Abstract
Objectives
To obtain a broad molecular epidemiological characterization of plasmid-mediated AmpC β-lactamase CMY-2 in Escherichia coli isolates from food animals in China.
Methods
A total of 1083 E. coli isolates from feces, viscera, blood, drinking water, and sub-surface soil were examined for the presence of CMY-2 β-lactamases. CMY-2-producing isolates were characterized as follows: the bla CMY-2 genotype was determined using PCR and sequencing, characterization of the bla CMY-2 genetic environment, plasmid sizing using S1 nuclease pulsed-field gel electrophoresis (PFGE), PCR-based replicon typing, phylogenetic grouping, XbaI-PFGE, and multi-locus sequence typing (MLST).
Results
All 31 CMY-2 producers were only detected in feces, and presented with multidrug resistant phenotypes. All CMY-2 strains also co-harbored genes conferring resistance to other antimicrobials, including extended spectrum β-lactamases genes (bla CTX-M-14 or bla CTX-M-55), plasmid-mediated quinolone resistance determinants (qnr, oqxA, and aac-(6′)-Ib-cr), floR and rmtB. The co-transferring of bla CMY-2 with qnrS1 and floR (alone and together) was mainly driven by the Inc A/C type plasmid, with sizes of 160 or 200 kb. Gene cassette arrays inserted in the class 1 or class 2 integron were amplified among 12 CMY-2 producers. CMY-2 producers belonged to avirulent groups B1 (n = 12) and A (n = 11), and virulent group D (n = 8). There was a good correlation between phylogenetic groups and sequence types (ST). Twenty-four STs were identified, of which the ST complexes (STC) 101/B1 (n = 6), STC10/A (n = 5), and STC155/B1 (n = 3) were dominant.
Conclusions
CMY-2 is the dominant AmpC β-lactamase in food animals and is associated with a transferable replicon IncA/C plasmid in the STC101, STC10, and STC155 strains.
Introduction
The prevalence of plasmid-encoded AmpC (pAmpC) β-lactamases, which confer resistance to extended-spectrum cephalosporins in Gram-negative bacilli, has increased in both humans and livestock isolates worldwide, and is a global problem [1]. CMY-2 is the most common pAmpC in E. coli from different geographical areas including Asia, North America, and Europe [2]–[5], and has now been reported in Salmonella and Escherichia coli isolates from a variety of food animals and products on all continents except Australia [6]. Some studies reported that the increased incidence of infections in humans with S. enterica serovar Newport possessing CMY-2 in North America was associated with exposure to dairy cattle as well as the consumption of raw milk, raw or improperly cooked beef mince, and the cross-contamination of raw meat with other foods [7], [8]. CMY-2-bearing plasmids, predominantly the A/C, I1, or K/B replicon types [9], [10], were readily transferable between Salmonella and E. coli from food animals and humans [10], [11]. Therefore, livestock-associated plasmid-encoded CMY-2 has posed increasing concerns to public health worldwide6.
E. coli is one of most common pathogens of nosocomial, healthcare-associated, and community infections [12]. According to CHINET (antimicrobial resistance surveillance networks in China), bacteria isolated from various samples from 14 hospitals in 10 regions or Provinces were predominantly E. coli, comprising 61.8% of the total isolates [13]. CMY-2 AmpC in Chinese pediatric patients was detected first in E.coli between 2003 and 2005; the occurrence of AmpC β-lactamase in E. coli and K. pneumoniae had the highest prevalence [13]. The resistance to multiple drugs, including third-generation cephalosporins, and the carriage of both pAmpC and extended spectrum β-lactamase (ESBL) genes in E. coli isolates from food animals has increased rapidly [14]-[17]. The detection rate of bla CMY-2 in isolates from chickens, which emerged between 2000 and 2003, increased rapidly from 2004–2007 [16]. However, limited information is available regarding the characteristics of the CMY-2 plasmid type or the clonal dissemination of CMY-2 in E.coli of food animal origin in China. The aim of this study was to assess the molecular epidemiology and characteristics of CMY-2-bearing plasmid-producing E. coli in food animals including pigs, chickens, ducks, and geese.
Materials and Methods
Bacterial isolates and antimicrobial susceptibility
A total of 1083 unique E. coli isolates from a range of food animal species (pigs, n = 424; chickens, n = 306; ducks, n = 175; geese, n = 178) were recovered between October 2010 and January 2012 from 58 fixed farms described previously [15]. Of the 1083 E. coli isolates, 587 were cultured from feces, 456 from the viscera, 14 from blood samples, and 26 from drinking water and sub-surface soil of duck farms. These isolates were collected partially from the Guangdong Province Surveillance Program on Antibiotic Resistance in bacteria isolated from animals. The program was carried out by the Laboratory of Clinical Microbiology, Veterinary Research Institute, Guangdong Academy of Agricultural Sciences. Methods for sample collection and bacterial isolation were described previously [18]. Susceptibility testing was determined for all isolates using the standard agar dilution method on Mueller-Hinton agar according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [19], [20]. The 13 antimicrobials tested were ampicillin (Amp), ceftiofur (Cft), cefotaxime (Ctx), ceftazidime (Caz), cefoxitin (Cxt), ceftriaxone (Ctr), gentamicin (Gen), kanamycin (Kan), amikacin (Ami), florfenicol (Flf), tetracycline (Tet), ciprofloxacin (Cip), and olaquindox (Oqx). Cefoxitin-resistant strains (MIC ≥8 mg/L) were used for selecting CMY-2-producing strains via PCR amplification of the bla CMY gene and DNA sequencing, molecular characterization of drug resistance, and epidemiology of bla CMY-2-harboring strains. E. coli ATCC 25922 was used as the control strain.
