho M, Vreden SG. Status of potential PfATP6 molecular markers for artemisinin resistance in Suriname. Malar J. 2012;11(1):322. doi: 10.1186/1475-2875-11-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jambou R, Legrand E, Niang M, et al. Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet. 2005;366(9501):1960–1963. doi: 10.1016/S0140-6736(05)67787-2. [DOI] [PubMed] [Google Scholar]
- 15.Vasconcelos KF, Plowe CV, Fontes CJ, et al. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase of isolates from the Amazon Region of Brazil. Mem Inst Oswaldo Cruz. 2000;95(5):721–728. doi: 10.1590/s0074-02762000000500020. [DOI] [PubMed] [Google Scholar]
- 16.Contreras CE, Cortese JF, Caraballo A, Plowe CV. Genetics of drug-resistant Plasmodium falciparum malaria in the Venezuelan state of Bolivar. Am J Trop Med Hyg. 2002;67(4):400–405. doi: 10.4269/ajtmh.2002.67.400. [DOI] [PubMed] [Google Scholar]
- 17.Peterson DS, Milhous WK, Wellems TE. Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc Natl Acad Sci U S A. 1990;87(8):3018–3022. doi: 10.1073/pnas.87.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giraldo LE, Acosta MC, Labrada LA, et al. Frequency of the Asn-108 and Thr-108 point mutations in the dihydrofolate reductase gene in Plasmodium falciparum from southwest Colombia. Am J Trop Med Hyg. 1998;59(1):124–128. doi: 10.4269/ajtmh.1998.59.124. [DOI] [PubMed] [Google Scholar]
- 19.Adhin MR, Labadie-Bracho M, Bretas G. Molecular surveillance as monitoring tool for drug-resistant Plasmodium falciparum in Suriname. Am J Trop Med Hyg. 2013;89(2):311–316. doi: 10.4269/ajtmh.12-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mehlotra RK, Mattera G, Bockarie MJ, et al. Discordant patterns of genetic variation at two chloroquine resistance loci in worldwide populations of the malaria parasite Plasmodium falciparum. Antimicrob Agents Chemother. 2008;52(6):2212–2222. doi: 10.1128/AAC.00089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zalis MG, Pang L, Silveira MS, Milhous WK, Wirth DF. Characterization of Plasmodium falciparum isolated from the Amazon region of Brazil: evidence for quinine resistance. Am J Trop Med Hyg. 1998;58(5):630–637. doi: 10.4269/ajtmh.1998.58.630. [DOI] [PubMed] [Google Scholar]
- 22.Griffing S, Syphard L, Sridaran S, et al. pfmdr1 Amplification and fixation of pfcrt chloroquine resistance alleles in Plasmodium falciparum in Venezuela. Antimicrob Agents Chemother. 2010;54(4):1572–1579. doi: 10.1128/AAC.01243-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson AL, Purfield A, Mcdaniel P, et al. pfmdr1 Genotyping and in vivo Mefloquine resistance on the Thai-Myanmar border. Am J Trop Med Hyg. 2005;72(5):586–592. [PubMed] [Google Scholar]
- 24.Vieira PP, das Graças Alecrim M, da Silva LH, González-Jiménez I, Zalis MG. Analysis of the PfCRT K76T Mutation in Plasmodium falciparum Isolates from the Amazon Region of Brazil. J Infect Dis. 2001;183(12):1832–1833. doi: 10.1086/320739. [DOI] [PubMed] [Google Scholar]
- 25.Liu DQ, Liu RJ, Ren DX, et al. Alteration in resistance of Plasmodium falciparum to chloroquine after cessation of chloroquine medication for twelve years. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 1992;10(4):241–244. Chinese. [PubMed] [Google Scholar]
- 26.Mwai L, Ochong E, Abdirahman A, et al. Chloroquine resistance before and after its withdrawal in Kenya. Malar J. 2009;8:106. doi: 10.1186/1475-2875-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labadie-Bracho M, Adhin MR. Increased pfmdr1 copy number in Plasmodium falciparum isolates from Suriname. Trop Med Int Health. 2013;18(7):796–799. doi: 10.1111/tmi.12118. [DOI] [PubMed] [Google Scholar]
- 28.Nacher M, Guérin PJ, Demar-Pierre M, Djossou F, Nosten F, Carme B. Made in Europe: will artemisinin resistance emerge in French Guiana? Malar J. 2013;12(1):152. doi: 10.1186/1475-2875-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans L, Coignez V, Barojas A, et al. Quality of anti-malarials collected in the private and informal sectors in Guyana and Suriname. Malar J. 2012;11(1):203. doi: 10.1186/1475-2875-11-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hastings IM, Watkins WM. Tolerance is the key to understanding antimalarial drug resistance. Trends Parasitol. 2006;22(2):71–77. doi: 10.1016/j.pt.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 31.Vreden SGS, Jitan JK, Bansie RD, Adhin MR. Evidence of an increased incidence of day 3 parasitaemia in Suriname: an indicator of the emerging resistance of Plasmodium falciparum to artemether. Mem Inst Oswaldo Cruz. 2013;108(8):968–973. doi: 10.1590/0074-0276130167. [DOI] [PMC free article] [PubMed] [Google Scholar]