Abstract
Background
Enteric Redmouth Disease (ERM), caused by Yersinia ruckeri, is one of the most important infectious diseases in rainbow trout (Oncorhynchus mykiss) aquaculture in Europe. More recently, non-motile vaccine resistant isolates appear to have evolved and are causing disease problems throughout Europe, including Germany. The aim of this study was to analyse the variation of biochemical and molecular characteristics of Y. ruckeri isolates collected in north west Germany as a basis for strain differentiation. The isolates originated mainly from rainbow trout and were characterised by biochemical profiling, 16S rDNA sequencing, repetitive sequence-based PCRs, including (GTG)5-PCR, BOX-PCR, ERIC-PCR and REP-PCR, and pulsed-field gel electrophoresis (PFGE).
Results
In total, 83 isolates were characterised, including 48 isolates collected during a field study in north west Germany. All isolates were confirmed as Y. ruckeri by the API 20E system. Five isolates were additionally confirmed as Y. ruckeri by Y. ruckeri-specific PCR and 16S rDNA sequencing. Only 17 isolates hydrolyzed Tween 80/20. Sixty-six isolates (79.5%) were non-motile. Two different patterns were obtained by REP-PCR, five patterns by ERIC-PCR, four patterns by (GTG)5-PCR and three patterns by BOX-PCR. NotI-directed PFGE resulted in 17 patterns that differed from each other by 25–29 fragments. Isolates from the field study clustered together as PFGE type C. According to the results of API 20E, repetitive sequence-based PCRs and PFGE, these isolates could be subdivided into 27 different groups.
Conclusions
The detailed molecular and phenotypic characterisation scheme developed in this study could be used to help trace the dissemination of Y. ruckeri isolates, and thus may represent part of improved disease monitoring plans in the future.
Keywords: Enteric Red mouth Disease, REP-PCR, ERIC-PCR, BOX-PCR, PFGE, Non-motile strains
Background
Enteric Red mouth Disease (ERM), caused by Yersinia ruckeri, is one of the most important infectious diseases in rainbow trout (Oncorhynchus mykiss) aquaculture in Europe. Signs of a clinical infection of rainbow trout may include haemorrhages in various tissues and organs, particularly around the mouth, in the gills, muscles, peritoneum and the lower intestine. The bacterium has been isolated from various fish species, including rainbow trout, steelhead (Oncorhynchus mykiss), lake trout (Salvelinus namaycush), cutthroat (O. clarkii), brown (Salmo trutta) and brook trout (S. fontinalis), coho (O. kisutch), sockeye (O. nerka), Chinook (O. tshawytscha) and Atlantic salmon (Salmo salar) [1]. In Europe, it is endemic in many trout farms and can cause high economic losses [2-6]. The disease most commonly affects younger rainbow trout at temperatures above 10°C. Outbreaks are often related to adverse situations or stressed carrier fish which shed the pathogen and facilitate the onset of an infection. In infected trout populations, ERM can be controlled effectively by antibiotic treatment and by the application of inactivated whole cell vaccines [7], which are available commercially and are considered to be successful [8]. Nevertheless, outbreaks of ERM are still periodically observed, especially in endemic areas.
Among Y. ruckeri isolates, several serological variations have been reported [9-11]. While initially most outbreaks of ERM were caused by Y. ruckeri isolates of serotype O1 (Hagerman strain), Y. ruckeri isolates of different serotypes have been more recently obtained from sick fish during outbreaks of ERM in the UK, Spain and the U.S.A. [12-14]. Moreover, several of the commercial ERM vaccines seemed to not provide sufficient protection against infections caused by some of these isolates [13,14]. All these ‘vaccine-tolerant’ isolates lacked motility (assigned as biotype 2, BT2 isolates), while isolates from previous outbreaks were all motile (biotype 1) [13]. It was hypothesised that vaccination resulted in a selective pressure that enabled the emergence of non-motile isolates that are tolerant to the commercial vaccines. Therefore, there is a high risk that these non-motile vaccine-tolerant isolates spread and cause severe outbreaks of disease in trout farms. Wheeler et al.[15] found that the BT2 isolates in mainland Europe have arisen independently from the local BT1 isolates. Recently, Welch et al.[16] indicated that the European BT2 isolates were not from the same clonal group. In Germany, the earliest report about BT2 isolates was published in 1994 [17]. Recently, ‘vaccine-tolerant’ Y. ruckeri were isolated from outbreaks in trout hatcheries in north west Germany, in particular in the federal state of North Rhine-Westphalia (NRW).