Molecular characterization of drug resistance
Among the bla CMY-harboring strains, β-lactamase genes (bla CTX-M, bla OXA, bla SHV, bla TEM) and plasmid-mediated quinolone resistance (PMQR) genes (qnrABCDS, aac(6′)-Ib-cr, qepA, oqxA) were detected by PCR amplification using specific primers and conditions (Table S1). The 16S rRNA methylase genes rmtB and the plasmid-borne florfenicol resistance gene, floR, were similarly detected using primers listed in Table S1.
Genetic environment of bla CMY-2 gene and detection of integrons
The genetic environment of the bla CMY-2 genes was investigated using PCR and sequencing. The ISEcp1 forward primer and CMY-2 reverse primer were used to investigate regions upstream of the bla CMY-2 genes. The CMY-2 forward primer and reverse primers for IS903, IS26, orf447, mucA, or orf513 were used to characterize regions downstream of the bla CMY-2 genes. Integrons of class 1 or class 2 as well inserted gene cassettes were detected. Sequences of the above primers are listed in Tables S2 and S3.
Conjugation and plasmid analysis
Conjugation experiments were performed on bla CMY-2-containing strains using streptomycin-resistant E. coli C600 as the recipient [4]. Transconjugants were selected on MacConkey agar plates supplemented with streptomycin (1000 mg/L) and cefoxitin (8 mg/L). Transconjugants were tested for the presence of drug resistance genes and antimicrobial susceptibility, as described above.
Plasmids were typed using PCR-based replicon typing (PBRT) [21]. PFGE with S1 nuclease (TakaRa Biotechnology, Dalian, China) digestion of whole genomic DNA was performed for all 15 donor strains and transconjugants, as described previously [22]. After Southern transfer to a Hybond-N+ membrane (GE Healthcare, Little Chalfont, United Kingdom), the plasmids were probed with the bla CMY-2 gene and respective replicons (DIG High Prime DNA Labeling and Detection Starter Kit I, Roche Applied Science, Mannheim, Germany).
Population structure analysis
All CMY-2-producing E. coli isolates were classified according to E. coli phylogenetic groups A, B1, B2, and D using multiplex PCR [23]. XbaI-pulsed-field gel electrophoresis (PFGE) patterns were typed using a CHEF-MAPPER System (Bio-Rad Laboratories, Hercules, CA) as described previously [15]. For multi-locus sequence typing (MLST) analysis, seven conserved housekeeping genes (adk, fumC, gyrB, icd, purA, mdh, and recA) were analyzed by PCR amplification using specific primers (Table S4) and sequencing. Allelic profiles and sequence type (ST) determinations were performed according to the E. coli MLST website (http://mlst.ucc.ie/mlst/dbs/E.coli) scheme.
Results
Antimicrobial susceptibility
A total of 233 (21.5%) out of 1083 E. coli isolates were resistant to cefoxitin with MIC values ranging from 8 mg/L to >512 mg/L. Only 33 of the cefoxitin-resistant isolates produced CMY, including 19 from pigs, 10 from chickens, and two each from ducks and geese. Of the 33 bla CMY bearing isolates, 31 carried bla CMY-2 from feces (Table 1), and two from the feces and viscera of geese carried bla CMY-41 and bla CMY-64, respectively (data not shown). All 33 CMY-producing isolates exhibited multi-drug resistance profiles, and showed resistance to both β-lactam drugs and more than two non-β-lactam drugs. The most common resistance pattern was Amp-Ctx-Caz-Ctr-Flf-Gen-Kan-Tet-Cip-Oqx (20/33, 61%). The presence of resistance to olaquindox (a growth promoter used extensively in pig and poultry farms) was detected in the majority of bla CMY-2-bearing strains (28/33, 85%). Twenty-three bla CMY-2-bearing isolates were resistant to ceftiofur, a newly approved β-lactam for veterinary use in China. Resistance to amikacin was relatively low (8/32, 25%).
Table 1. Overall results of co-resistance, phylogenetic grouping, MLST and plasmid replicon analysis of CMY-2producing E. coli isolates of food producing animals in China.
