Abstract
Background
We previously reported a genomewide significant linkage for major psychosis in chromosome 13q13-q14.
Methods
An association analysis in 247 unrelated DSM-IV schizophrenia (SZ) patients and 250 unrelated controls from the Eastern Quebec population genotyped with 2150 single nucleotide polymorphism (SNPs) in 13q13-q14. We also used the kindred sample where linkage was detected (125 SZ, 120 bipolar (BP) and 36 schizoaffective disorder (SAD) patients vs. 467 unaffected adult relatives) for replication.
Results
An association of the T allele of rs1156026 found in the case-control sample (odds ratio (OR) = 1.81, p = 4×10−6, false discovery rate = 0.01) was replicated in the kindred sample (OR=1.54, p=0.01), strengthening the overall association evidence (p=8×10−7). The effect size increased in the subset of unrelated patients with a family history (OR = 2.28) and in the 15 families where SZ was predominant (OR=2.03). In the kindred sample, onset of either SZ or BP was on average 5 years earlier for T/T compared to C/C homozygotes leading to stronger association in patients with onset before 26 years of age (SZ: OR=2.40, p = 1.3×10−4, SZ, BP and SAD combined: OR=1.87 p=8×10−5).
Conclusions
Case-control and family-based association provided evidence of a locus at 13q13-q14 related to SZ. The proximity of the associated SNP with the linkage signal and the extension of the associated phenotype to major psychosis with younger age of onset indicate congruence between the linkage and association signals. The rs1156026 association is novel and factors explaining its non-detection in previous studies are discussed.
Keywords: age of onset, bipolar disorder, genetics, prior linkage evidence, schizophrenia, single nucleotide polymorphisms
Introduction
Our investigation of the chromosome 13q13-q14 region evolved in three steps. In a first step we reported in 2005 a genomewide suggestive linkage signal (maximum LOD score of 2.96) in the chromosome 13q13-q14 region that is common to schizophrenia (SZ) and bipolar disorder (BP) in a large kindred sample from Eastern Quebec [1]. Remarkably, in a second step we reproduced and enhanced this linkage signal in a second sample of kindred from the same population with the same microsatellite markers and the same SZ and BP combined phenotype, yielding a −log10 (p-value) of 5.21 in the combined sample [2]. This genomic region has been previously implicated in multiple linkage studies of SZ and BP (reviewed in [2]). Previous association studies in other populations suggested two candidate genes in the region. Diacylglycerol kinase eta (DGKH), located 500 kb from D13S1297 where the linkage signal peaked, was initially detected in a genomewide association study (GWAS) of BP [3] and was later found associated in both BP and SZ samples of Han Chinese origin [4]. The serotonin receptor 2A (HTR2A), located at 4 Mb from the peak linkage, has been a long standing candidate for both SZ and BP, and a current meta-analysis indicated a modest but significant association between the single nucleotide polymorphism (SNP) rs6311, located in the 5′ region of HTR2A, and SZ in Caucasians (http://www.schizophreniaforum.org/res/sczgene/meta.asp?geneID=293). Intriguingly the largest mega-analyses of SZ and BP GWAS did not report any association signal to this region [5, 6].
In a third step, in search of the origin of the linkage signal, we now report a SNP association and copy number variant (CNV) detection study in the 13q13-q14 region in unrelated SZ patients and normal controls using data from a medium-scale genomewide SNP array. An association detected in the case-control sample was then replicated in the kindred sample with SNPs close to marker D13S1297 where our previously reported linkage signal peaked [2].
Methods
Study subjects and phenotype definition
The case-control sample consisted of 247 unrelated SZ patients and 250 unrelated normal controls from the Eastern Quebec population. All subjects were Caucasian of French-Canadian ancestry. The proportion of males was 79 percent among the cases and 78 percent among the controls. Controls were adults, with a median age of 45 at the time of psychiatric evaluation.
The kindred sample consisted of 845 members of 48 multigenerational families of which 21 were mainly affected by BP (<15% of the affected family members had SZ), 15 mainly affected by a SZ spectrum disorder (< 15% had BP) and 12 were mixed pedigrees, i.e. affected almost equally by SZ and BP. In the kindred sample, narrow and broad definitions of the SZ and BP phenotypes were used in the analyses. We also defined a narrow and broad “common locus” (CL) phenotype including SZ, BP and schizoaffective disorder (SAD). More details on the phenotype definitions are given elsewhere [1, 2] and in the online supplement. The numbers of affected subjects for each phenotype definition are in Table 1. For the association analyses in the kindred sample we formed a comparison group with the 467 genotyped non affected subjects from our family sample satisfying the following criteria: A) no diagnosis in the broad definition of CL, B) age greater or equal to 25 years and C) not the parent of a CL patient. We refer to these subjects as non-affected adult relatives (NAARs).
Table 1.