In order to avoid further spreading of these ‘vaccine-tolerant’ isolates, an epidemiological survey on the occurrence and further phenotypic and molecular characterisation of Y. ruckeri from trout hatcheries was performed. In this study, interspecies differences of Y. ruckeri isolates from north west Germany were detected via biochemical and molecular traits to determine how related isolates from different origins are. For this, several typing methods were compared for their ability to differentiate Y. ruckeri isolates from rainbow trout and other fish species.
Results
Biochemical and physiological characterisation
Among the variable biochemical characteristics of all 83 Y. ruckeri isolates (Table 1), gelatine hydrolysis, Voges-Proskauer (VP) reaction and Tween hydrolysis were the most variable biochemical reactions. The results of VP, nitrate production, citrate utilization and gelatine hydrolysis from both, conventional biochemical analysis and testing by the API 20E system, were listed also in Table 1. For motility, Tween 80/20 hydrolysis and citrate utilization, the isolates obtained in Lower Saxony (LS) from fish species other than rainbow trout were more similar to the reference strain DSM 18,506 than the isolates obtained from rainbow trout hatcheries in NRW and LS. Only 16 isolates hydrolysed Tween 80/20. For these isolates motility was confirmed by microscopical inspection as well as by cultivation in API M medium. Those isolates were recognized as non-motile isolates. In particular, isolates collected from trout hatcheries in NRW were lacking flagella and motility; 92% of the isolates from this federal state were non-motile (Table 1).
Table 1.
Biochemical characteristics of Yersinia ruckeri isolates from fish in aquaculture in north west Germany
|
Test |
Reference strain (DSM 18,506) |
Number of the field isolates tested positive (%) |
||||
|---|---|---|---|---|---|---|
|
Total |
Isolates from rainbow trouts in H |
Isolates from rainbow trouts in N |
Isolates from rainbow trouts in L |
Isolates from other fish in L |
||
| (n = 82) | (n = 8) | (n = 49) | (n = 19) | (n = 6) | ||
| Motility |
+ |
16 (19.5) |
1 (12.5) |
4 (8.2) |
6 (31.6) |
5 (83.3) |
| Nitrate reduction |
+ |
77 (93.9) |
8 (100.0) |
47 (95.9) |
17 (89.5) |
5 (83.3) |
| Nitrate reduction * |
- |
72 (87.8) |
8 (100) |
45 (91.8) |
14 (73.7) |
5 (83.3) |
| Citrate utilization |
+ |
59 (72.0) |
6 (75.0) |
32 (65.3) |
15 (78.9) |
6 (100) |
| Citrate utilization * |
+ |
50 (61.7) |
6 (75.0) |
29 (59.2) |
10 (52.6) |
5 (83.3) |
| Voges-Proskauer |
- |
48 (58.5) |
5 (62.5) |
29 (59.2) |
10 (52.6) |
4 (66.7) |
| Voges-Proskauer* |
- |
52 (63.4) |
4 (50.0) |
37 (75.5) |
7 (36.8) |
4 (66.7) |
| Gelatine hydrolysis |
- |
81 (98.8) |
8 (100.0) |
49 (100.0) |
19 (100.0) |
5 (83.3) |
| Gelatine hydrolysis* |
- |
77 (93.9) |
8 (100.0) |
48 (98.0) |
16 (84.2) |
5 (83.3) |
| Methyl-red |
+ |
76 (92.7) |
7 (87.5) |
48 (98.0) |
18 (94.7) |
3 (50.0) |
| Tween 20 hydrolysis |
+ |
16 (19.5) |
1 (12.5) |
4 (8.2) |
6 (31.6) |
5 (83.3) |
| Tween 80 hydrolysis |
+ |
16 (19.5) |
1 (12.5) |
4 (8.2) |
6 (31.6) |
5 (83.3) |
| Acid from sorbitol | - | 5 (6.1) | 1 (12.5) | 1 (2.0) | 0 (0.0) | 3 (50.0) |
* API 20E test system.