| Strains | Source | Other resistance genes | Phylogen. group | MLST | blaCMY-2 linked-element upstream | Gene cassettes inserted in class1 and 2 integrons | Plasmid transfer | Co-transferred resistant gene | Plasmid replicon types and approx. size (kb) | |
| ST | STCa | |||||||||
| L145 | Pig | blaOXA-1,oqxA,floR | A | 10 | STC10 | ISEcp1 | ND | - | - | - |
| L147 | Pig | oqxA | A | 10 | STC10 | ISEcp1 | ND | - | - | - |
| L393 | Chicken | blaTEM-1,qnrS1,oqxA,floR,aac-(6′)-Ib-cr | A | 2690 | STC10 | ISEcp1 | ND | + | qnrS1,floR,aac-(6′)-Ib-cr | A/C,160 |
| L699 | Chicken | blaTEM-1,qnrS1,floR | A | 48 | STC10 | ISEcp1 | class2: sat1+aadA1 | + | qnrS1,floR | A/C,200 |
| L78 | Pig | blaCTX-M-14,blaTEM-1,qnrS1,oqxA,floR | A | 3244 | STC10 | ISEcp1 | ND | + | blaTEM-1,qnrS1,floR | A/C,160 |
| L667 | Pig | blaTEM-1,oqxA | B1 | 3403 | STC101 | ISEcp1 | class1: drfA17+aadA5 | - | - | - |
| L671 | Pig | blaTEM-1,blaOXA-1,oqxA,qnrS1 | B1 | 359 | STC101 | ISEcp1 | class1: drfA17+aadA5 | + | qnrS1 | K,200 |
| L669 | Pig | blaTEM-1,oqxA | B1 | 359 | STC101 | ISEcp1 | class1: drfA17+aadA5 | - | - | - |
| L670 | Pig | blaTEM-1,oqxA,aac-(6′)-Ib-cr | B1 | 359 | STC101 | ISEcp1 | class1: drfA17+aadA5 | - | - | - |
| L679 | Pig | blaTEM-1,qnrS1,oqxA,floR | B1 | 101 | STC101 | ISEcp1 | ND | - | - | - |
| L1119 | Pig | blaTEM-1,qnrS1,aac-(6′)-Ib-cr,oqxA,floR | B1 | 101 | STC101 | ISEcp1 | ND | + | qnrS1,floR | A/C,160 |
| L518 | Pig | blaTEM-1,oqxA,rmtB | D | 648 | none | ISEcp1 | ND | - | - | - |
| L461 | Duck | blaCTX-M-55,blaTEM-1,qnrS1,oqxA,floR,rmtB | D | 648 | none | ISEcp1 | class1: aadA22 | + | blaCTX-M-55, qnrS1 | FIB,40 |
| L351 | Chicken | floR,rmtB | B1 | 155 | ST155C | ISEcp1 | ND | + | floR | A/C,200 |
| L391 | Chicken | blaTEM-1,qnrS1,floR | B1 | 155 | ST155C | ISEcp1 | class1: dfrA1+aadA1 | + | qnrS1,floR | A/C,160 |
| L392 | Chicken | blaTEM-1,qnrB6,floR | B1 | 2294 | ST155C | ISEcp1 | class1: dfrA1+aadA1 | + | floR | A/C,160 |
| L813 | Pig | blaTEM-1,qnrS1,rmtB,aac-(6′)-Ib-cr | B1 | 156 | none | ISEcp1 | ND | + | qnrS1 | FIB,40 |
| L361 | Duck | blaTEM-1,qnrS1,aac-(6′)-Ib-cr | D | 156 | none | ISEcp1 | ND | - | - | - |
| T117 | Pig | blaTEM-1 | A | 1114 | none | ISEcp1 | ND | - | - | - |
| A10-2 | Pig | blaTEM-1,blaOXA-1,qnrS1,oqxA,floR | A | 1114 | none | ISEcp1 | ND | - | - | - |
| L1039 | Pig | blaTEM-1,oqxA,floR | D | 457 | ST457C | ISEcp1 | ND | - | - | - |
| T43 | Pig | oqxA | D | 3376 | ST457C | ND | ND | - | - | - |
| C42 | Pig | floR | A | 3402 | none | ISEcp1 | ND | - | - | - |
| T26 | Pig | blaTEM-1,oqxA,floR | A | 3404 | none | ND | ND | + | blaTEM-1,floR | A/C,160 |
| L1166 | Pig | floR | A | 3269 | none | ISEcp1 | ND | + | floR | A/C,160 |
| L653 | Pig | blaTEM-1,oqxA,floR | A | 3014 | none | ISEcp1 | ND | - | - | - |
| L215 | Chicken | blaTEM-1 | D | 362 | none | ISEcp1 | class1: drfA17+aadA5 | - | - | - |
| L349 | Chicken | qnrS1,floR | D | 354 | none | ISEcp1 | class1: drfA17+aadA5 | + | qnrS1,floR | A/C,200 |
| L394 | Chicken | qnrS1,blaTEM-1,floR | B1 | 3245 | none | ISEcp1 | class1: orf+aadA2 | + | blaTEM-1,qnrS1,floR | HI2,220 |
| L398 | Chicken | - | B1 | 1431 | none | ND | class1: dfrA17+aadA5 | - | - | - |
| L399 | Chicken | qnrS1 | D | 69 | none | ISEcp1 | ND | + | qnrS1 | K,160 |
STC, ST complex.
ND, not detected.
Characterization of bla CMY-2 harboring isolates
All except one CMY-2-producing isolate harbored more than one resistance gene conferring resistance to different antimicrobial drugs (Table 1). Of the detected ESBLs genes, bla CTX-M type alone (bla CTX-M-14 and bla CTX-M-55) was identified in two strains; no isolate harbored the bla SHV gene. The narrow β-lactamase-encoding genes bla TEM-1 and bla OXA-1 were found in 22 and three strains, respectively. Among the detected PMQR determinants, oqxA, qnr, and aac-(6′)-Ib-cr were detected in 17, 15, and five strains, respectively (Table 1). Of the 15 qnr genes, 14 were qnrS1, and one was qnrB6. No qnrA, qnrD, qnrC, or qepA genes were detected. The floR and rmtB genes were identified in 18 and four strains, respectively. Both ESBL producers also carried the qnrS1, oqxA, and floR genes. Remarkably, one strain (L461) contained five genes conferring resistance to five antimicrobial drug classes (Table 1). A CMY-41 producer from goose liver concurrently carried bla CTX-M-15 and bla CTX-M-65 (data not shown).
Genetic environment of bla CMY-2 and detection of integrons
Twenty-eight of 31 bla CMY-2 were linked to an upstream ISEcp1 element; bla CMY-41 was also associated with the upstream ISEcp1 element. No IS elements were detected upstream of bla CMY-64 or downstream of bla CMY. Of the 31 CMY-2 producers, 27 contained class 1 integrases, and one harbored a class 2 integrase. Of the 27 class 1 integrase-positive isolates examined, 11 were found to possess cassettes inserted within the integrons, including the four-gene cassette arrays dfrA17+aadA5, dfrA1+aadA1, orfF+aadA2, and aadA22 (Table 1). dfrA17+aadA5 was most common (7/11, 64%), followed by dfrA1+aadA1 (2/11, 18%); orfF+aadA2 and aadA22 were found in single isolates. The class 2 integron present in one strain (L699) harbored the 1.0 kb sat1+aadA1 arrays. One CMY-41 producer from geese also contained a dfrA17+aadA5 cassette array.
