Abstract
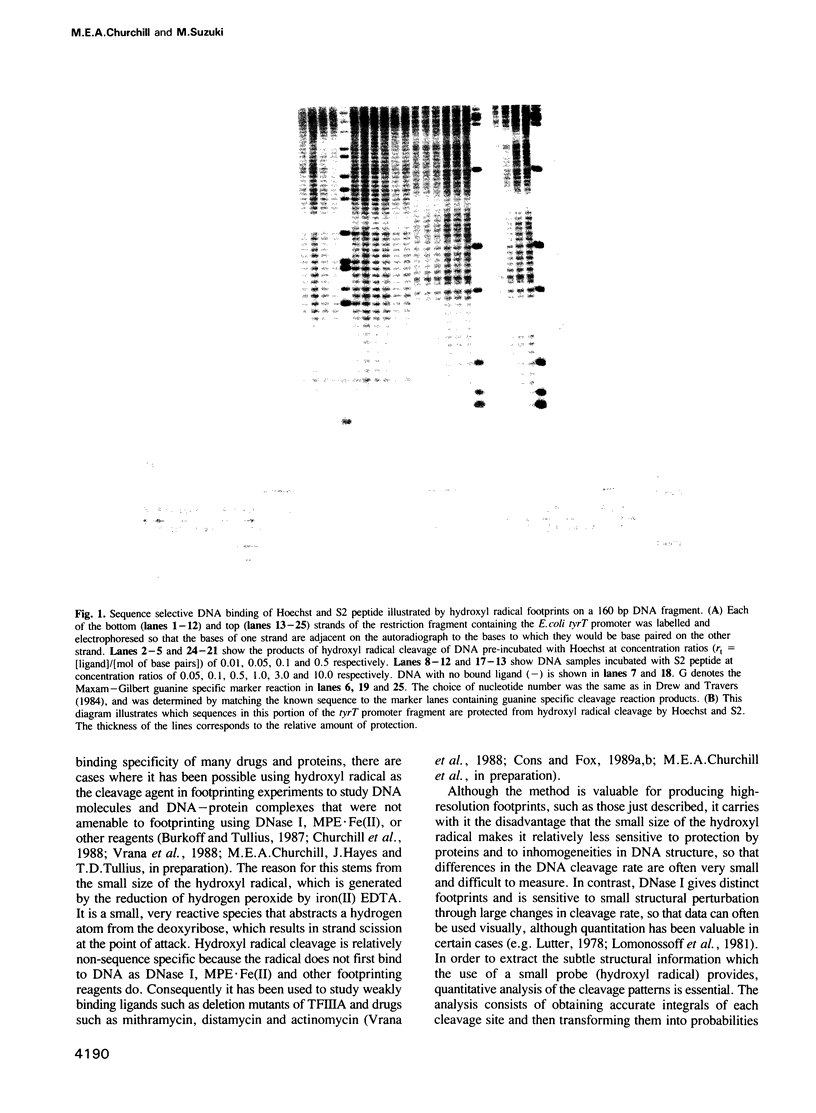
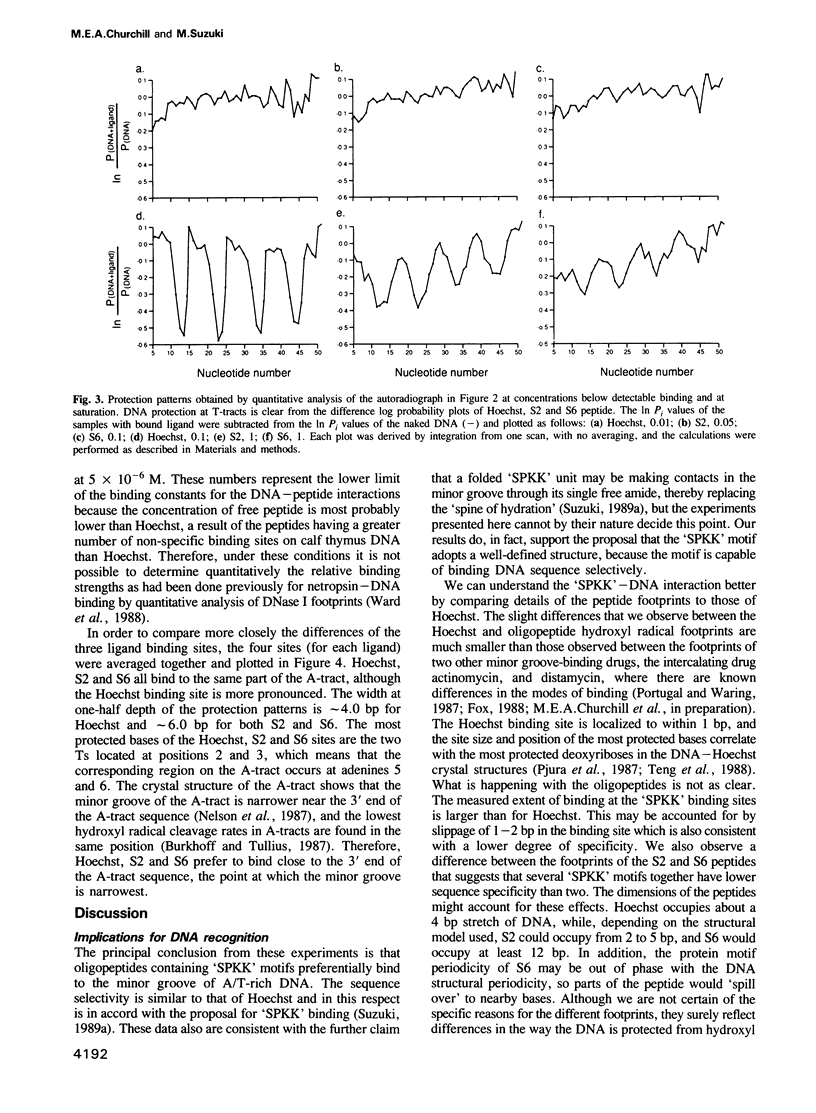
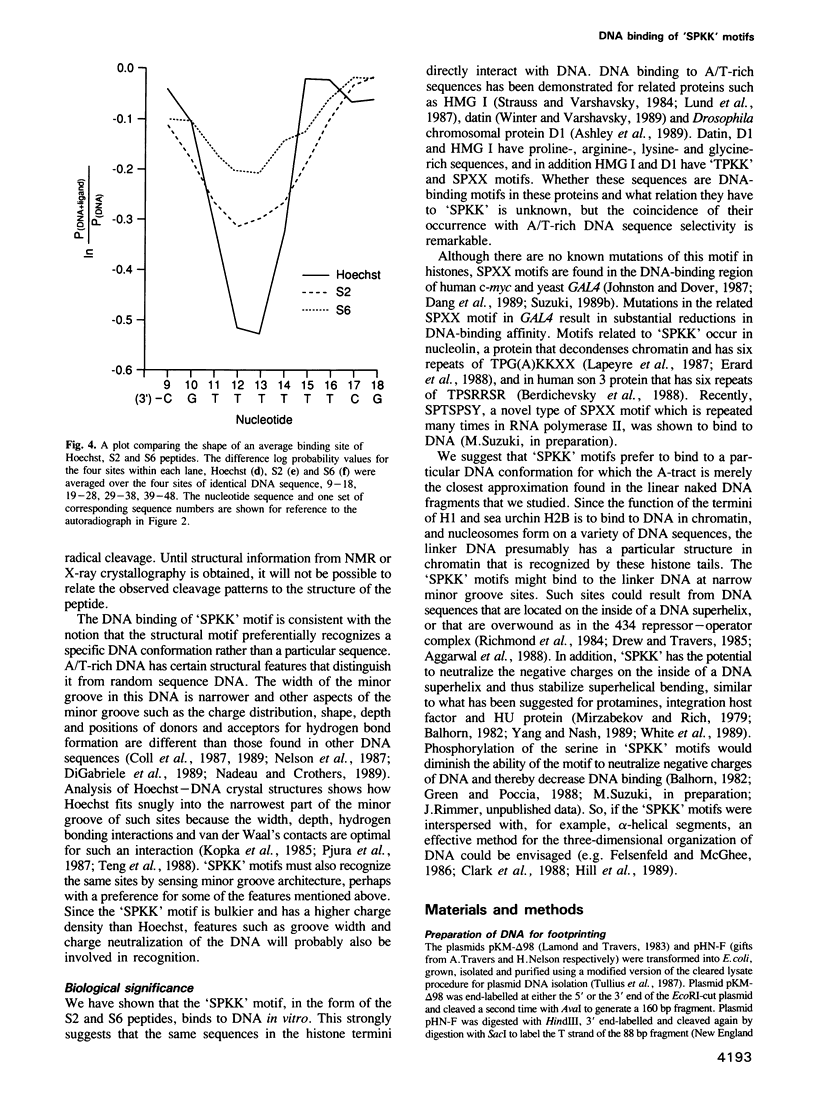
The termini of histone H1 and sea urchin spermatogenous H1 and H2B, which are essential for correct chromatin condensation, often contain repeats of the sequence SPK(R)K(R). A special type of beta-turn structural motif has been proposed for this sequence, and it has been shown that a segment of the sea urchin sperm H1 N terminus, which has six repeats of the motif (S6 peptide), binds to DNA and competes with the DNA binding drug Hoechst 33258. Here, we demonstrate by quantitative analysis of hydroxyl radical footprints that the synthetic oligopeptide, SPRKSPRK (S2), and the S6 peptide prefer to bind to the minor groove of DNA at the same A/T-rich sites. The locations of these binding sites are similar to Hoechst, but the sequence specificity of the oligopeptides is lower than that of Hoechst, and the detailed protection patterns differ slightly. We suggest that these small peptides and Hoechst recognize similar sequence-dependent features of the local architecture of DNA.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Ashley C. T., Pendleton C. G., Jennings W. W., Saxena A., Glover C. V. Isolation and sequencing of cDNA clones encoding Drosophila chromosomal protein D1. A repeating motif in proteins which recognize at DNA. J Biol Chem. 1989 May 15;264(14):8394–8401. [PubMed] [Google Scholar]
- Balhorn R. A model for the structure of chromatin in mammalian sperm. J Cell Biol. 1982 May;93(2):298–305. doi: 10.1083/jcb.93.2.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan R. G., Matthews B. W. The helix-turn-helix DNA binding motif. J Biol Chem. 1989 Feb 5;264(4):1903–1906. [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Churchill M. E., Tullius T. D., Kallenbach N. R., Seeman N. C. A Holliday recombination intermediate is twofold symmetric. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4653–4656. doi: 10.1073/pnas.85.13.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. J., Hill C. S., Martin S. R., Thomas J. O. Alpha-helix in the carboxy-terminal domains of histones H1 and H5. EMBO J. 1988 Jan;7(1):69–75. doi: 10.1002/j.1460-2075.1988.tb02784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole R. D. Microheterogeneity in H1 histones and its consequences. Int J Pept Protein Res. 1987 Oct;30(4):433–449. doi: 10.1111/j.1399-3011.1987.tb03352.x. [DOI] [PubMed] [Google Scholar]