L: Lower Saxony, H: Hessen, N: North Rhine-Westphalia.
In the API 20E test, 27 isolates showed the numeric profile 5,107,100, 23 isolates with the numeric profile 5,306,100. These profiles differed from that of the reference strain DSM 18,506 with the profile 5,304,100. For a biochemical typing of the isolates, typing numbers from “a” to “g” were assigned to individual isolates according to their profile number (Table 2).
Table 2.
Different genetic groups of Y. ruckeri
|
Patterns from different genotypic method
a
|
API 20E profile b | No. of isolates | Typing Group (tp) | ||||
|---|---|---|---|---|---|---|---|
| REP-PCR | BOX-PCR | (GTG) 5 -PCR | ERIC-PCR | PFGE | |||
| R1 |
B1 |
G1 |
E1 |
Pt C1 |
a |
5 |
tp1 |
| b |
21 |
tp2 |
|||||
| c |
1 |
tp3 |
|||||
| d |
1 |
tp4 |
|||||
| h |
2 |
tp5 |
|||||
| Pt C10 |
a |
8 |
tp6 |
||||
| b |
1 |
tp7 |
|||||
| c |
7 |
tp8 |
|||||
| d |
1 |
tp9 |
|||||
| g |
1 |
tp10 |
|||||
| Pt C4 |
c |
1 |
tp11 |
||||
| Pt C6 |
g |
1 |
tp12 |
||||
| Pt C8 |
b |
2 |
tp13 |
||||
| Pt C9 |
a |
1 |
tp14 |
||||
| Pt C3 |
b |
1 |
tp15 |
||||
| Pt C7 |
c |
3 |
tp16 |
||||
| Pt C5 |
d |
1 |
tp17 |
||||
| Pt C2 |
a |
7 |
tp18 |
||||
| b |
2 |
tp19 |
|||||
| c |
7 |
tp20 |
|||||
| Pt C11 |
a |
2 |
tp21 |
||||
| Pt C12 |
c |
1 |
tp22 |
||||
| G 2 |
E 3 |
Pt A1 |
e |
1 |
tp23 |
||
| B 2 |
G4 |
E 4 |
Pt A2 |
e |
1 |
tp24 |
|
| B 3 |
G3 |
E 2 |
Pt B1 |
f |
1 |
tp25 |
|
| R 2 |
B 1 |
G1 |
E 5 |
Pt B2 |
e |
2 |
tp26 |
| R 3 | B 1 | G 1 | E 6 | Pt D1 | g | 1 (DSM18506)b | tp27 |
aPCR-based typing methods: REP-PCR, repetitive extragenic palindromic-PCR; BOX-PCR, BOX-A1R-based repetitive extragenic palindromic-PCR; (GTG)5-PCR, polytrinucleotide (GTG)5-PCR; ERIC-PCR, enterobacterial repetitive intergenic consensus-PCR. Macrorestriction-based typing method, PFGE macrorestriction analyis by pulsed-field gel electrophoresis.
bReference strain DSM 18506.
cAPI 20E profiles were as follows: a (5,306,100, n = 23), b (5,107,100, n = 27), c (5,307,100, n = 20), d (5,106,100, n = 3), e (5,307,500, n = 4), f (5,305,700, n = 1), g (5,304,100, n = 2 + reference strain DSM 18,506), h (5,104,100, n = 2).