Population structure analysis
All 31 blaCMY-2 strains were distributed into groups B1 (n = 12) and A (n = 11) of the commensal strains, strains associated with enterotoxigenic and enterohemorrhagic infections, and the potentially virulent phylogenetic group D (n = 7). The strain carrying bla CMY-41 from the liver of a diseased goose belonged to group D, and the strain containing bla CMY-64 from a goose intestine belonged to group A.
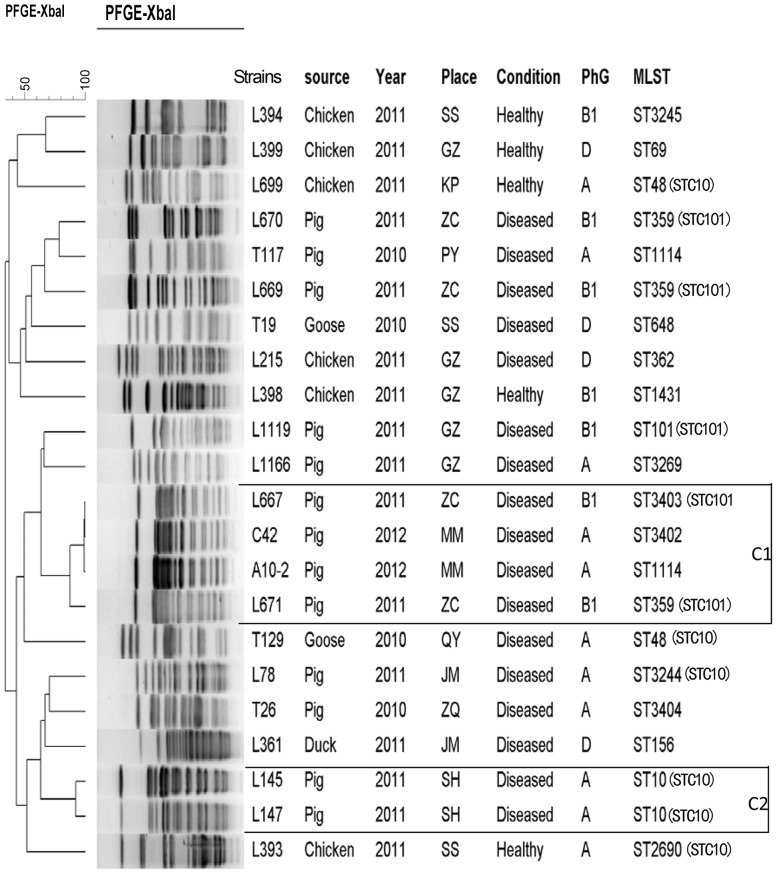
A total of 18 different PFGE-types were detected among 22 typeable CMY isolates (20 CMY-2, one CMY-41, and one CMY-64), including 19 single types, and two clusters (>90% similarity) containing two (Cluster 2) and four isolates each (Cluster1) (Fig. 1). Interestingly, the four isolates in Cluster1 were obtained from two different locations and further divided into two phylogenetic groups, each containing two isolates. The Cluster 2 isolates belonged to the same phylogenetic group and originated from the same place.
Figure 1. Dendrogram of XbaI-PFGE patterns of CMY-producing E. coli strains recovered from food-producing animals.
All the strains were CMY-2 producers, except for T19 (CMY-41) and T129 (CMY-64). Similarity analysis was performed using the Dice coefficient, and clustering was performed by following the unweighted-pair group method using average linkages (UGPMA). A total of 16 PFGE patterns were identified, and the two clusters with highly similar PFGE patterns were labeled C1 and C2. Abbreviations for the place column: SS, Sanshui; GZ, Guangzhou; KP, Kaiping; ZC, Zengcheng; PY, Panyu; MM, Maoming; QY, Qingyuan; JM, Jiangmen; ZQ, Zhaoqing; SH, Sihui. Abbreviation for PhG: Phylogenetic Group.
MLST analysis of the 31 CMY-2 producing isolates identified 24 different STs, including seven novel ones (ST3244, ST3245, ST3403, ST3269, ST3402, ST3404, and ST3376) (Table S5). A total of 20 isolates were related to six STs or ST-complexes (STCs) described previously: 10, 101, 155, 156, 648, and 1114. The prevalent ST/STCs were 101 (n = 6, 19%), 10 (n = 5, 16%), 155 (n = 3, 10%), 156 (n = 2, 6%), 457 (n = 2, 6%), 1114 (n = 2, 6%), and ST648 (n = 2, 6%). The remaining isolates were each of a single ST type, including four novel ST types (Table S5).
STC 101 included ST101, the single locus variant (SLV; isolate ST359), and one double locus variant (DLV; isolate ST3403). STC 10 included ST10, one SLV isolate (ST48), one DLV isolate (ST2690), and one triple locus variant (TLV) isolate (the novel ST3244). STC155 included two ST155 and one SLV isolate (ST2294).