- Coll M., Aymami J., van der Marel G. A., van Boom J. H., Rich A., Wang A. H. Molecular structure of the netropsin-d(CGCGATATCGCG) complex: DNA conformation in an alternating AT segment. Biochemistry. 1989 Jan 10;28(1):310–320. doi: 10.1021/bi00427a042. [DOI] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cons B. M., Fox K. R. High resolution hydroxyl radical footprinting of the binding of mithramycin and related antibiotics to DNA. Nucleic Acids Res. 1989 Jul 25;17(14):5447–5459. doi: 10.1093/nar/17.14.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cons B. M., Fox K. R. Interaction of mithramycin with metal ions and DNA. Biochem Biophys Res Commun. 1989 Apr 28;160(2):517–524. doi: 10.1016/0006-291x(89)92463-7. [DOI] [PubMed] [Google Scholar]
- Dang C. V., van Dam H., Buckmire M., Lee W. M. DNA-binding domain of human c-Myc produced in Escherichia coli. Mol Cell Biol. 1989 Jun;9(6):2477–2486. doi: 10.1128/mcb.9.6.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGabriele A. D., Sanderson M. R., Steitz T. A. Crystal lattice packing is important in determining the bend of a DNA dodecamer containing an adenine tract. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1816–1820. doi: 10.1073/pnas.86.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R. Structural specificities of five commonly used DNA nucleases. J Mol Biol. 1984 Jul 15;176(4):535–557. doi: 10.1016/0022-2836(84)90176-1. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Erard M. S., Belenguer P., Caizergues-Ferrer M., Pantaloni A., Amalric F. A major nucleolar protein, nucleolin, induces chromatin decondensation by binding to histone H1. Eur J Biochem. 1988 Aug 15;175(3):525–530. doi: 10.1111/j.1432-1033.1988.tb14224.x. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G., McGhee J. D. Structure of the 30 nm chromatin fiber. Cell. 1986 Feb 14;44(3):375–377. doi: 10.1016/0092-8674(86)90456-3. [DOI] [PubMed] [Google Scholar]
- Fox K. R. Footprinting studies on the interactions of nogalamycin, arugomycin, decilorubicin and viriplanin with DNA. Anticancer Drug Des. 1988 Dec;3(3):157–168. [PubMed] [Google Scholar]