Number of affected subjects for each phenotype definition.
| Phenotypes
| |||||
|---|---|---|---|---|---|
| CL
|
SZ
|
BP
|
|||
| narrow | broad | narrow | broad | narrow | broad |
|
|
|
|
|||
| 281 | 378 | 125 | 136 | 120 | 205 |
Abbreviations: SZ, schizophrenia; BP, bipolar; CL refers to the “common locus” phenotype as defined in the Methods
Age of onset in the kindred sample was defined as the age of the first definitive episode or of the first probable episode of a disorder from the spectrum of SZ and BP. The former definition was used for the genetic analyses. In the unrelated SZ patients sample, age at which could be made a definitive lifetime diagnosis of DSM SZ was the only available definition of age of onset.
Ethics
The study was explained to each family member, unrelated cases and controls, and a signed consent was obtained, as reviewed by our University Ethics Committee.
Candidate region definition
We opted to cover a wide region given the uncertainty in the boundaries of linkage signals for complex traits. We took as anchor points the two markers delimiting the linkage region with a −log10 (p-value) above 3.0 in our kindred sample, D13S1491 and D13S1272, [2] and defined the candidate region as extending 5 Mb proximal from D13S1491 to 5 Mb distal from D13S1272. The interval corresponds to coordinates 33.564 to 50.086 Mb in human genome assembly build GRCH37.3.
Genotyping and CNV detection
The case-control sample was genotyped using a customized Illumina genomewide SNP array. CNVs were inferred from the genotyping probe signals using the hidden Markov model implemented in PennCNV [7]. Our region of interest derived from the linkage evidence in our kindred sample contained 2,150 SNPs. For replication in the kindred sample, SNPs were analyzed using an in house minisequencing approach [8]. Application of standard quality control procedures left 2,081 SNPs to be included in the analysis. Details can be found in the online supplement.
Association analysis in the case-control sample
Population structure was investigated using principal component (PC) analysis. Association analysis between phenotype and genotype was performed using standard allelic, trend and genotypic tests under a dominant and an additive model. Allelic and genotypic odds ratios were estimated on the genotyped SNPs and on untyped SNPs imputed using genotype data from the 1000 Genomes Project (www.1000genomes.org). Association to both genotyped and imputed SNPs was also tested conditionally on genotypes and on allele counts of the genotyped SNPs showing the strongest association. Haplotype association tests were performed on sliding windows of three and five consecutive SNPs. The analysis is described in greater details in the online supplement.
Association analysis in the family sample
Allelic log-odds ratios (ORs) were estimated and Wald tests of association were performed under a logistic model estimated using generalized estimating equations (GEEs) with an independence working correlation structure between the subjects in the same family and an empirical variance estimate robust to intra-familial correlation [9]. The same approach was then applied to the combined case-control and family samples. The generalized disequilibrium test (GDT) [10], a score test robust to population stratification, was also applied to confirm the GEE Wald test results. The haplotype analysis is described in the online supplement.
Association with age of onset
First, we compared the mean age of onset between genotypes in patients. Second, we tested genetic association in the kindred sample by comparing the patients with onset before age 26 to the NAARs. We chose age 26 based on the median age of first definitive episode of SZ in our kindred sample. In the case-control sample we compared the SZ patients with onset before age 26 to the controls.
Results
Matching of cases to controls on ancestry
Comparison of the unrelated case and control groups along the first ten PCs revealed no important difference (p-values between 0.033 and 0.72). Supplementary figure 1 shows the near perfect overlap between the two groups on the first two PCs. The inflation factor estimated by the genomic control method applied to the genotype data was only 1.006. The good fit of the observed distribution of p-values to the expected one in the candidate region can be seen on a quantile-quantile plot (Supplementary figure 2). In consideration of this absence of evidence of population structure differences between cases and controls, we decided to not apply any correction.
