Abstract
Circulating tumor DNA (ctDNA) represents a promising biomarker for noninvasive assessment of cancer burden, but existing methods have insufficient sensitivity or patient coverage for broad clinical applicability. Here we introduce CAncer Personalized Profiling by deep Sequencing (CAPP-Seq), an economical and ultrasensitive method for quantifying ctDNA. We implemented CAPP-Seq for non-small cell lung cancer (NSCLC) with a design covering multiple classes of somatic alterations that identified mutations in >95% of tumors. We detected ctDNA in 100% of stage II–IV and 50% of stage I NSCLC patients, with 96% specificity for mutant allele fractions down to ~0.02%. Levels of ctDNA significantly correlated with tumor volume, distinguished between residual disease and treatment-related imaging changes, and provided earlier response assessment than radiographic approaches. Finally, we explored biopsy-free tumor screening and genotyping with CAPP-Seq. We envision that CAPP-Seq could be routinely applied clinically to detect and monitor diverse malignancies, thus facilitating personalized cancer therapy.
Introduction
Analysis of ctDNA has the potential to revolutionize detection and monitoring of tumors. Noninvasive access to malignant DNA is particularly attractive for solid tumors, which cannot be repeatedly sampled without invasive procedures. In NSCLC, PCR-based assays have been used to detect recurrent point mutations in genes such as KRAS (kirsten rat sarcoma viral oncogene homolog) or EGFR (epidermal growth factor receptor) in plasma DNA1–4, but the majority of patients lack mutations in these genes. Recently, approaches employing massively parallel sequencing have been used to detect ctDNA5–12. However, the methods reported to date have been limited by modest sensitivity13, applicability to only a minority of patients, the need for patient-specific optimization, and/or cost. To overcome these limitations, we developed a novel strategy for analysis of ctDNA. Our method, called CAPP-Seq, combines optimized library preparation methods for low DNA input masses with a multi-phase bioinformatics approach to design a “selector” consisting of biotinylated DNA oligonucleotides that target recurrently mutated regions in the cancer of interest. To monitor ctDNA, the selector is applied to tumor DNA to identify a patient’s cancer-specific genetic aberrations and then directly to circulating DNA to quantify them (Fig. 1a). Here we demonstrate the technical performance and explore the clinical utility of CAPP-Seq in patients with early and advanced stage NSCLC.
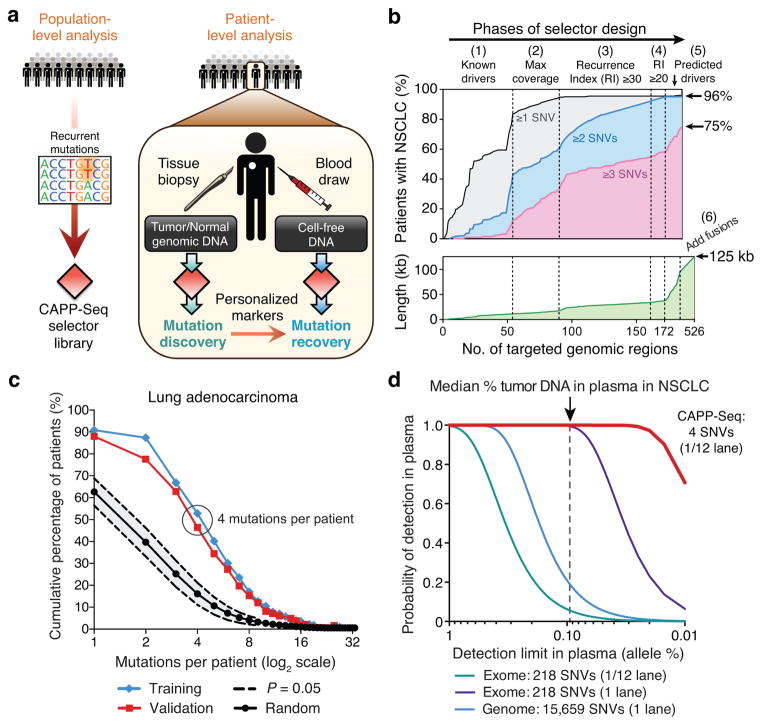
Figure 1. Development of CAncer Personalized Profiling by deep Sequencing (CAPP-Seq).
(a) Schematic depicting design of CAPP-Seq selectors and their application for assessing circulating tumor DNA. (b) Multi-phase design of the NSCLC selector. Phase 1: Genomic regions harboring known and suspected driver mutations in NSCLC are captured. Phases 2–4: Addition of exons containing recurrent SNVs using WES data from lung adenocarcinomas and squamous cell carcinomas from TCGA (n = 407). Regions were selected iteratively to maximize the number of mutations per tumor while minimizing selector size. Recurrence index = total unique patients with mutations covered per kb of exon. Phases 5,6: Exons of predicted NSCLC drivers15,16 and introns and exons harboring breakpoints in rearrangements involving ALK, ROS1, and RET were added. Bottom: increase of selector length during each design phase. (c) Analysis of the number of SNVs per lung adenocarcinoma covered by the NSCLC selector in the TCGA WES cohort (Training; n = 229) and an independent lung adenocarcinoma WES data set (Validation; n = 183)20. Results are compared to selectors randomly sampled from the exome (P < 1.0 × 10−6 for the difference between random selectors and the NSCLC selector). (d) Analytical modeling of CAPP-Seq, whole exome and whole genome sequencing for different detection limits of tumor circulating DNA in plasma. Calculations are based on the median number of mutations detected per NSCLC for CAPP-Seq (i.e., 4) and the reported number of mutations in NSCLC exomes and genomes21. Additional details are described in Methods. The vertical dotted line represents the median fraction of tumor-derived circulating DNA detected in plasma from patients in this study.