Transferability of bla CMY-2 and plasmid analysis
The transferability of bla CMY-2 was observed in 15 out of 31 isolates at transfer frequencies ranging from 10−3 to 10−7 transconjugants per recipient. Of the 15 transferable bla CMY-2, 10 were located on the IncA/C type plasmid with sizes of 160 to 200 kb, two 40 kb IncFIB plasmids, two IncK plasmids sized 160 kb and 200 kb, and one 220 kb HI2 plasmid. Genes encoding resistance to other antimicrobials also co-transferred in some strains. Remarkably, qnrS1 and floR co-transferred with bla CMY-2 in eight of 15 isolates. The co-transfer of bla CMY-2, bla CTX-M-55, and qnrS1 was observed in one isolate from a duck.
Discussion
In the present study, we performed a broad molecular epidemiological characterization of CMY-2-producing E. coli collected from the Guangdong Province Surveillance Program on antibiotic resistance in bacteria isolated from animals between 2010 and 2012. Compared with earlier surveillance on E. coli antibiotic resistance from fixed food animal farms, resistance to a panel of cephalosporins was increased significantly from <7% to between 20 and 60% (data not shown) for all tested drugs in 2003 to 2005 [14], [15]; resistance to cefotaxime, ceftriaxone, cefoxitin, ceftiofur, and ceftazidime was 55.8, 59.7, 21.5, 60, and 20.8%, respectively. The rapid increase in resistance to third-generation cephalosporins, particularly for ceftiofur, which is newly approved for use in veterinary clinical settings, was consistent with previous reports from other Provinces in China [16], [17].
Cefoxitin/ceftiofur resistant isolates from E. coli and Salmonella occur frequently worldwide, which is likely to be associated with the production of CMY enzyme(s) [24]. The use of ceftiofur in the veterinary clinical setting has encouraged selection of CMY-2 AmpC in both Salmonella and E. coli [6]. In China, the occurrence of CMY-2 in E. coli originating in chickens increased rapidly [16]; the detection rate of bla CMY-2 in chicken isolates was higher than in those from pigs [25], which is consistent with reports from Japan [26]. In the present study, all CMY-2 producers were isolated from feces, consistent with a previous study [14], suggesting that the gastro-intestinal tract of animals is a reservoir for CMY-2 producers. Only half of the samples were collected from animal feces in the present study, which might explain the lower occurrence of CMY-2. Of note, many CMY-2 producers were isolated from pigs, which was an increased prevalence compared with previous reports [14], [25]. The use of ceftiofur was approved for pigs in 2005 in China, which may have contributed to this rapid increase.
Salmonella and E. coli isolates carrying bla CMY-2 have been associated with community-acquired infections [1], [6], [27]. Plasmids carrying AmpC genes often carry other genes that confer resistance to non-β-lactams, but rarely ESBL genes [28]. Previous studies demonstrated that CMY-2-producing E. coli easily acquired other resistance genes, resulting in a multidrug resistant profile. The floR gene was the most common gene acquired, which conferred resistance to florfenicol. Some studies have shown that the increasing occurrence of CMY-2 was associated with the use of florfenicol in the veterinary clinic [29]. In the present study, most CMY-2 producers harbored not only genes encoding β-lactamases (including ESBLs), but also many diverse genes encoding resistance to other antimicrobials. In addition to floR, variants of qnr, a plasmid-mediated quinolone resistant gene, were detected frequently. The presence of qnr or floR, either alone or in combination, was detected frequently in CMY-2 isolates. Transconjugation experiments confirmed that the bla CMY-2 gene could co-transfer with multiple antibiotic resistance genes, frequently with qnrS1+floR or qnrS1, driven by 160 and 200 kb-sized IncA/C plasmids. Previous studies demonstrated that plasmids carrying CMY-2 could self-transfer between different strains alone, but rarely co-transferred with multiple genes [4], [27], [30]. The spread of bla CMY-2 was driven mainly by the IncA/C, IncI1, or IncK plasmids [31].
Co-localization of bla CMY-2 and floR on an IncA/C plasmid was detected commonly. A close relationship between the IncA/C plasmid and the multidrug resistance (MDR) of plasmid bearing isolates from human, animal, and environmental origins has been reported [9]. Considering the high occurrence of MDR genes (such as floR and qnr) in IncA/C plasmids and the co-transfer of these genes within the same plasmids in the present study, we suggest that there should be a stable association of these resistance genes with A/C plasmids. In addition, bla CMY-2 co-transferred with qnrS1 located on the IncK plasmid, with sizes of 160 and 200 kb. These results highlight the potential risk for co-selection of isolates carrying bla CMY-2 via the use of florfenicol or fluoroquinolones in the raising of food animals [32], [33]. Interestingly, the co-transfer of bla CMY-2 with qnrS1+floR located on the IncHI2 plasmid was first reported in a chicken isolate. The IncHI2 plasmid was most prevalent in Salmonella isolates of food animal origin, and could readily capture PMQR [34]. Our results suggest that the dissemination of IncHI2 plasmids carrying CMY-2, PMQR, and floR between E. coli and Salmonella might have occurred.
Although the occurrence of OqxAB was surprisingly high in China (39%) [35] compared with Denmark (1.8%), and Korea (0.4%) [36], [37], the encoded genes rarely co-transferred with blaCMY-2 on the same plasmid. Previous findings demonstrated that rmtB was the most prevalent 16S rRNA methylase gene in the Enterobacteriaceae isolates that produced ESBLs in China [23], [38]. Similarly, only the rmtB gene was detected in five strains in this study. However, in contrast with the previously described co-transfer of rmtB and ESBL-encoding genes, the co-transfer of rmtB with bla CMY-2 was not observed in the current study, as demonstrated by a conjugation experiment.