Association with genotyped SNPs in the case-control sample
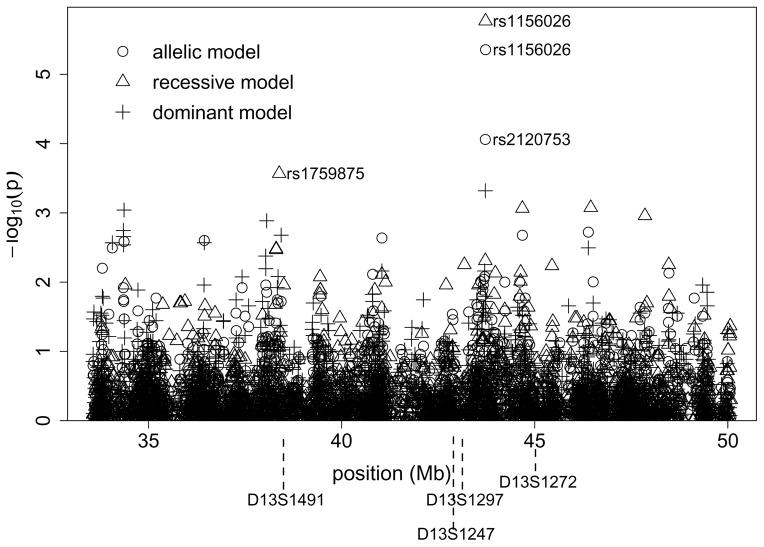
The SNP rs1156026 was the only SNP associated to SZ with a FDR < 0.05 in our primary analysis of the SNPs individually using Fisher’s exact test, with an OR of 1.81 for the T allele (Figure 1 and Table 2) and 2.63 (95% confidence interval (CI) [1.74, 4.00]) for the T/T genotype against the others. Results with the Cochran-Armitage trend test were nearly identical to the Fisher exact allelic test (not shown). When the subset of 60 patients with positive family history of SZ, psychosis or paranoia in first, second or third degree relatives was compared to the control group, rs1156026 remained the SNP with the lowest p-value in the region. The SNP effect size was larger in this familial subset (T allele OR = 2.28, T/T genotype OR = 3.10, 95% CI [1.64, 5.86]) but the greatly reduced number of cases resulted in a less significant association (Table 2). There was no significant difference in mean age of onset between the three rs1156026 genotypes (results not shown). When the case sample was restricted to the 133 patients with onset before age 26 in the genetic comparison with the control group, rs1156026 was again the SNP with the lowest p-value in the region. The ORs remained about the same as in the analysis of the full sample and the p-value was less significant due to the lower power in a smaller sample. SNP rs1156026 is located about 500 kb from D13S1297, the marker where the linkage signal peaked in the kindred sample [2].
Figure 1.
Association to genotyped SNPs in the region defined around the linkage signal at 13q13-q14. Results of the Fisher exact test are shown.
Table 2.
Association of SNP alleles and haplotypes with the narrow definition of schizophrenia.
| Marker | Alleles
|
Case-control sample (discovery)
|
Kindred sample (replication)
|
Combined analyses
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAFa
|
ORb (95% CI) | Pc | FDRd | MAF
|
ORf (95% CI) | Pg | ORf (95% CI) | Pg | |||||
| Min | Maj | cases | ctrls | cases | NAARe | ||||||||
| All cases (247 cases, 250 controls) | All families (119 cases, 467 NAARs) | (366 cases, 717 ctrls+NAARs) | |||||||||||
| rs2120753 | G | A | 0.356 | 0.480 | 0.60 (0.46, 0.78) | 8.7e-5 | 0.179 | 0.429 | 0.484 | 0.80 (0.58, 1.11) | 0.18 | 0.68 (0.56, 0.84) | 2.5e-4 |
| rs1156026 | T | C | 0.622 | 0.476 | 1.81 (1.39, 2.35) | 4.4e-6 | 0.014 | 0.538 | 0.436 | 1.54 (1.1, 2.17) | 0.012 | 1.69 (1.37, 2.08) | 8.5e-7 |
| SZ families (64 cases, 127 NAARs) | (311 cases, 377 ctrls+NAARs) | ||||||||||||
| rs2120753 | G | A | 0.356 | 0.480 | 0.60 (0.46, 0.78) | 8.7e-5 | 0.179 | 0.516 | 0.610 | 0.65 (0.41, 1.04) | 0.074 | 0.63 (0.51, 0.77) | 1.1e-5 |
| rs1156026 | T | C | 0.622 | 0.476 | 1.81 (1.39, 2.35) | 4.4e-6 | 0.014 | 0.523 | 0.374 | 2.03 (1.34, 3.09) | 8.8e-4 | 1.85 (1.48, 2.31) | 6.4e-8 |
| Cases with positive family history (60 cases vs 250 controls) | (124 cases, 377 ctrls+NAARs) | ||||||||||||
| rs2120753 | G | A | 0.292 | 0.480 | 0.45 (0.28, 0.70) | 2.1e-4 | 0.516 | 0.61 | 0.65 (0.41, 1.04) | 0.074 | 0.53 (0.39, 0.72) | 6.2e-5 | |
| rs1156026 | T | C | 0.675 | 0.476 | 2.28 (1.48, 3.58) | 1.0e-4 | 0.523 | 0.374 | 2.03 (1.34, 3.09) | 8.8e-4 | 2.24 (1.64, 3.05) | 3.6e-7 | |
| rs2120753-rs1156026 (r2=0.30) | |||||||||||||
| haplotype | Case-control sample (discovery)
|
Kindred sample (replication)
|
||||||
|---|---|---|---|---|---|---|---|---|
| frequencies
|
ORh (95% CI) | Pi | frequencies
|
ORj (95% CI) | Pk | |||
| cases | ctrls | cases | NAAR | |||||
| All cases | All families | |||||||
| G - C | 0.292 | 0.389 | baseline | 5.5e-4 | 0.335 | 0.416 | baseline | 0.038 |
| A - C | 0.086 | 0.135 | 0.87 (0.55, 1.37) | 0.033 | 0.138 | 0.151 | 1.11 (0.68, 1.82) | 0.66 |
| G - T | 0.064 | 0.091 | 0.92 (0.54, 1.59) | 0.205 | 0.106 | 0.067 | 2.05 (1.15, 3.68) | 0.052 |
| A - T | 0.558 | 0.385 | 1.94 (1.45, 2.60) | 1.4e-7 | 0.421 | 0.365 | 1.47 (1.02, 2.12) | 0.14 |
| Cases with positive family history | SZ families | |||||||
| G - C | 0.265 | 0.389 | baseline | 0.0045 | 0.394 | 0.496 | baseline | 0.0023 |
| A - C | 0.060 | 0.135 | 0.68 (0.29, 1.62) | 0.057 | 0.099 | 0.132 | 0.97 (0.44, 2.14) | 0.45 |
| G - T | 0.027 | 0.091 | 0.43 (0.12, 1.51) | 0.041 | 0.134 | 0.113 | 1.64 (0.89, 3.03) | 0.51 |
| A - T | 0.648 | 0.385 | 2.84 (1.72, 4.68) | 1.7e-7 | 0.373 | 0.259 | 1.98 (1.24, 3.19) | 0.027 |
Minor allele frequency.