Results
Design of a CAPP-Seq selector for NSCLC
For the initial implementation of CAPP-Seq we focused on NSCLC, although our approach is generalizable to any cancer for which recurrent mutations have been identified. To design a selector for NSCLC (Fig. 1b, Supplementary Table 1, and Methods), we began by including exons covering recurrent mutations in potential driver genes from COSMIC14 and other sources15,16. Next, using whole exome sequencing (WES) data from 407 patients with NSCLC profiled by The Cancer Genome Atlas (TCGA), we applied an iterative algorithm to maximize the number of missense mutations per patient while minimizing selector size (Supplementary Fig. 1 and Supplementary Table 1).
Approximately 8% of NSCLCs harbor rearrangements involving the receptor tyrosine kinases, ALK (anaplastic lymphoma receptor tyrosine kinase), ROS1 (c-ros oncogene 1 tyrosine kinase) or the RET proto-oncogene17–19. To utilize the personalized nature and lower false detection rate inherent in the unique junctional sequences of structural rearrangements5,6, we included the introns and exons spanning recurrent fusion breakpoints in these genes in the final design phase (Fig. 1b). To detect fusions in tumor and plasma DNA, we developed a breakpoint-mapping algorithm (Methods). Application of this algorithm to next generation sequencing (NGS) data from two NSCLC cell lines known to harbor fusions with previously uncharacterized breakpoints22,23 readily identified the breakpoints at nucleotide resolution (Supplementary Fig. 2).
Collectively, the NSCLC selector design targets 521 exons and 13 introns from 139 recurrently mutated genes, in total covering ~125 kb (Fig. 1b). Within this small target (0.004% of the human genome), the selector identifies a median of 4 single nucleotide variants (SNVs) and covers 96% of patients with lung adenocarcinoma or squamous cell carcinoma. To validate the number of mutations covered per tumor, we examined the selector region in WES data from an independent cohort of 183 lung adenocarcinoma patients20. The selector covered 88% of patients with a median of 4 SNVs per patient, ~4-fold more than would be expected from random sampling of the exome (P < 1.0 × 10−6; Fig. 1c), thus validating our selector design algorithm.
Methodological optimization and performance assessment
We performed deep sequencing with the NSCLC selector to achieve ~10,000x coverage (pre-duplication removal) based on considerations of sequencing depth, median number of reporters, and ctDNA detection limit (Fig. 1d). We profiled a total of 90 samples, including two NSCLC cell lines, 17 primary tumor samples and matched peripheral blood leukocytes (PBLs), and 40 plasma samples from 18 human subjects, including five healthy adults and 13 patients with NSCLC (Supplementary Table 2). To assess and optimize selector performance, we first applied it to circulating DNA purified from healthy control plasma, observing efficient and uniform capture of genomic DNA (Supplementary Table 2). Sequenced plasma DNA fragments had a median length of ~170 bp (Fig. 2a), closely corresponding to the length of DNA contained within a chromatosome24. By optimizing library preparation from small quantities of plasma DNA, we increased recovery efficiency by >300% and decreased bias for libraries constructed from as little as 4 ng (Supplementary Fig. 3). Consequently, fluctuations in sequencing depth were minimal (Fig. 2b,c).
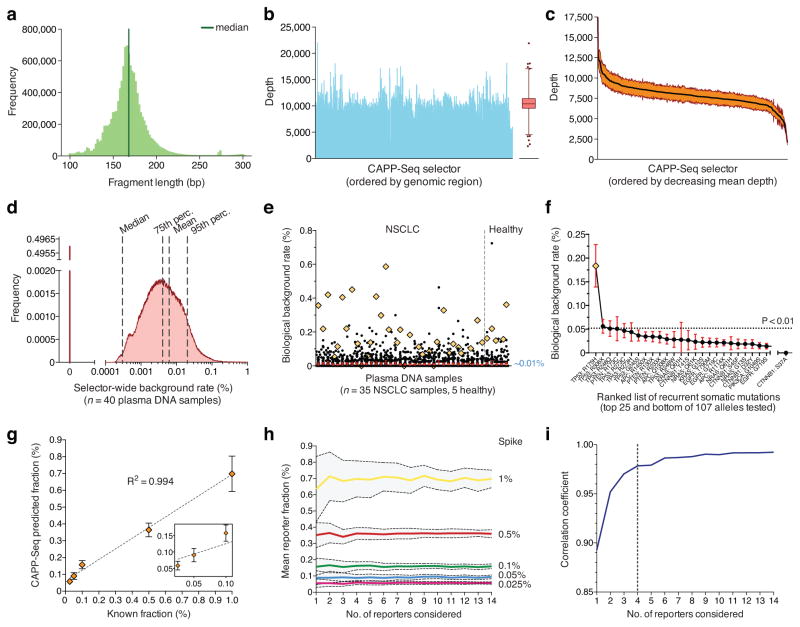
Figure 2. Analytical performance.
(a–c) Quality parameters from a representative CAPP-Seq analysis of plasma DNA, including length distribution of sequenced circulating DNA fragments (a), and depth of sequencing coverage across all genomic regions in the selector (b). (c) Variation in sequencing depth across plasma DNA samples from four patients. Orange envelope represents s.e.m. (d) Analysis of background rate for 40 plasma DNA samples collected from 13 patients with NSCLC and five healthy individuals (Supplementary Methods). (e) Analysis of biological background in d focusing on 107 recurrent somatic mutations from a previously reported SNaPshot panel25. Mutations found in a given patient’s tumor were excluded. The mean frequency over all subjects was ~0.01%. A single outlier mutation (TP53 R175H) is indicated by an orange diamond. (f) Individual mutations from e ranked by most to least recurrent, according to mean frequency across the 40 plasma DNA samples. The p-value threshold of 0.01 (horizontal line) corresponds to the 99th percentile of global selector background in d. (g) Dilution series analysis of expected versus observed frequencies of mutant alleles using CAPP-Seq. Dilution series were generated by spiking fragmented HCC78 DNA into control circulating DNA. (h) Analysis of the effect of the number of SNVs considered on the estimates of fractional abundance (95% confidence intervals shown in gray). (i) Analysis of the effect of the number of SNVs considered on the mean correlation coefficient between expected and observed cancer fractions (blue dashed line) using data from panel h. 95% confidence intervals are shown for e,f. Statistical variation for g is shown as s.e.m.