Plasmids and integrons can contain or capture a variety of resistance genes that are beneficial for survival of the bacterial host and help them adapt to changing environments [39], [40]. Integrons are genetic platforms involved in the spread of different previously captured gene cassettes that encode determinants of antimicrobial-resistance and represent a fundamental resource for bacterial evolution [41]. Integrons are divided into five classes based on integrase gene sequence; class 1 integrons are by far the most common in clinical isolates of Gram-negative bacteria [41]. In this study, we found five types of integrons encompassing eight different genes: aadA1, aadA2, aadA5, aadA22, dfrA1, dfrA17, sat1, and orfF. The most common integron profile (dfrA17-aadA5) was found in Salmonella and other Enterobacteriaceae, and recently in two Staphylococcus species isolated in China, suggesting the successful spread of this integron around the world and across bacterial phyla [40]. Different types of integrons contained by CMY-2 producers represented a diverse trend in strain evolution. DNA fingerprints also revealed that most CMY-2 producers were unlikely to be derived from a single E. coli clone. However, the clonal dissemination of bla CMY-2 between different farms at the same and different geographical locations was also found.
There was strong correlation between phylogenetic groups and STs in this study. One of the most common ST lineages, STC101, was isolated from pigs, and belonged to the avirulent phylogroup B1. Most of these harbored class 1 integron-containing drfA17+aadA5 cassette arrays. The global spread of bla CTX-M-9G ESBLs and bla NDM-1 was driven by ST101/B1 E. coli, and could be explained by the accumulation of a large number of virulence genes [42]. Similarly ST101C/B1 CMY-2, which is prevalent dominantly and consistently at pig farms from different geographical regions, could be explained by its ability for acquiring class 1 integrons with the most common cassettes arrays and antibiotic resistant genes. Capturing and acquiring these genes could help establish the dissemination of STC101/B1 CMY-2 isolates between pig farms. The other most common ST, STC10, belonged to the avirulent phylogroup A, and was distributed widely between pigs, chickens, and geese. The significant contribution of ST10C to the spread of resistance in humans was reported in Europe and Canada [43]. Interestingly, a chicken origin ST48 (belonging to ST10C) contained qnrS1+floR, as well a class 2 integron carrying a sat1+aadA1 cassette array. Compared with ST10, its SLV or DLV contained more antibiotic resistant genes, which helps the host bacteria adapt to their surroundings under antibiotic selective pressures. Three strains isolated from a diverse range of animal species corresponded to ST648, which belongs to virulent phylogroup D. It was worthy noting that two of these three strains contained class 1 integrons with different cassette arrays, which might allow the potential persistence between animals. E.coli strains of ST648/D clones were reported to cause most cases of ESBL-producing E. coli bacteremia in the Netherlands [44]. Additional concerns arise from this ST belonging to phylogroup D, due to its ability to produce New Delhi metallo (NDM)-type carbapenemases in hospitalized patients in Pakistan and the United Kingdom [45], [46]. In a recent study, ST648/D clones were the main vectors that allowed the spread of bla CMY-2 between dogs in the Republic of Korea [47].
This study demonstrated the presence of bla CMY-2 in broad host-range conjugative plasmids. To our knowledge, this represents the first comprehensive analysis of CMY-2 plasmids in E. coli isolated from food animals in China. bla CMY-2 was co-transferred with qnrS1 and/or floR, linked to diverse lineages of E. coli STCs (including 101, 10, and 155), and disseminated among different food producing animals in China. The acquisition of multiple antimicrobial resistant genes and integrons might have allowed CMY-2-positive isolates to persist in the environment and evolve under antibiotic selective pressure. Continued surveillance of CMY-2 in animal reservoirs is necessary to curb the spread of multidrug resistant pathogens from animals to humans.
Supporting Information
Primers used for the PCR amplification of antimicrobial resistance genes.
(DOC)
Primers used for the PCR amplification of genetic environment of bla CMY-2 gene.
(DOC)
Primers used for the PCR amplification of integrons.
(DOC)
MLST Primers used for the PCR amplification of E. coli .
(DOC)
The number of alleles and ST results for thirty-one CMY-2 producing strains.
(DOC)
Acknowledgments
The authors thank Prof. Qijing Zhang from Department of Veterinary Microbiology and Preventive Medicine, Iowa State University, Ames, IA, USA, for critically reviewing and revising the manuscript.