Maximum likelihood odds ratio (OR) estimate and its exact 95% confidence interval (CI).
P-values of Fisher’s Exact Test.
Benjamini-Hochberg FDR taking into account the p-values from 6243 tests (there are 2081 SNPs in the chromosome 13 candidate region and each SNP is tested using three different models: allelic, dominant and recessive).
Non-affected adult relatives
GEE OR estimate and its 95% CI calculated using the empirical variance estimate.
P-value of Wald test calculated using the empirical variance estimate.
Maximum likelihood OR estimate of the haplotype versus the reference G-C haplotype and its asymptotic 95% CI taking into account missing phase information.
P-value from score test of the haplotype against all others.
GEE OR estimate of the haplotype versus the reference G-C haplotype obtained using the inferred individual haplotype pairs and its 95% CI calculated using the empirical variance estimate.
P-value of Wald test of the haplotype against all others calculated using the inferred individual haplotype pairs and the empirical variance estimate.
Association with untyped variants
Imputation of SNPs located within 200 kb of rs1156026 using the 1000 Genome Project data revealed no other SNP with stronger association. The lowest p-value obtained was 2.9 × 10−5, with rs2657116. Over the same interval, the two windows of three consecutive SNPs showing the strongest association to SZ in global haplotype association tests included rs1156026 (Supplementary figure 3). Windows of five consecutive SNPs yielded no p-value below 10−4 (results not shown). However, the examination of the association of individual haplotypes in the top two three-SNP windows revealed that the rs2120753 - rs2657100 - rs1156026 AAT and AGT haplotypes were associated to SZ (p = 2 × 10−5 for AAT after correction for testing eight haplotypes), indicating that the association was driven by the rs2120753 - rs1156026 AT haplotype. We therefore tested association with haplotypes formed by these two correlated SNPs and obtained an odds ratio of 1.94 for the AT haplotype compared to the reference GC haplotype (Table 2). When the subset of 60 cases with positive family history was compared to the control group, the odds ratio for the AT haplotype raised to 2.84. In both instances, the uncorrected p-value of the score test of the AT haplotype versus all others is close to 10−7.
Conditional analysis
We tested association of the genotyped and imputed SNPs conditional on the T/T genotype and the allele count of rs1156026 to determine whether other SNPs were associated to SZ independently of rs1156026. All p-values were non significant considering multiple testing (p > 0.0005). We obtained the same conclusion when conditioning on the allele count at rs2120753 (p > 0.001), but we note that rs1156026 ranked in first place.
Linkage disequilibrium
We examined LD between rs1156026 and other neighboring frequent SNPs in subjects of European ancestry in the 1000 Genomes Project to interpret the association results in our dataset and other studies. Only 8 SNPs and one insertion/deletion (indel) are correlated at a r2 > 0.1 with rs1156026 and all are within 25 kb (Supplementary Figure 5). Among them, we have genotyped only rs2120753, and the r2 with rs1156026 in our control sample was similar to the 1000 Genomes estimate.
CNVs
We detected CNVs in 245 cases and 137 controls genotyped in the same batch, to insure that genotyping signal intensities were comparable. The region from rs1998697 to rs9574453 spanning 6 kb is deleted in one case and duplicated in another. A deletion spanning 10 kb from rs1407608 to rs9646096 has been detected in a control. Overlapping duplications and deletions have been reported in the Database of Genomic Variants (DGV) ([11] projects.tcag.ca/variation). A duplication spanning 9 kb from rs11619167 to rs2234211 detected in a case has not been reported in the DGV.