The detection limit of CAPP-Seq is affected by (i) the input number and recovery rate of circulating DNA molecules, (ii) sample cross-contamination, (iii) potential allelic bias in the capture reagent, and (iv) PCR or sequencing errors. We examined each of these elements in turn. First, by comparing the number of input DNA molecules per sample with estimates of library complexity (Supplementary Fig. 4a and Supplementary Methods), we calculated a circulating DNA molecule recovery rate of ≥49% (Supplementary Table 2). This was in agreement with molecule recovery yields calculated following PCR (Supplementary Fig. 4b). Second, by analyzing patient-specific homozygous SNPs across samples, we found cross-contamination of ~0.06% in multiplexed plasma DNA (Supplementary Fig. 4c and Supplementary Methods), prompting us to exclude any tumor-derived SNV from further analysis if found as a germline SNP in another profiled patient. Next, we evaluated the allelic skew in heterozygous germline SNPs within patient PBL samples and observed minimal bias toward capture of reference alleles (Supplementary Fig. 4d). Finally, we analyzed the distribution of non-reference alleles across the selector for the 40 plasma DNA samples, excluding tumor-derived SNVs and germline SNPs. We found mean and median background rates of 0.006% and 0.0003%, respectively (Fig. 2d), both considerably lower than previously reported NGS-based methods for ctDNA analysis8,10.
In addition to technical reasons, non-germline plasma DNA could be present in the absence of cancer due to contributions from pre-neoplastic cells from diverse tissues, and such “biological” background may impact sensitivity. We hypothesized that biological background, if present, would be particularly high for recurrently mutated positions in known cancer driver genes and therefore analyzed mutation rates of 107 cancer-associated SNVs25 in all 40 plasma samples, excluding somatic mutations found in a patient’s tumor. Though the median fractional abundance was comparable to the global selector background (~0%), the mean was marginally higher at ~0.01% (Fig. 2e). Strikingly, we detected one mutational hotspot (tumor suppressor TP53, R175H) at a median frequency of ~0.18% across all plasma DNA samples, including patients and healthy subjects (Fig. 2f). Since this TP53 mutant allele is observed significantly above global background (P < 0.01), we hypothesize that it reflects true biological clonal heterogeneity, and thus excluded it as a potential reporter. To address background more generally, we also normalized for allele-specific differences in background rate when assessing the significance of ctDNA detection (Supplementary Methods). As a result, we found that biological background is not a major factor for ctDNA quantitation at detection limits above ~0.01%.
Next, we empirically benchmarked the detection limit and linearity of CAPP-Seq (Fig. 2g and Supplementary Fig. 5a). We accurately detected defined inputs of NSCLC DNA at fractional abundances between 0.025% and 10% with high linearity (R2 ≥ 0.994). We observed only marginal improvements in error metrics above a threshold of 4 SNP reporters (Fig. 2h,i and Supplementary Fig. 5b,c), equivalent to the median number of SNVs per tumor identified by the selector. Moreover, the fractional abundance of fusion breakpoints, indels (insertions and deletions), and CNVs (copy number variants) correlated highly with expected concentrations (R2 ≥ 0.97; Supplementary Fig. 5d).
Somatic mutation detection and tumor burden quantitation
We next applied CAPP-Seq to the discovery of somatic mutations in tumors collected from 17 patients with NSCLC (Table 1 and Supplementary Table 3), including formalin fixed surgical resection or needle biopsy specimens and malignant pleural fluid. At a mean sequencing depth of ~5,000x (pre-duplicate removal) in tumor and paired germline samples (Supplementary Table 2), we detected 100% of previously identified SNVs and fusions and discovered many additional somatic variants (Table 1 and Supplementary Table 3). Moreover, we characterized breakpoints at base-pair resolution and identified partner genes for each of eight known fusions involving ALK or ROS1 (Supplementary Fig. 2). Tumors containing fusions were almost exclusively from never smokers and contained fewer SNVs than those lacking fusions, as expected21 (Supplementary Fig. 2). Excluding patients with fusions, we identified a median of 6 SNVs (3 missense) per patient (Table 1), in line with our selector design-stage predictions (Fig. 1b,c).
Table 1. Patient characteristics and pre-treatment CAPP-Seq monitoring results.
ND, mutant DNA was not detected above background (see Methods). NA, tumor volume could not be reliably assessed. Dashes, plasma sample not available. Smoking history, ≥20 pack years (Heavy), >0 and <20 pack years (Light). Additional details are provided in Supplementary Table 3 and Supplementary Table 4.