Genetic characterisation
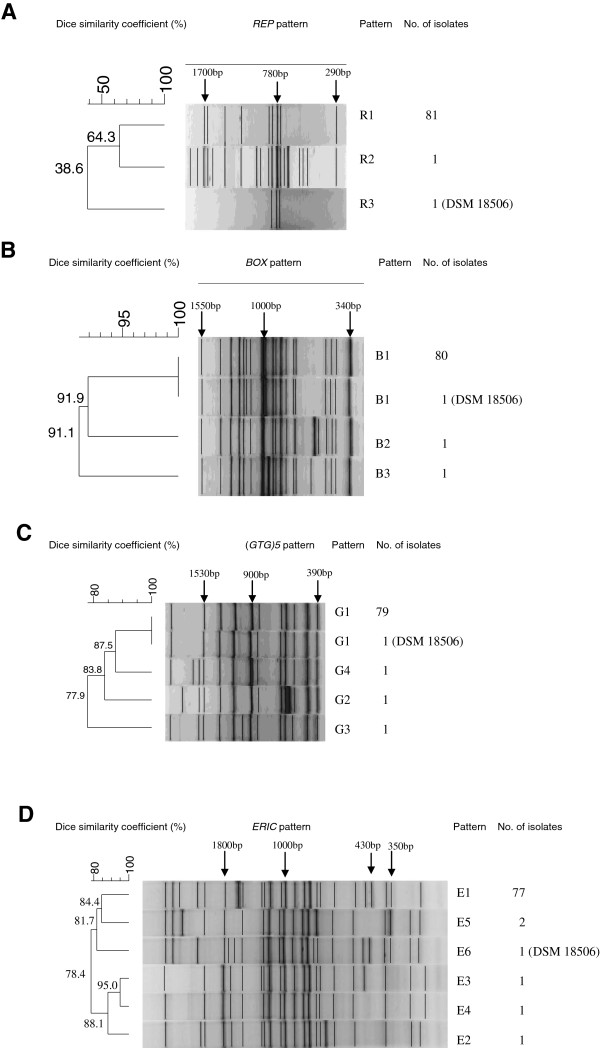
When the isolates were analysed by repetitive sequence-based PCR assays, two amplicon patterns were obtained after REP-PCR, five patterns by ERIC-PCR, four patterns by (GTG)5-PCR and three patterns by BOX-PCR (Figure 1). The similarity rates in REP-PCR were clearly lower than in the other repetitive sequence-based PCRs. In BOX-PCR and (GTG)5-PCR, the reference strain DSM 18,506 exhibited the same amplicon patterns as most of the isolates (Figure 1). By REP-PCR, 3 or 19 bands were seen in the size range of 200–2000 bp; by BOX-PCR, 18–19 bands were found distributed between 340–1550 bp; by (GTG)5-PCR, 11–13 bands were detected from 390–2500 bp; and by ERIC-PCR, 20–25 bands were present in the range from 200–2000 bp (Figure 1). Five isolates, which were positive in sorbitol fermentation, did not exhibit the 430 bp amplicon in ERIC-PCR. Only five isolates from fish species other than rainbow trout showed some differences in repetitive PCRs. These five isolates were additionally confirmed as Y. ruckeri (identities = 100%, data not shown) by 16S rDNA sequencing; while the nucleotide sequence of the reference strain DSM 18,506 was not completely identical to the sequence reported from the strain ATCC 29,473 (identities = 99%, data not shown). Conversely, all the isolates from rainbow trout showed rather uniform patterns of repetitive sequence-based PCR.
Figure 1.
A dendrogram of Y. ruckeri isolates constructed using the UPGMA method (tolerance 1%) using Gel Compar II (Applied Maths), based on fingerprints of different repetitive sequence-based PCRs: A, REP-PCR; B, BOX-PCR; C, (GTG)5-PCR; D, ERIC-PCR.