Replication of the SNP association in the kindred sample
The SNPs rs1156026 and rs2120753 were selected for genotyping in the kindred sample. A significant association of the T allele of SNP rs1156026 to SZ was detected. Table 2 shows the results of the GEE analysis for the narrow SZ phenotype (results for the broad SZ phenotype were similar and are not shown). The estimated OR of 1.54 increased to 2.03 when the analysis was restricted to the sample of families where SZ was the predominant disorder, and the statistical significance of the GEE Wald test also improved from 0.012 to 8.8 × 10−4. The GDT with the narrow SZ phenotype gave a result similar to the GEE Wald test in the full kindred sample (p=0.027) but a less significant one in the sub-sample of SZ families (p=0.072). The combination of the case-control sample with the sub-sample of SZ families gave the strongest overall evidence of association (p = 6.4 × 10−8). Contrary to the case-control sample, little association was observed with the rs2120753 A allele, and the rs2120753 - rs1156026 AT haplotype was no more strongly associated to SZ than the T allele of SNP rs1156026 by itself. Also, there was less evidence of association under the recessive model in the kindred sample than in the case-control sample (results not shown). We also examined association to the narrow BP and CL phenotype, which previously gave linkage signals in our kindred sample [2]. Neither rs2120753 nor rs1156026 were significantly associated (Table 3).
Table 3.
Association of SNP alleles considering either all cases or early-onset cases only for the narrow definition of schizophrenia, bipolar disorder and the common locus phenotype in the kindred sample
| Marker | Alleles
|
MAFa
|
All cases vs NAARs
|
Early-onset cases vs NAARs
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Min. | Maj. | cases
|
NAARsb (n=467) | ORc (95% CI) | Pd | ORc (95% CI) | Pd | ||
| All | Early-onsete | ||||||||
| Schizophrenia (119 cases, 58 early-onset cases) | |||||||||
| rs2120753 | G | A | 0.429 | 0.377 | 0.484 | 0.80 (0.58, 1.11) | 0.18 | 0.65 (0.42, 1.01) | 0.054 |
| rs1156026 | T | C | 0.538 | 0.638 | 0.436 | 1.54 (1.1, 2.17) | 0.012 | 2.40 (1.53, 3.76) | 1.3e-4 |
| Bipolar disorder (117 cases, 40 early-onset cases) | |||||||||
| rs2120753 | G | A | 0.461 | 0.382 | 0.484 | 0.92 (0.65, 1.29) | 0.61 | 0.66 (0.38, 1.16) | 0.15 |
| rs1156026 | T | C | 0.449 | 0.512 | 0.436 | 1.06 (0.75, 1.48) | 0.76 | 1.37 (0.76, 2.44) | 0.29 |
| Common locus phenotype (273 cases, 118 early-onset cases) | |||||||||
| rs2120753 | G | A | 0.458 | 0.396 | 0.484 | 0.90 (0.73, 1.12) | 0.37 | 0.71 (0.52, 0.95) | 0.023 |
| rs1156026 | T | C | 0.487 | 0.589 | 0.436 | 1.24 (0.98, 1.56) | 0.075 | 1.87 (1.37, 2.55) | 8.1e-5 |
Minor allele frequency.
Non-affected adult relatives
GEE OR estimate and its 95% CI calculated using the empirical variance estimate.
P-value of Wald test calculated using the empirical variance estimate.
onset before age 26
In the kindred sample the mean age at the first probable episode was 26.2 years (standard deviation (SD) = 7.8) for narrow SZ, which is close to reports in general samples of SZ [12–14]. For narrow BP the mean onset age was 30.8 years (SD = 11.8). Mean age of onset depended on rs1156026 genotype for the narrow BP (p = 0.017) and CL (p = 1.1 × 10−4) phenotypes in 2 df F tests comparing the three genotypes, driven mainly by an earlier onset in T/T carriers (Supplementary figure 4A and C). For SZ, we rather observed a linear decreasing trend in the mean age of onset with the number of T alleles (−2 years per copy of the T allele, p = 0.034, Supplementary figure 4B). The age of onset in carriers of the T/T genotype was on average approximately 5 years earlier than in carriers of the C/C genotype for SZ, BP and CL (comprising also SAD). We then restricted the case sample to the patients with onset of their disorder before age 26 and repeated the comparison of allele frequency with the NAARs. For SZ, the OR for the T allele of rs1156026 increased to 2.40 (Table 3). The enrichment in T alleles in BP with onset before age 26 was insufficient to obtain a significant association. Grouping together SZ, BP and SAD cases with onset before age 26 however revealed a significant association with the T allele (p = 8.1 × 10−5), with an OR = 1.87.
Analysis of the haplotypes formed by rs2120753 and rs1156026 did not improve the signal in any of the above analyses, the association being driven by the rs1156026 T allele (results not shown).
Discussion
The consistent genomewide significant linkage signal previously observed in our kindred sample in 13q13-q14 [2] made us target a precise region to be tested in association analysis in a case-control sample. This analysis congruently yielded a strong association signal with the rs1156026 T allele (OR = 1.81) and the rs2120753 - rs1156026 AT haplotype (OR = 1.94). The detected effect size for that haplotype had an observed p-value of 1.4 × 10−7 very close to the commonly used threshold of 5 × 10−8 for genomewide significance. These two SNPs were then tested in the kindred sample and a replication was obtained for the rs1156026 T allele (OR = 1.54), which better captured the association in that sample than the rs2120753 - rs1156026 AT haplotype. Furthermore, the analysis comparing cases with positive family history to controls and the analysis of the sub-sample of predominantly SZ families both led to an increase in effect size for the rs1156026 T allele (OR = 2.28 in the case-control sample, OR = 2.03 in the SZ families).