| Case | Age | Sex | Histology | Stage | TNM | Smoking history | No. of SNVs (non- silent) | Indels | Fusion
|
Pre-treatment
|
|||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALK/ROS1 | Partner | ctDNA (%) | ctDNA (pg mL−1) | Tumor (cc) | |||||||||
| P12 | 86 | F | SCC | IA | T1bN0M0 | Heavy | 6 (3) | 1 | ND | ND | 5.5 | ||
|
| |||||||||||||
| P1 | 66 | M | Adeno | IB | T2aN0M0 | Heavy | 12 (3) | 4 | 0.025 | 1.9 | 23.1 | ||
|
| |||||||||||||
| P16 | 82 | F | Adeno | IB | T2aN0M0 | Heavy | 26 (5) | 2 | 0.019 | 2.5 | 22.5 | ||
|
| |||||||||||||
| P17 | 85 | F | Adeno | IB | T2aN0M0 | Heavy | 2 (2) | 0 | ND | ND | 10.2 | ||
|
| |||||||||||||
| P13 | 90 | F | SCC | IIB | T3N0M0 | Heavy | 5 (4) | 0 | 1.78 | 269.8 | 339.3 | ||
|
| |||||||||||||
| P2 | 61 | M | Large Cell | IIIA | T3N1M0 | Heavy | 12 (3) | 1 | 0.896 | 64.7 | 23.1 | ||
|
| |||||||||||||
| P3 | 67 | F | Adeno | IIIB | T1bN3M0 | Light | 1 (1) | 0 | 0.095 | 16.2 | 7.9 | ||
|
| |||||||||||||
| P14 | 55 | M | Adeno | IIIB | T1aN3M0 | Heavy | 8 (5) | 0 | 0.05 | 10.2 | 5.2 | ||
|
| |||||||||||||
| P15 | 41 | M | Adeno | IIIB | T3N3M0 | Light | 25 (10) | 1 | 0.58 | 108.1 | 121.8 | ||
|
| |||||||||||||
| P4 | 47 | F | Adeno | IV | T2aN2M1b | Heavy | 3 (2) | 0 | 0.039 | 2.1 | 12.4 | ||
|
| |||||||||||||
| P5 | 49 | F | Adeno | IV | T1bN0M1a | None | 4 (3) | 0 | 3.2 | 143.8 | 82.1 | ||
|
| |||||||||||||
| P6 | 54 | M | Adeno | IV | T3N2M1b | None | 3 (2) | 0 | ALK | KIF5B | 1.0 | 350.2 | NA |
|
| |||||||||||||
| P9 | 49 | M | Adeno | IV | T4N3M1a | None | 0 | 0 | ALK | EML4 | 0.04 | 3.8 | 66.2 |
| ROS1 | MKX, FYN | ||||||||||||
|
| |||||||||||||
| P10 | 35 | F | Adeno | IIIA | T4N0M0 | None | 0 | 0 | ROS1 | SLC34A2 | – | – | – |
|
| |||||||||||||
| P11 | 38 | F | Adeno | IIIA | T3N2M0 | None | 2 (1) | 0 | ROS1 | CD74 | – | – | – |
|
| |||||||||||||
| P7 | 50 | M | Adeno | IV | T1aN2M1b | Light | 0 | 0 | ALK | EML4 | – | – | – |
|
| |||||||||||||
| P8 | 48 | F | Adeno | IV | T4N0M1b | None | 1 (0) | 0 | ALK | EML4 | – | – | – |
Next, we assessed the sensitivity and specificity of CAPP-Seq for disease monitoring and minimal residual disease detection using plasma samples from five healthy controls and 35 samples collected from 13 patients with NSCLC (Table 1 and Supplementary Table 4). We integrated information content across multiple instances and classes of somatic mutations into a ctDNA detection index. This index is analogous to a false positive rate and is based on a decision tree in which fusion breakpoints take precedence due to their nonexistent background and in which p-values from multiple reporter types are integrated (Methods). Applying this approach in an ROC analysis, CAPP-Seq achieved an area under the curve (AUC) of 0.95, with maximal sensitivity and specificity of 85% and 96%, respectively, for all plasma DNA samples from pretreated patients and healthy controls. Sensitivity among stage I tumors was 50% and among stage II–IV patients was 100% with a specificity of 96% (Fig. 3a,b). Moreover, when considering both pre and post-treatment samples, CAPP–Seq exhibited robust performance, with AUC values of 0.89 for all stages and 0.91 for stages II–IV (P < 0.0001; Supplementary Fig. 6). Furthermore, by adjusting the ctDNA detection index, we could increase specificity up to 98% while still capturing 2/3 of all cancer-positive samples and 3/4 of stages II–IV cancer-positive samples (Supplementary Fig. 6). Thus, CAPP-Seq can achieve robust assessment of tumor burden and can be tuned to deliver a desired sensitivity and specificity.
Figure 3. Sensitivity and specificity analysis.
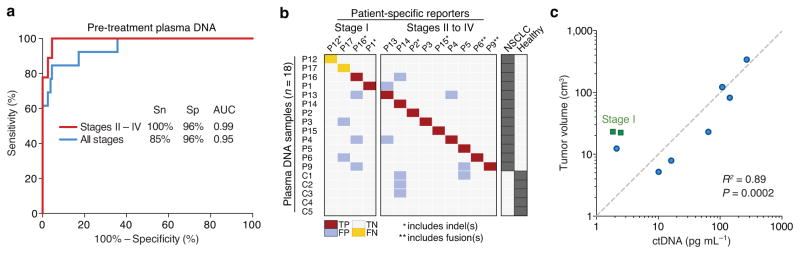
(a) Receiver Operating Characteristic (ROC) analysis of plasma DNA samples from pre-treatment samples and healthy controls, divided into all stages (n = 13 patients) and stages II–IV (n = 9 patients). Area Under the Curve (AUC) values are significant at P < 0.0001. Sn, sensitivity; Sp, specificity. (b) Raw data related to a. TP, true positive; FP, false positive; TN, true negative; FN, false negative. (c) Concordance between tumor volume, measured by CT or PET/CT, and pg mL−1 of ctDNA from pretreatment samples (n = 9), measured by CAPP-Seq. Patients P6 and P9 were excluded due to inability to accurately assess tumor volume and differences related to the capture of fusions, respectively (see Supplementary Methods). Of note, linear regression was performed in non-log space; the log-log axes and dashed diagonal line are for display purposes only.