- Green G. R., Poccia D. L. Interaction of sperm histone variants and linker DNA during spermiogenesis in the sea urchin. Biochemistry. 1988 Jan 26;27(2):619–625. doi: 10.1021/bi00402a019. [DOI] [PubMed] [Google Scholar]
- Green G. R., Poccia D. L. Phosphorylation of sea urchin sperm H1 and H2B histones precedes chromatin decondensation and H1 exchange during pronuclear formation. Dev Biol. 1985 Mar;108(1):235–245. doi: 10.1016/0012-1606(85)90026-0. [DOI] [PubMed] [Google Scholar]
- Harshman K. D., Dervan P. B. Molecular recognition of B-DNA by Hoechst 33258. Nucleic Acids Res. 1985 Jul 11;13(13):4825–4835. doi: 10.1093/nar/13.13.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. S., Martin S. R., Thomas J. O. A stable alpha-helical element in the carboxy-terminal domain of free and chromatin-bound histone H1 from sea urchin sperm. EMBO J. 1989 Sep;8(9):2591–2599. doi: 10.1002/j.1460-2075.1989.tb08398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Dover J. Mutations that inactivate a yeast transcriptional regulatory protein cluster in an evolutionarily conserved DNA binding domain. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2401–2405. doi: 10.1073/pnas.84.8.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight K. L., Bowie J. U., Vershon A. K., Kelley R. D., Sauer R. T. The Arc and Mnt repressors. A new class of sequence-specific DNA-binding protein. J Biol Chem. 1989 Mar 5;264(7):3639–3642. [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond A. I., Travers A. A. Requirement for an upstream element for optimal transcription of a bacterial tRNA gene. Nature. 1983 Sep 15;305(5931):248–250. doi: 10.1038/305248a0. [DOI] [PubMed] [Google Scholar]
- Lapeyre B., Bourbon H., Amalric F. Nucleolin, the major nucleolar protein of growing eukaryotic cells: an unusual protein structure revealed by the nucleotide sequence. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1472–1476. doi: 10.1073/pnas.84.6.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennard A. C., Thomas J. O. The arrangement of H5 molecules in extended and condensed chicken erythrocyte chromatin. EMBO J. 1985 Dec 16;4(13A):3455–3462. doi: 10.1002/j.1460-2075.1985.tb04104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonossoff G. P., Butler P. J., Klug A. Sequence-dependent variation in the conformation of DNA. J Mol Biol. 1981 Jul 15;149(4):745–760. doi: 10.1016/0022-2836(81)90356-9. [DOI] [PubMed] [Google Scholar]