The Schizophrenia Psychiatric GWAS consortium recently published a mega-analysis of GWAS of SZ [5] with sufficient power to detect at genomewide significance levels odds ratios around 1.15 with frequent SNPs. Most SZ cases and controls included in the mega-analysis were genotyped using Affymetrix SNP chips, which do not include rs1156026. Among all SNPs and indels found in the 1000 Genomes Project data, only two moderately correlated with rs1156026 (rs1156027 and rs2589312, Supplementary Figure 5) are on the 6.0 version of the Affymetrix SNP chip, and none on earlier versions. There is therefore little information to impute rs1156026 genotypes in the vast majority of subjects included in the mega-analysis. In order to evaluate the effect of poor coverage of rs1156026 on the association of that SNP with SZ, we deleted the rs1156026 genotypes from our sample and repeated the imputation process using the other SNPs on our Illumina chip, among which only rs2120753 is correlated with rs1156026. The allelic OR dropped from 1.81 to 1.44, for a p = 9.5 × 10−4. The association to rs1156026 would therefore have gone undetected had that SNP not been genotyped.
The T allele of rs1156026 was associated to an earlier age of onset in SZ, BP and in the SZ, BP and SAD combined phenotype CL in the kindred sample. An onset of illness occurring 5 years earlier on average in rs1156026 T/T carriers compared to C/C carriers may have considerable implications for the life course and outcome. This earlier age of onset resulted in detectable association of the T allele to the narrow CL phenotype with onset before age 26, while no association was detected when ignoring age of onset. Our finding an association in the younger age of onset subgroup may be congruent with previous repetitive observations that linkage sites on 13q would be shared by SZ and BP in our kindred sample and in other samples [2, 15, 16]. An interpretation could be that the commonality of the underlying genetic effect might be partly expressed through a younger age of onset in major psychosis. The difference in age of onset assessment in the two samples may explain that the association to earlier onset was found only in the kindred sample, not in the unrelated SZ patient sample. We had available in the former sample the onset of first episode whereas in the latter sample we could analyze only the age at which the first definite lifetime DSM diagnosis could be made. It is probable that the occurrence of first episode represents a more valid or sensitive measure to detect genetic effects than age of first definite diagnosis.
The minimal age of 25 for inclusion of adult relatives in the non-affected comparison group within the kindred sample allowed us to maximize the size of that group, at the expense of including a few subjects who could develop major psychosis at a later age. We re-estimated SNP allele frequencies in a comparison sample restricted to the 378 NAARs older than 39 years, corresponding to the 90th percentile of the age of first definitive episode of SZ in that sample. The frequency of the T allele of rs1156026 increased slightly compared to the larger comparison group, a change compatible with random sampling variation (Supplementary tables 1 and 2). This is evidence that contamination by the potential appearance of future major psychosis cases among NAARs below 39 years of age had no significant impact on the T allele frequency, which supports inclusion of these younger NAARs in the comparison sample.
We found very few CNVs in the investigated region, none of them exceeding 10 kb in length. This is much shorter than the CNVs reported to cause genomic disorders with an SZ phenotype [17]. CNVs of the length that we detected are likely to be benign. Our SNP array had a low resolution for CNV detection: the CNVs that we did detect contained few SNPs, and for that reason false positives are likely, and shorter CNVs probably remained undetected. Given the short length and low frequency of CNVs in the region, very few SNP genotypes were likely to be miscalled due to the presence of CNVs and their impact on the SNP association results is probably negligible.
Our study design presented important advantages. First the focus on a linkage region reduced the penalty for multiple testing in the present case-control association analysis, allowing a FDR less than 0.05 for the T allele of SNP rs1156026. Second, population stratification confounding was minimized in our study. Indeed the recruitment from the same population permitted an excellent match on ancestry between our cases and controls as evidenced by the principal component analysis of their genotypes.
Our study did not however escape the upward bias in the effect size of the marker ranking first among a large number of tested markers [18]. The allelic ORs for the T allele of rs1156026 observed in our replication sample matched well those predicted by the bootstrap bias-reduction method implemented in BR-squared [19, 20], which were 1.43 (95% CI [1.12 – 2.38]) for the analysis of all SZ cases and 1.51 (95% CI [1.10 – 2.16]) for the analysis of SZ cases with positive family history, indicating that these are the effect sizes to be expected in replication studies.