Monitoring of NSCLC tumor burden in plasma samples
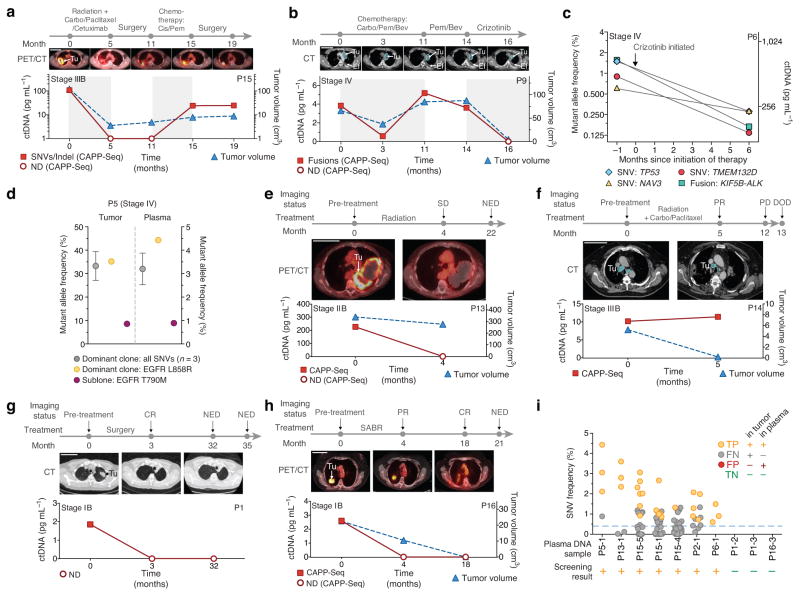
We next asked whether significantly detectable levels of ctDNA correlate with radiographically measured tumor volumes and clinical responses to therapy. Fractions of ctDNA detected in plasma by SNV and/or indel reporters ranged from ~0.02% to 3.2% (Table 1), with a median of ~0.1% in pre-treatment samples. Absolute levels of ctDNA in pre-treatment plasma were significantly correlated with tumor volume as measured by computed tomography (CT) and positron emission tomography (PET) imaging (R2 = 0.89, P = 0.0002; Fig. 3c).
To determine whether ctDNA concentrations reflect disease burden in longitudinal samples, we analyzed plasma DNA from three patients with advanced NSCLC undergoing distinct therapies (Fig. 4a–c). As in pre-treatment samples, ctDNA levels were highly correlated with tumor volumes during therapy (R2 = 0.95 for P15; R2 = 0.85 for P9). This behavior was observed whether the mutation type measured was a collection of SNVs and an indel (P15, Fig. 4a), multiple fusions (P9, Fig. 4b), or SNVs and a fusion (P6, Fig. 4c). Of note, in one patient (P9) we identified both a classic EML4-ALK fusion and two previously unreported fusions involving ROS1: FYN-ROS1 and ROS1-MKX (Supplementary Fig. 2). All fusions were confirmed by qPCR amplification of genomic DNA and were independently recovered in plasma samples (Supplementary Table 4). To the best of our knowledge this is the first observation of ROS1 and ALK fusions in the same individual with NSCLC.
Figure 4. Noninvasive detection and monitoring of circulating tumor DNA.
(a–h) Disease monitoring using CAPP-Seq. (a,b) Disease burden changes in response to treatment in a patient with stage III NSCLC using SNVs and an indel (a), and a patient with stage IV NSCLC using three rearrangement breakpoints (b). (c) Concordance between different reporters (SNVs and a fusion) in a patient with stage IV NSCLC. (d) Detection of a subclonal EGFR T790M resistance mutation in a patient with stage IV NSCLC. The fractional abundance of the dominant clone and T790M-containing clone are shown in the primary tumor (left) and plasma samples (right). (e,f) CAPP-Seq results from post-treatment plasma DNA samples are predictive of clinical outcomes in a patient with stage IIB NSCLC (e) and a patient with stage IIIB NSCLC (f). (g,h) Monitoring of tumor burden following complete tumor resection (g) and Stereotactic Ablative Radiotherapy (SABR) (h) for two patients with stage IB NSCLC. (i) Exploratory analysis of the potential application of CAPP-Seq for biopsy-free tumor genotyping or cancer screening. All plasma DNA samples from patients in Table 1 were examined for the presence of mutant allele outliers without knowledge of the primary tumor mutations (see Supplementary Methods); samples with detectable mutations are shown, along with three samples assumed to be cancer-negative (P1-2, P1-3 and P16-3; Supplementary Methods). The lowest fraction of ctDNA among positive samples was ~0.4% (dashed horizontal line). Error bars in d represent s.e.m. (a,b,e–h) Scale bars, 10 cm. Tu, tumor; Ef, pleural effusion; SD, stable disease; PD, progressive disease; PR, partial response; CR, complete response; DOD, dead of disease.
We designed the NSCLC CAPP-Seq selector to detect multiple SNVs per tumor. In one patient (P5), this design allowed us to identify a dominant clone with an activating EGFR mutation as well as an erlotinib-resistant subclone with a “gatekeeper” EGFR T790M mutation26. The ratio between clones was identical in a tumor biopsy and simultaneously sampled plasma (Fig. 4d), demonstrating that our method has potential for detecting and quantifying clinically relevant subclones.
Patients with stages II–III NSCLC undergoing definitive radiotherapy often have surveillance CT or PET/CT scans that are difficult to interpret due to radiation-induced inflammatory and fibrotic changes in the lung and surrounding tissues. For patient P13, who was treated with radiotherapy for stage IIB NSCLC, follow-up imaging showed a large mass that was felt to represent residual disease. However, ctDNA at the same time point was undetectable (Fig. 4e) and the patient remained disease free 22 months later, supporting the ctDNA result. Another patient (P14) was treated with chemoradiotherapy for stage IIIB NSCLC and follow-up imaging revealed a near complete response (Fig. 4f). However, the ctDNA concentration slightly increased following therapy, suggesting progression of occult microscopic disease. Indeed, clinical progression was detected 7 months later and the patient ultimately succumbed to NSCLC. These data highlight the promise of ctDNA analysis for identifying patients with residual disease after therapy.
We next asked whether the low detection limit of CAPP-Seq would allow monitoring in early stage NSCLC. Patients P1 (Fig. 4g) and P16 (Fig. 4h) underwent surgery and stereotactic ablative radiotherapy (SABR), respectively, for stage IB NSCLC. We detected ctDNA in pre-treatment plasma of P1 but not at 3 or 32 months following surgery, suggesting this patient was free of disease and likely cured. For patient P16, the initial surveillance PET-CT scan following SABR showed a residual mass that was interpreted as representing either residual tumor or post-radiotherapy inflammation. We detected no evidence of residual disease by ctDNA, supporting the latter, and the patient remained free of disease at last follow-up 21 months after therapy. Taken together, these results demonstrate the potential utility of CAPP-Seq for measuring tumor burden in early and advanced stage NSCLC and for monitoring ctDNA during distinct types of therapy.