- Lund T., Dahl K. H., Mørk E., Holtlund J., Laland S. G. The human chromosomal protein HMG I contains two identical palindrome amino acid sequences. Biochem Biophys Res Commun. 1987 Jul 31;146(2):725–730. doi: 10.1016/0006-291x(87)90589-4. [DOI] [PubMed] [Google Scholar]
- Lutter L. C. Kinetic analysis of deoxyribonuclease I cleavages in the nucleosome core: evidence for a DNA superhelix. J Mol Biol. 1978 Sep 15;124(2):391–420. doi: 10.1016/0022-2836(78)90306-6. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzabekov A. D., Rich A. Asymmetric lateral distribution of unshielded phosphate groups in nucleosomal DNA and its role in DNA bending. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1118–1121. doi: 10.1073/pnas.76.3.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Murray V., Martin R. F. Sequence specificity of 125I-labelled Hoechst 33258 damage in six closely related DNA sequences. J Mol Biol. 1988 Sep 5;203(1):63–73. doi: 10.1016/0022-2836(88)90091-5. [DOI] [PubMed] [Google Scholar]
- Nadeau J. G., Crothers D. M. Structural basis for DNA bending. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2622–2626. doi: 10.1073/pnas.86.8.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pjura P. E., Grzeskowiak K., Dickerson R. E. Binding of Hoechst 33258 to the minor groove of B-DNA. J Mol Biol. 1987 Sep 20;197(2):257–271. doi: 10.1016/0022-2836(87)90123-9. [DOI] [PubMed] [Google Scholar]
- Portugal J., Waring M. J. Hydroxyl radical footprinting of the sequence-selective binding of netropsin and distamycin to DNA. FEBS Lett. 1987 Dec 10;225(1-2):195–200. doi: 10.1016/0014-5793(87)81156-0. [DOI] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Strauss F., Varshavsky A. A protein binds to a satellite DNA repeat at three specific sites that would be brought into mutual proximity by DNA folding in the nucleosome. Cell. 1984 Jul;37(3):889–901. doi: 10.1016/0092-8674(84)90424-0. [DOI] [PubMed] [Google Scholar]
- Suck D., Lahm A., Oefner C. Structure refined to 2A of a nicked DNA octanucleotide complex with DNase I. Nature. 1988 Mar 31;332(6163):464–468. doi: 10.1038/332464a0. [DOI] [PubMed] [Google Scholar]