We observed weak linkage to rs1156026 and rs2120753 in parametric linkage analysis using the same model parameters as in previous reports [1, 2] (results not shown), implying that these two SNPs would not explain the previous linkage signal (unfortunately we could not formally test that hypothesis because the usual procedures now applied in nuclear families or small pedigrees [21] cannot be implemented in large kindreds such as ours). The weak linkage makes it unlikely that either of these SNPs is the single causal variant at the locus. A few common variants are moderately correlated to rs1156026 (Supplementary Figure 5) and could be causal variants, assuming that the imputation procedure failed to reveal their strong association with SZ. These correlated variants are in an intergenic region; there is no evidence that correlation with rs1156026 extends to SNPs in the neighboring genes DnaJ (Hsp40) homolog, subfamily C, member 15 (DNAJC15) and ecto-NOX disulfide-thiol exchanger 1 (ENOX1), nor to the nearby DGKH and HTR2A genes previously found associated to SZ and/or BP (not shown). An alternative explanation to reconcile the association and linkage results is that multiple rare causal variants lie with the rs1156026 T allele on haplotypes extending beyond the LD region defined by correlation between frequent SNPs, in what has been called a synthetic association [22].
In summary, we reported a new significant association of the T allele of SNP rs1156026 with SZ in a case-control sample, which we then replicated in the kindred sample where we initially detected linkage at the 13q13-q14 locus[1, 2]. In the kindred sample, the rs1156026 T allele was also found to be associated with an earlier age of onset not only for SZ but also for BP, making this allele a marker of earlier onset of major psychosis. The location of the SNP rs1156026 close to the peak of the previously reported linkage signal and its association to the major psychosis CL phenotype with onset before age 26 in the kindred sample suggest an overlap between the genetic variants responsible for the linkage and the association signals in 13q13-q14 in that sample. The absence of linkage to rs1156026 makes it unlikely to be the single causal variant at that locus, and further investigations will be required to identify the variant or variants in the region involved in the aetiology of major psychosis.
Supplementary Material
Figure 2.
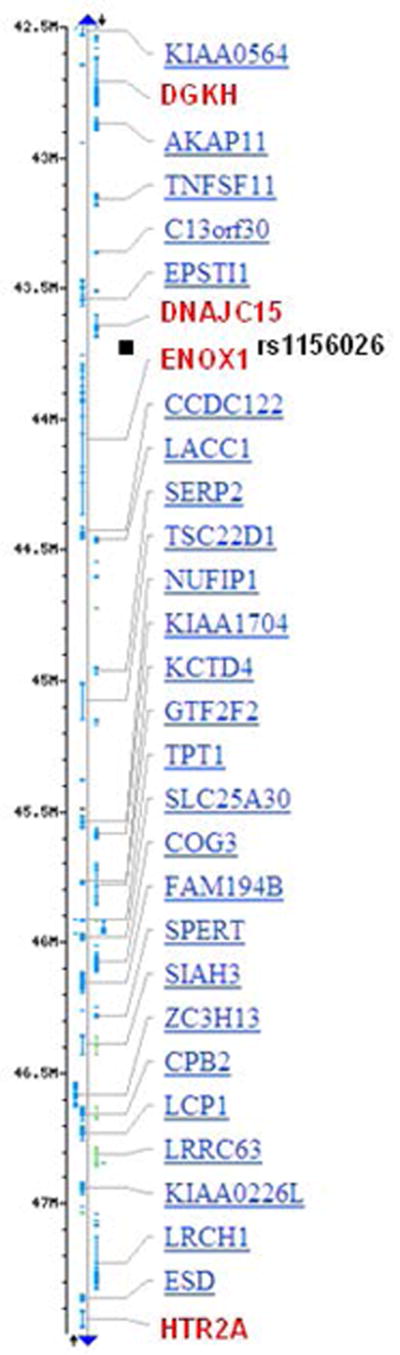
Genes (in red) at 13q14.11-13q14.2 that showed association with schizophrenia or surrounding rs1156026 (black square). Positions are in Mb (Build 37.3). DGKH: diacylglycerol kinase, eta; DNAJC15: DnaJ (Hsp40) homolog, subfamily C, member 15; ENOX1: ecto-NOX disulfide-thiol exchanger 1; HTR2A: 5-hydroxytryptamine (serotonin) receptor 2A.
Acknowledgments
We are grateful to our professional research assistants: Louise Bélanger, Marie-Claude Boisvert, Linda René, Lisette Gagnon, Claudie Poirier, Nicole Leclerc, Julie Lamarche, Pierrette Boutin, David Demers, Lise St-Germain, Anne-Marie Simard, Claudia Émond, Isabel Moreau and to the family members, adults and children, who participated in this study. This work was funded by the Canadian Institutes of Health research (CIHR, grants MT-12854 and MOP-74430) and by a Canada Research Chair (# 950-200810) in the genetics of neuropsychiatric disorders of which M. Maziade is the Chair. The data management system was funded by the Canada Foundation for Innovation Leaders Opportunity Fund (grant 27592). A. Bureau is supported by a research fellowship from the Fonds de recherche du Québec - Santé.