Biopsy-free cancer screening and tumor genotyping
Finally, we explored whether CAPP-Seq analysis of ctDNA could potentially be used for cancer screening and biopsy-free tumor genotyping. As proof-of-principle, we blinded ourselves to the mutations present in each patient’s tumor and applied a novel statistical method to test for the presence of cancer DNA in each plasma sample in our cohort (Supplementary Fig. 7). By implementing our cancer screening method for high specificity, we correctly classified 100% of patient plasma samples with ctDNA above fractional abundances of 0.4% with a false positive rate of 0% (Fig. 4i and Supplementary Methods). CAPP-Seq could therefore potentially improve upon the low positive predictive value of low-dose CT screening in patients at high risk of developing NSCLC29.
Separately, when we specifically examined the ability to non-invasively detect actionable mutations in EGFR and KRAS25, we correctly identified 100% of mutations at allelic fractions greater than 0.1% with 99% specificity. CAPP-Seq may therefore have utility for biopsy-free tumor genotyping in locally advanced or metastatic patients. However, methodological improvements will be required to detect and genotype stage I tumors without prior knowledge of tumor genotype.
Discussion
In this study, we present CAPP-Seq as a new method for ctDNA quantitation. Key features include high sensitivity and specificity, coverage of nearly all patients with NSCLC, lack of patient-specific optimization, and low cost. By incorporating optimized library construction and bioinformatics methods, CAPP-Seq achieves the lowest background error rate and lowest detection limit of any NGS-based method used for ctDNA analysis to date. Our approach also reduces the potential impact of stochastic noise and biological variability (e.g., mutations near the detection limit or subclonal tumor evolution) on tumor burden quantitation by integrating information content across multiple instances and classes of somatic mutations. These features facilitated the detection of minimal residual disease, and the first report of ctDNA quantitation from stage I NSCLC tumors using deep sequencing. Although we focused on NSCLC, our method could be applied to any malignancy for which recurrent mutation data are available.
In many patients, levels of ctDNA are considerably lower than the detection thresholds of previously described sequencing-based methods13. For example, pre-treatment ctDNA concentration is <0.5% in the majority of patients with lung and colorectal carcinomas1,30,31. Following therapy, ctDNA concentrations typically drop, thus requiring even lower detection thresholds. Previously published methods employing amplicon8,10,11, whole exome12, or whole genome9,32,33,24 sequencing would not be sensitive enough to detect ctDNA in most patients with NSCLC, even at 10-fold or greater sequencing costs (Fig. 1d and Supplementary Fig. 8).
To further expand the potential clinical applications of ctDNA quantitation, additional gains in the detection threshold are desirable. Potential approaches include using barcoding strategies that suppress PCR errors resulting from library preparation34,35 and increasing the amount of plasma used for ctDNA analysis above the average of ~1.5mL used in our study. A second limitation of CAPP-Seq is the potential for inefficient capture of fusions, which could lead to underestimates of tumor burden (e.g., P9; Supplementary Methods). However, this bias can be analytically addressed when other reporter types are present (e.g., P6; Supplementary Table 4). Finally, while we found that CAPP-Seq could quantitate CNVs, our current selector design did not prioritize these types of aberrations. We anticipate that adding coverage for certain CNVs will prove useful for monitoring various types of cancers.
In summary, targeted hybrid capture and high-throughput sequencing of plasma DNA allows for highly sensitive and non-invasive detection of ctDNA in the vast majority of patients with NSCLC at low cost. CAPP-Seq could therefore be routinely applied clinically and has the potential for accelerating the personalized detection, therapy, and monitoring of cancer. We anticipate that CAPP-Seq will prove valuable in a variety of clinical settings, including the assessment of cancer DNA in alternative biological fluids and specimens with low cancer cell content.
Online Methods
Patient selection
Between April 2010 and June 2012, patients undergoing treatment for newly diagnosed or recurrent NSCLC were enrolled in a study approved by the Stanford University Institutional Review Board and provided informed consent. Enrolled patients had not received blood transfusions within 3 months of blood collection. Patient characteristics are in Supplementary Table 3. All treatments and radiographic examinations were performed as part of standard clinical care. Volumetric measurements of tumor burden were based on visible tumor on CT and calculated according to the ellipsoid formula: (length/2) × (width^2).
Sample collection and processing
Peripheral blood from patients was collected in EDTA Vacutainer tubes (BD). Blood samples were processed within 3 h of collection. Plasma was separated by centrifugation at 2,500 × g for 10 min, transferred to microcentrifuge tubes, and centrifuged at 16,000 × g for 10 min to remove cell debris. The cell pellet from the initial spin was used for isolation of germline genomic DNA from PBLs (peripheral blood leukocytes) with the DNeasy Blood & Tissue Kit (Qiagen). Matched tumor DNA was isolated from FFPE specimens or from the cell pellet of pleural effusions. Genomic DNA was quantified by Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen).
Cell-free DNA purification and quantification
Circulating DNA was isolated from 1–5 mL plasma with the QIAamp Circulating Nucleic Acid Kit (Qiagen). The concentration of purified plasma DNA was determined by quantitative PCR (qPCR) using an 81 bp amplicon on chromosome 124 and a dilution series of intact male human genomic DNA (Promega) as a standard curve. Power SYBR Green was used for qPCR on a HT7900 Real Time PCR machine (Applied Biosystems), using standard PCR thermal cycling parameters.
NGS library construction
Indexed Illumina NGS libraries were prepared from plasma DNA and shorn tumor, germline, and cell line genomic DNA. For patient plasma DNA, 7–32 ng DNA were used for library construction without additional fragmentation. For tumor, germline, and cell line genomic DNA, 69–1000 ng DNA was sheared prior to library construction with a Covaris S2 instrument using the recommended settings for 200 bp fragments. See Supplementary Table 2 for details.