Funding Statement
This work was supported in part by the National Natural Science Foundation of China (U1201214) and 973 Program (2013CB127203). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Liebana E, Carattoli A, Coque TM, Hasman H, Magiorakos AP, et al. (2013) Public Health Risks of Enterobacterial Isolates Producing Extended-Spectrum β-Lactamases or AmpC β-Lactamases in Food and Food-Producing Animals: An EU Perspective of Epidemiology, Analytical Methods, Risk Factors, and Control Options. Clin Infect Dis 56: 1030–1037. [DOI] [PubMed] [Google Scholar]
- 2. Alvarez M, Tran JH, Chow N, Jacoby GA (2004) Epidemiology of conjugative plasmid-mediated AmpC beta-lactamases in the United States. Antimicrob Agents Chemother 48: 533–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mulvey MR, Bryce E, Boyd DA, Ofner-Agostini M, Land AM, et al. (2005) Molecular characterization of cefoxitin-resistant Escherichia coli from Canadian hospitals. Antimicrob Agents Chemother 49: 358–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li Y, Li Q, Du Y, Jiang X, Tang J, et al. (2008) Prevalence of plasmid-mediated AmpC beta-lactamases in a Chinese university hospital from 2003 to 2005: first report of CMY-2-Type AmpC beta-lactamase resistance in China. J Clin Microbiol 46: 1317–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Woodford N, Reddy S, Fagan EJ, Hill RL, Hopkins KL, et al. (2007) Wide geographic spread of diverse acquired AmpC beta-lactamases among Escherichia coli and Klebsiella spp. in the UK and Ireland. J Antimicrob Chemother 59: 102–105. [DOI] [PubMed] [Google Scholar]
- 6. Trott D (2013) β-lactam resistance in gram-negative pathogens isolated from animals. Curr Pharm Des 19: 239–249. [PubMed] [Google Scholar]
- 7. Pitout JD, Reisbig MD, Mulvey M, Chui L, Louie M, et al. (2003) Association between handling of pet treats and infection with Salmonella enterica serotype newport expressing the AmpC beta-lactamase, CMY-2. J Clin Microbiol 41: 4578–4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gupta A, Fontana J, Crowe C, Bolstorff B, Stout A, et al. (2003) Emergence of multidrug-resistant Salmonella enterica serotype Newport infections resistant to expanded-spectrum cephalosporins in the United States. J Infect Dis 188: 1707–1716. [DOI] [PubMed] [Google Scholar]
- 9. Mataseje LF, Baudry PJ, Zhanel GG, Morckc DW, Readd RR, et al. (2010) Comparison of CMY-2 plasmids isolated from human, animal, and environmental Escherichia coli and Salmonella spp. from Canada. Diagn Microbiol Infect Dis 67: 387–391. [DOI] [PubMed] [Google Scholar]
- 10. Mulvey MR, Susky E, McCracken M, Morck DW, Read RR (2009) Similar cefoxitin-resistance plasmids circulating in Escherichia coli from human and animal sources. Vet Microbiol 134: 279–287. [DOI] [PubMed] [Google Scholar]
- 11. Winokur PL, Vonstein DL, Hoffman LJ, Uhlenhopp EK, Doern GV (2001) Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob Agents Chemother 45: 2716–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pitout JD, Laupland KB (2008) Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 8: 159–166. [DOI] [PubMed] [Google Scholar]
- 13. Zhuo C, Su DH, Wei LH, Wu L, Zhang ZX, et al. (2009) CHINET 2007 surveillance of antimicrobial resistance in Escherichia. coli and Klebsiella spp . Chin J Infect Chemother 9: 185–191. [Google Scholar]
- 14. Liu JH, Wei SY, Ma JY, Zeng ZL, Lu DH, et al. (2007) Detection and characterisation of CTX-M and CMY-2-lactamases among Escherichia coli isolates from farm animals in Guangdong Province of China. J Antimicrob Agents 29: 576–581. [DOI] [PubMed] [Google Scholar]
- 15. Jiang HX, Lu DH, Chen ZL, Wang XM, Chen JR, et al. (2011) High prevalence and widespread distribution of multi-resistant Escherichia coli isolates in pigs and poultry in China. Vet J 187: 99–103. [DOI] [PubMed] [Google Scholar]
- 16. Li L, Jiang ZG, Xia LN, Shen JZ, Dai L, et al. (2010) Characterization of antimicrobial resistance and molecular determinants of beta-lactamase in Escherichia coli isolated from chickens in China during 1970–2007. Vet Microbiol 144: 505–510. [DOI] [PubMed] [Google Scholar]
- 17. Zheng H, Zeng Z, Chen S, Liu YH, Yao QF, et al. (2012) Prevalence and characterisation of CTX-M beta-lactamases amongst Escherichia coli isolates from healthy food animals in China. Int J Antimicrob Agents 39: 305–310. [DOI] [PubMed] [Google Scholar]
- 18. Smet A, Martel A, Persoons D, Dewulf J, Heyndrickx M, et al. (2008) Diversity of extended-spectrum β-lactamases and class C β-lactamases among cloacal Escherichia coli isolates in Belgian broiler farms. Antimicrob Agents Chemother 52: 1238–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute (2008) Performance standards for antimicrobial disk susceptibility tests for bacteria isolated from animals; approved standard. 3rd ed. Document M31-A3. Wayne, PA.
- 20.Clinical and Laboratory Standards Institute (2010) Performance standards for antimicrobial susceptibility testing; twentieth informational supplement. Document M100-S20. Wayne, PA.