- Sulston J., Mallett F., Staden R., Durbin R., Horsnell T., Coulson A. Software for genome mapping by fingerprinting techniques. Comput Appl Biosci. 1988 Mar;4(1):125–132. doi: 10.1093/bioinformatics/4.1.125. [DOI] [PubMed] [Google Scholar]
- Suzuki M. SPKK, a new nucleic acid-binding unit of protein found in histone. EMBO J. 1989 Mar;8(3):797–804. doi: 10.1002/j.1460-2075.1989.tb03440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M. SPXX, a frequent sequence motif in gene regulatory proteins. J Mol Biol. 1989 May 5;207(1):61–84. doi: 10.1016/0022-2836(89)90441-5. [DOI] [PubMed] [Google Scholar]
- Teng M. K., Usman N., Frederick C. A., Wang A. H. The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG). Nucleic Acids Res. 1988 Mar 25;16(6):2671–2690. doi: 10.1093/nar/16.6.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A., Churchill M. E., Kam L. Hydroxyl radical footprinting: a high-resolution method for mapping protein-DNA contacts. Methods Enzymol. 1987;155:537–558. doi: 10.1016/0076-6879(87)55035-2. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnell W. G., Satchwell S. C., Travers A. A. A decapeptide motif for binding to the minor groove of DNA. A proposal. FEBS Lett. 1988 May 23;232(2):263–268. doi: 10.1016/0014-5793(88)80750-6. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward B., Rehfuss R., Goodisman J., Dabrowiak J. C. Determination of netropsin-DNA binding constants from footprinting data. Biochemistry. 1988 Feb 23;27(4):1198–1205. doi: 10.1021/bi00404a020. [DOI] [PubMed] [Google Scholar]
- Warrant R. W., Kim S. H. alpha-Helix-double helix interaction shown in the structure of a protamine-transfer RNA complex and a nucleoprotamine model. Nature. 1978 Jan 12;271(5641):130–135. doi: 10.1038/271130a0. [DOI] [PubMed] [Google Scholar]
- White S. W., Appelt K., Wilson K. S., Tanaka I. A protein structural motif that bends DNA. Proteins. 1989;5(4):281–288. doi: 10.1002/prot.340050405. [DOI] [PubMed] [Google Scholar]
- Winter E., Varshavsky A. A DNA binding protein that recognizes oligo(dA).oligo(dT) tracts. EMBO J. 1989 Jun;8(6):1867–1877. doi: 10.1002/j.1460-2075.1989.tb03583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. C., Nash H. A. The interaction of E. coli IHF protein with its specific binding sites. Cell. 1989 Jun 2;57(5):869–880. doi: 10.1016/0092-8674(89)90801-5. [DOI] [PubMed] [Google Scholar]