The NGS libraries were constructed using the KAPA Library Preparation Kit (Kapa Biosystems) employing a DNA Polymerase possessing strong 3′-5′ exonuclease (or proofreading) activity and displaying the lowest published error rate (i.e. highest fidelity) of all commercially available B-family DNA polymerases36,37. The manufacturer’s protocol was modified to incorporate with-bead enzymatic and cleanup steps using Agencourt AMPure XP beads (Beckman-Coulter) 38. Ligation was performed for 16 h at 16 °C using 100-fold molar excess of indexed Illumina TruSeq adapters. Single-step size selection was performed by adding 40 μL (0.8X) of PEG buffer to enrich for ligated DNA fragments. The ligated fragments were then amplified using 500 nM Illumina backbone oligonucleotides and 4–9 PCR cycles, depending on input DNA mass. Library purity and concentration was assessed by spectrophotometer (NanoDrop 2000) and qPCR (KAPA Biosystems), respectively. Fragment length was determined on a 2100 Bioanalyzer using the DNA 1000 Kit (Agilent).
Library design for hybrid selection
Hybrid selection was performed with a custom SeqCap EZ Choice Library (Roche NimbleGen). This library was designed through the NimbleDesign portal (v1.2.R1) using genome build hg19 NCBI Build 37.1/GRCh37 and with Maximum Close Matches set to 1. Input genomic regions were selected according to the most frequently mutated genes and exons in NSCLC. These regions were identified from the COSMIC database, TCGA, and other published sources as described in the Supplementary Methods. Final selector coordinates are provided in Supplementary Table 1.
Hybrid selection and NGS
NimbleGen SeqCap EZ Choice was used according to the manufacturer’s protocol with modifications. Between 9 and 12 indexed Illumina libraries were included in a single capture hybridization. Following hybrid selection, the captured DNA fragments were amplified with 12 to 14 cycles of PCR using 1X KAPA HiFi Hot Start Ready Mix and 2 μM Illumina backbone oligonucleotides in 4 to 6 separate 50 μL reactions. The reactions were then pooled and processed with the QIAquick PCR Purification Kit (Qiagen). Multiplexed libraries were sequenced using 2 × 100 bp paired-end runs on an Illumina HiSeq 2000.
Mapping and quality control
Paired-end reads were mapped to the hg19 reference genome with BWA 0.6.2 (default parameters)39, and sorted and indexed with SAMtools40. QC was assessed using a custom Perl script to collect a variety of statistics, including mapping characteristics, read quality, and selector on-target rate (i.e., number of unique reads that intersect the selector space divided by all aligned reads), generated respectively by SAMtools flagstat, FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and BEDTools coverageBed41. Plots of fragment length distribution and sequence depth and coverage were automatically generated for visual QC assessment. To mitigate the impact of sequencing errors, analyses not involving fusions were restricted to properly paired reads, and only bases with Phred quality scores ≥30 (≤0.1% probability of a sequencing error) were further analyzed.
Detection thresholds
Two dilution series were performed to assess the linearity and accuracy of CAPP-Seq for quantitating ctDNA. In one experiment, shorn genomic DNA from a NSCLC cell line (HCC78) was spiked into circulating DNA from a healthy individual, while in a second experiment, shorn genomic DNA from one NSCLC cell line (NCI-H3122) was spiked into shorn genomic DNA from a second NSCLC line (HCC78). A total of 32 ng DNA was used for library construction. Following mapping and quality control, homozygous reporters were identified as alleles unique to each sample with at least 20x sequencing depth and an allelic fraction >80%. Fourteen such reporters were identified between HCC78 genomic DNA and plasma DNA (Fig. 2g,h), whereas 24 reporters were found between NCI-H3122 and HCC78 genomic DNA (Supplementary Fig. 5).
Bioinformatics pipeline
Details of bioinformatics methods are supplied in the Supplementary Methods. Briefly, for detection of SNVs and indels, we employed VarScan 242 with strict postprocessing filters to improve variant call confidence, and for fusion identification and breakpoint characterization we used a novel algorithm, called FACTERA (Supplementary Methods). To quantify tumor burden in plasma DNA, allele frequencies of reporter SNVs and indels were assessed using the output of SAMtools mpileup40, and fusions, if detected, were enumerated with FACTERA.
Statistical analyses
The NSCLC selector was validated in silico using an independent cohort of lung adenocarcinomas20 (Fig. 1c). To assess statistical significance, we analyzed the same cohort using 10,000 random selectors sampled from the exome, each with an identical size distribution to the CAPP-Seq NSCLC selector. The performance of random selectors had a normal distribution, and p-values were calculated accordingly. Of note, all identified somatic lesions were considered in this analysis.
Related to Fig. 1d, the probability P of recovering at least two reads of a single mutant allele in plasma for a given depth and detection limit was modeled by a binomial distribution. Given P, the probability of detecting all identified tumor mutations in plasma (e.g., median of 4 for CAPP-Seq) was modeled by a geometric distribution. Estimates are based on 250 million 100 bp reads per lane (e.g., using an Illumina HiSeq 2000 platform). Moreover, an on-target rate of 60% was assumed for CAPP-Seq and WES.
To evaluate the impact of reporter number on tumor burden estimates, we performed Monte Carlo sampling (1,000x), varying the number of reporters available {1,2,…,max n} in two spiking experiments (Fig. 2g–i and Supplemental Fig. 4).
To assess the significance of tumor burden estimates in plasma DNA using SNVs, we compared patient-specific SNV frequencies to the null distribution of selector-wide background alleles. Indels were analyzed separately using mutation-specific background rates and Z statistics. Fusion breakpoints were considered significant when present with >0 read support due to their ultra-low false detection rate.