- 21. Carattoli A, Lovari S, Franco A, Cordaro G, Di Matteo P, et al. (2005) Extended-spectrum beta-lactamases in Escherichia coli isolated from dogs and cats in Rome, Italy, from 2001 to 2003. Antimicrob Agents Chemother 49: 833–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barton BM, Harding GP, Zuccarelli AJ (1995) A general method for detecting and sizing large plasmids. Anal Biochem 226: 235–240. [DOI] [PubMed] [Google Scholar]
- 23. Clermont O, Bonacorsi S, Bingen E (2000) Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66: 4555–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li XZ, Mehrotra M, Ghimire S, Adewoye A (2007) beta-Lactam resistance and beta-lactamases in bacteria of animal origin. Vet Microbiol 121: 197–214. [DOI] [PubMed] [Google Scholar]
- 25. Xia LN, Tao XQ, Shen JZ, Dai L, Wang Y, et al. (2011) A survey of beta-lactamase and 16S rRNA methylase genes among fluoroquinolone-resistant Escherichia coli isolates and their horizontal transmission in Shandong, China. Foodborne Pathog Dis 8: 1241–1248. [DOI] [PubMed] [Google Scholar]
- 26. Hiroi M, Yamazaki F, Harada T, Takahashi N, Iida N, et al. (2012) Prevalence of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in food-producing animals. J Vet Med Sci 74: 189–195. [DOI] [PubMed] [Google Scholar]
- 27. Yan JJ, Hong CY, Ko WC, Chen YJ, Tsai SH, et al. (2004) Dissemination of bla CMY-2 among Escherichia coli isolates from food animals, retail ground meats, and humans in southern Taiwan. Antimicrob Agents Chemother 48: 1353–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jacoby GA (2009) AmpC β-lactamases. Clin Microbiol Rev 22: 161–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meunier D, Jouy E, Lazizzera C, Kobisch M, Cloeckaert A, et al. (2010) Plasmid-borne florfenicol and ceftiofur resistance encoded by the floR and bla CMY-2 genes in Escherichia coli isolates from diseased cattle in France. J Med Microbiol 59: 467–71. [DOI] [PubMed] [Google Scholar]
- 30. Chen S, Zhao S, White DG, Schroeder CM, Lu R, et al. (2004) Characterization of multiple-antimicrobial-resistant Salmonella serovars isolated from retail meats. Appl Environ Microbiol 70: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mata C, Miro E, Alvarado A, Garcillán-Barcia MP, Toleman M, et al. (2012) Plasmid typing and genetic context of AmpC beta-lactamases in Enterobacteriaceae lacking inducible chromosomal ampC genes: findings from a Spanish hospital 1999-2007. J Antimicrob Chemother 67: 115–122. [DOI] [PubMed] [Google Scholar]
- 32. Alcaine SD, Sukhnanand SS, Warnick LD, Su WL, McGann P, et al. (2005) Ceftiofur-resistant Salmonella strains isolated from dairy farms represent multiple widely distributed subtypes that evolved by independent horizontal gene transfer. Antimicrob Agents Chemother 49: 4061–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Egorova S, Timinouni M, Demartin M, Granier SA, Whichard JM, et al. (2008) Ceftriaxone-resistant salmonella enterica serotype Newport, France. Emerg Infect Dis 14: 954–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li L, Liao X, Yang Y, Sun J, Li L, et al. (2013) Spread of oqxAB in Salmonella enterica serotype Typhimurium predominantly by IncHI2 plasmids. J Antimicrob Chemother 68: 2263–2268. [DOI] [PubMed] [Google Scholar]
- 35. Zhao J, Chen Z, Chen S, Deng Y, Liu Y, et al. (2010) Prevalence and dissemination of oqxAB in Escherichia coli isolates from animals, farmworkers, and the environment. Antimicrob Agents Chemother 54: 4219–4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hansen LH, Sorensen SJ, Jorgensen HS, Jensen LB (2005) The prevalence of the OqxAB multidrug efflux pump amongst olaquindox-resistant Escherichia coli in pigs. Microb Drug Resist 11: 378–382. [DOI] [PubMed] [Google Scholar]
- 37. Park KS, Kim MH, Park TS, Nam YS, Lee HJ, et al. (2012) Prevalence of the plasmid-mediated quinolone resistance genes, aac(6′)-Ib-cr, qepA, and oqxAB in clinical isolates of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli and Klebsiella pneumoniae in Korea. Ann Clin Lab Sci 42: 191–197. [PubMed] [Google Scholar]
- 38. Deng Y, He L, Chen S, Zheng H, Zeng Z, et al. (2011) F33:A-:B- and F2:A-:B- plasmids mediate dissemination of rmtB-bla CTX-M-9 group genes and rmtB-qepA in Enterobacteriaceae isolates from pets in China. Antimicrob Agents Chemother 55: 4926–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carattoli A (2013) Plasmids and the spread of resistance. Int J Med Microbiol 303: 298–304. [DOI] [PubMed] [Google Scholar]
- 40. Wiesner M, Zaidi MB, Calva E, Fernández-Mora M, Calva JJ, et al. (2009) Association of virulence plasmid and antibiotic resistance determinants with chromosomal multilocus genotypes in Mexican Salmonella enterica serovar Typhimurium strains. BMC Microbiol 9: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mazel D (2006) integrons: agents of bacterial evolution. Nat Rev Microbiol 4: 608–620. [DOI] [PubMed] [Google Scholar]
- 42. Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, et al. (2011) Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J Antimicrob Chemother 66: 2002–2005. [DOI] [PubMed] [Google Scholar]
- 43. Izdebski R, Baraniak A, Fiett J, Adler A, Kazma M, et al. (2013) Clonal structure, extended-spectrum beta-lactamases, and acquired AmpC-type cephalosporinases of Escherichia coli populations colonizing patients in rehabilitation centers in four countries. Antimicrob Agents Chemother 57: 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van der Bij AK, Peirano G, Goessens WH, van der Vorm ER, van Westreenen M, et al. (2011) Clinical and molecular characteristics of extended-spectrum-beta-lactamase-producing Escherichia coli causing bacteremia in the Rotterdam Area, Netherlands. Antimicrob. Agents Chemother 55: 3576–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hornsey M, Phee L, Wareham DW (2011) A novel variant (NDM-5) of the New Delhi metallo-β-lactamase (NDM) in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother 55: 5952–5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, et al. (2011) Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J. Antimicrob. Chemother 66: 2002–2005. [DOI] [PubMed] [Google Scholar]
- 47. Tamang MD, Nam HM, Jang GC, Kim SR, Chae MH, et al. (2012) Molecular characterization of extended-spectrum-beta-lactamase-producing and plasmid-mediated AmpC beta-lactamase-producing Escherichia coli isolated from stray dogs in South Korea. Antimicrob Agents Chemother 56: 2705–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for the PCR amplification of antimicrobial resistance genes.
(DOC)
Primers used for the PCR amplification of genetic environment of bla CMY-2 gene.
(DOC)
Primers used for the PCR amplification of integrons.
(DOC)
MLST Primers used for the PCR amplification of E. coli .
(DOC)
The number of alleles and ST results for thirty-one CMY-2 producing strains.
(DOC)