Footnotes
Financial Disclosures
AB, YCC, MAR, CM and MM are inventors on a patent application filed by Laval University relating to novel markers for mental disorders, including SNP rs1156026. MM and MAR have been consultants for GlaxoSmithKline (GSK) and Eli Lilly and have received research funding from GSK, Eli Lilly and AstraZeneca that are not related to the material of this study. JC, AF and TP report no biomedical financial interests or potential conflicts of interest.
References
- 1.Maziade M, Roy MA, Chagnon YC, Cliche D, Fournier JP, Montgrain N, et al. Shared and specific susceptibility loci for schizophrenia and bipolar disorder : A dense genome scan in Eastern Quebec families. Molecular Psychiatry. 2005;10:486–499. doi: 10.1038/sj.mp.4001594. [DOI] [PubMed] [Google Scholar]
- 2.Maziade M, Chagnon YC, Roy MA, Bureau A, Fournier A, Merette C. Chromosome 13q13-q14 locus overlaps mood and psychotic disorders: the relevance for redefining phenotype. Eur J Hum Genet. 2009;17:1034–1042. doi: 10.1038/ejhg.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B, et al. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2008;13:197–207. doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng Z, Wang T, Li T, Li Y, Chen P, Zhao Q, et al. Common SNPs and haplotypes in DGKH are associated with bipolar disorder and schizophrenia in the Chinese Han population. Mol Psychiatry. 16:473–475. doi: 10.1038/mp.2010.86. [DOI] [PubMed] [Google Scholar]
- 5.Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun X, Ding H, Hung K, Guo B. A new MALDI-TOF based mini-sequencing assay for genotyping of SNPS. Nucleic Acids Research. 2000;28:e68. doi: 10.1093/nar/28.12.e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeger SL, Liang KY, Albert PS. Models for longitudinal data: a generalized estimating equation approach. Biometrics. 1988;44:1049–1060. [PubMed] [Google Scholar]
- 10.Chen WM, Manichaikul A, Rich SS. A generalized family-based association test for dichotomous traits. Am J Hum Genet. 2009;85:364–376. doi: 10.1016/j.ajhg.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, Feuk L, Duggan GE, Khaja R, Scherer SW. Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome. Cytogenet Genome Res. 2006;115:205–214. doi: 10.1159/000095916. [DOI] [PubMed] [Google Scholar]
- 12.Hafner H, Maurer K, Loffler W, Riecher-Rossler A. The influence of age and sex on the onset and early course of schizophrenia. Br J Psychiatry. 1993;162:80–86. doi: 10.1192/bjp.162.1.80. [DOI] [PubMed] [Google Scholar]
- 13.Kirkbride JB, Errazuriz A, Croudace TJ, Morgan C, Jackson D, Boydell J, et al. Incidence of schizophrenia and other psychoses in England 1950 –2009: a systematic review and meta-analyses. PLoS One. 7:e31660. doi: 10.1371/journal.pone.0031660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajji TK, Ismail Z, Mulsant BH. Age at onset and cognition in schizophrenia: meta-analysis. Br J Psychiatry. 2009;195:286–293. doi: 10.1192/bjp.bp.108.060723. [DOI] [PubMed] [Google Scholar]
- 15.Goes FS, Zandi PP, Miao K, McMahon FJ, Steele J, Willour VL, et al. Mood-incongruent psychotic features in bipolar disorder: familial aggregation and suggestive linkage to 2p11-q14 and 13q21-33. Am J Psychiatry. 2007;164:236–247. doi: 10.1176/ajp.2007.164.2.236. [DOI] [PubMed] [Google Scholar]
- 16.Badner JA, Gershon ES. Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry. 2002;7:405–411. doi: 10.1038/sj.mp.4001012. [DOI] [PubMed] [Google Scholar]
- 17.Bassett AS, Scherer SW, Brzustowicz LM. Copy number variations in schizophrenia: critical review and new perspectives on concepts of genetics and disease. Am J Psychiatry. 2010;167:899–914. doi: 10.1176/appi.ajp.2009.09071016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garner C. Upward bias in odds ratio estimates from genome-wide association studies. Genet Epidemiol. 2007;31:288–295. doi: 10.1002/gepi.20209. [DOI] [PubMed] [Google Scholar]
- 19.Faye LL, Sun L, Dimitromanolakis A, Bull SB. A flexible genome-wide bootstrap method that accounts for ranking and threshold-selection bias in GWAS interpretation and replication study design. Stat Med. 2011;30:1898–1912. doi: 10.1002/sim.4228. [DOI] [PubMed] [Google Scholar]
- 20.Sun L, Dimitromanolakis A, Faye LL, Paterson AD, Waggott D, Bull SB. BR-squared: a practical solution to the winner’s curse in genome-wide scans. Hum Genet. 2011;129:545–552. doi: 10.1007/s00439-011-0948-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen MH, Van Eerdewegh P, Vincent QB, Alcais A, Abel L, Dupuis J. Evaluation of approaches to identify associated SNPs that explain the linkage evidence in nuclear families with affected siblings. Hum Hered. 2010;69:104–119. doi: 10.1159/000264448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.