For each patient, we calculated a ctDNA detection index (akin to a false positive rate) based on p-value integration from his or her array of reporters (Table 1 and Supplementary Table 4). Specifically, for cases where only a single reporter type was present in a patient’s tumor, the corresponding p-value was used. If SNV and indel reporters were detected, and if each independently had a p-value <0.1, we combined their respective p-values using Fisher’s method43. Otherwise, given the prioritization of SNVs in the selector design, the SNV p-value was used. If a fusion breakpoint identified in a tumor sample (i.e., involving ROS1, ALK, or RET) was recovered in plasma DNA from the same patient, it trumped all other mutation types, and its p-value (~0) was used. If a fusion detected in the tumor was not found in corresponding plasma (potentially due to hybridization inefficiency; see Supplementary Methods), the p-value for any remaining mutation type(s) was used. The ctDNA detection index was considered significant if the metric was ≤0.05 (≈FPR ≤5%), the threshold that maximized CAPP-Seq sensitivity and specificity in ROC analyses (determined by Euclidean distance to a perfect classifier; i.e., TPR = 1 and FPR = 0; Fig. 3, Fig. 4, Table 1, and Supplementary Table 4).
Additional details are presented in the Supplementary Methods.
Supplementary Material
Acknowledgments
We would like to thank S. Quake and members of his lab for suggestions and N. Neff for technical assistance. This work was supported by the Department of Defense (MD, AAA, AMN), US National Institutes of Health Director’s New Innovator Award Program (MD; 1-DP2-CA186569), the Ludwig Institute for Cancer Research (MD, AAA), Radiological Society of North America (SVB; #RR1221), Association of American Cancer Institutes Translational Cancer Research Fellowship (SVB), and a grant from the Siebel Stem Cell Institute and the Thomas and Stacey Siebel Foundation (AMN). AAA and MD are supported by Doris Duke Clinical Scientist Development Awards.
Footnotes
Author Contributions
AMN, SVB, AAA, and MD developed the concept, designed the experiments, analyzed the data, and wrote the manuscript. SVB performed the molecular biology experiments and AMN performed the bioinformatics analyses. CLL helped develop analytical pipeline software. SVB, JT, JFW, NCWE, LAM, JWL, HAW, REM, JBS, BWL, and MD provided patient specimens. AAA and MD contributed equally as senior authors. All authors commented on the manuscript at all stages.
References
- 1.Taniguchi K, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res. 2011;17:7808–7815. doi: 10.1158/1078-0432.CCR-11-1712. [DOI] [PubMed] [Google Scholar]
- 2.Rosell R, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 3.Kuang Y, et al. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin Cancer Res. 2009;15:2630–2636. doi: 10.1158/1078-0432.CCR-08-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gautschi O, et al. Origin and prognostic value of circulating KRAS mutations in lung cancer patients. Cancer Lett. 2007;254:265–273. doi: 10.1016/j.canlet.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Leary RJ, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2:20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McBride DJ, et al. Use of cancer-specific genomic rearrangements to quantify disease burden in plasma from patients with solid tumors. Genes, Chromosomes & Cancer. 2010;49:1062–1069. doi: 10.1002/gcc.20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He J, et al. IgH gene rearrangements as plasma biomarkers in Non- Hodgkin’s lymphoma patients. Oncotarget. 2011;2:178–185. doi: 10.18632/oncotarget.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forshew T, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra168. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 9.Leary RJ, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4:162ra154. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Narayan A, et al. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res. 2012;72:3492–3498. doi: 10.1158/0008-5472.CAN-11-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson SJ, et al. Analysis of Circulating Tumor DNA to Monitor Metastatic Breast Cancer. N Engl J Med. 2013 doi: 10.1056/NEJMoa1213261. [DOI] [PubMed] [Google Scholar]
- 12.Murtaza M, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 13.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nature Rev Clinical Oncol. 2013;10:472–484. doi: 10.1038/nrclinonc.2013.110. [DOI] [PubMed] [Google Scholar]
- 14.Forbes SA, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Youn A, Simon R. Identifying cancer driver genes in tumor genome sequencing studies. Bioinformatics. 2011;27:175–181. doi: 10.1093/bioinformatics/btq630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergethon K, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30:863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwak EL, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pao W, Hutchinson KE. Chipping away at the lung cancer genome. Nat Med. 2012;18:349–351. doi: 10.1038/nm.2697. [DOI] [PubMed] [Google Scholar]
- 20.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Govindan R, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–1134. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koivunen JP, et al. EML4-ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res. 2008;14:4275–4283. doi: 10.1158/1078-0432.CCR-08-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rikova K, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 24.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci U S A. 2008;105:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su Z, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13:74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi S, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 27.Iyengar P, Timmerman RD. Stereotactic ablative radiotherapy for non-small cell lung cancer: rationale and outcomes. Journal of the National Comprehensive Cancer Network : JNCCN. 2012;10:1514–1520. doi: 10.6004/jnccn.2012.0157. [DOI] [PubMed] [Google Scholar]
- 28.Nesbitt JC, Putnam JB, Jr, Walsh GL, Roth JA, Mountain CF. Survival in early-stage non-small cell lung cancer. Annals Thoracic Surg. 1995;60:466–472. doi: 10.1016/0003-4975(95)00169-l. [DOI] [PubMed] [Google Scholar]
- 29.Aberle DR, et al. Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. N Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diehl F, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc Natl Acad Sci U S A. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diehl F, et al. Analysis of mutations in DNA isolated from plasma and stool of colorectal cancer patients. Gastroenterology. 2008;135:489–498. doi: 10.1053/j.gastro.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan KC, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2013;59:211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 33.Heitzer E, et al. Tumor associated copy number changes in the circulation of patients with prostate cancer identified through whole-genome sequencing. Genome Med. 2013;5:30. doi: 10.1186/gm434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitt MW, et al. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiroguchi K, Jia TZ, Sims PA, Xie XS. Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes. Proc Natl Acad Sci U S A. 2012;109:1347–1352. doi: 10.1073/pnas.1118018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quail MA, et al. Optimal enzymes for amplifying sequencing libraries. Nat Methods. 2012;9:10–11. doi: 10.1038/nmeth.1814. [DOI] [PubMed] [Google Scholar]
- 37.Oyola SO, et al. Optimizing Illumina next-generation sequencing library preparation for extremely AT-biased genomes. BMC Genomics. 2012;13:1. doi: 10.1186/1471-2164-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fisher S, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12:R1. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fisher RA. Statistical methods for research workers. Oliver and Boyd; Edinburgh, London: 1925. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.