NotI-directed PFGE yielded four major PFGE groups A-D. A total of 17 different PFGE types (A1, A2, B1, B2, C1-C12, D) were detected among the 83 Y. ruckeri isolates examined (Figure 2, Table 2). Cluster analysis of these PFGE types showed that the isolates from north west Germany were highly uniform, whereby 30 isolates (36.1%) belonged to PFGE type C1 and 18 isolates to PFGE type C10 (Figure 2). Compared with the reference strain DSM 18,506, all the isolates were different. The isolates from rainbow trout were clustered together (PFGE group C). Isolates from pike, koi and brown trout (PFGE groups A and B) showed the greatest variation when compared to rainbow trout isolates. All isolates previously assigned to biotype 2 clustered in PFGE group C. Isolates from this PFGE group were found in the federal states of LS, Hessen (H) and NRW (Figure 2). Most of the isolates were distributed in all three federal states (PFGE type C1 and C10), while isolates of PFGE type C2 were found only in LS and NRW (Figure 2).
Figure 2.
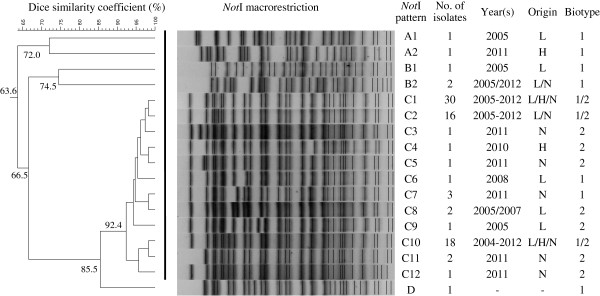
Dendrogram of Y. ruckeri isolates based on NotI-macrorestriction (PFGE) patterns. The similarity analysis was performed using the Dice coefficient and UPGMA method (tolerance 1%). Pulsotype D: reference strain DSM18,506. The similarity of the isolates obtained from Lower Saxony (L), Hessen (H) and North Rhine-Westphalia (N) are given as percentages at the major junctions in the dendrogram. The NotI-patterns, number of isolates, year(s) of isolation, geographical origin and biotype (biotype 1: motile, biotype 2: non-motile) are shown.
Discriminatory indices of the typing methods applied
As a discriminatory index, Simpson’s index of diversity was calculated for each typing method used to differentiate all Y. ruckeri isolates. Discriminatory indices varied distinctly between the different typing methods. REP-PCR had the lowest D value of 0.048. This means that two randomly selected isolates of this test population had a probability of 4.8% of showing a different REP-PCR pattern. Slightly higher D values were calculated for BOX-PCR (D = 0.071), (GTG)5-PCR (D = 0.095) and ERIC-PCR (D = 0.140). Highest D values were seen for biotyping via API 20E (D = 0.763) and NotI-directed PFGE (D = 0790). According to their biochemical and molecular characteristics as obtained by testing with API 20E, PFGE and various PCR-based methods, the Y. ruckeri isolates from north west Germany could be subdivided into 27 different groups (Table 2). Most of the isolates from north west Germany (30, 36.6%) belonged to the group (R1/B1/G1/E1/PFGE type C1). Additional information about the origins and isolated years were shown in Additional file 1: Table S1.
Discussion
Since the first isolation of Y. ruckeri in Idaho, USA in 1950s, outbreaks of ERM were frequently reported [7,18,19], especially from the aquaculture industry in countries such as Croatia [3], France [4,20], Finland [2], Germany [5], Italy [21], Switzerland [6] and the UK [14,22]. The bacterium was isolated from different host species [2,23,24], including several salmonid species, channel catfish, goldfish, common carp, European eel, coalfish, and perch. Typing of Y. ruckeri isolates from various geographic locations or different fish species has been performed for taxonomic or epidemiological purposes and initially it was based on phenotypical characteristics including the results of biochemical and serological tests [10,25,26]. In addition, molecular techniques, including random amplification of polymorphic DNA (RAPD), multilocus sequence typing (MLST) or pulsed-field gel electrophoresis (PFGE) have been applied to study the genetic diversity of Y. ruckeri isolates [15,27-29]. In a study on isolates from various geographic origins, including Chile, Peru, U.S.A., UK, and mainland Europe, a considerable intraspecies diversity of Y. ruckeri was found on the basis of the phenotypical characteristics obtained from API 20E, lipopolysaccharide (LPS) and outer membrane profiles [30] as well as by genetic analysis using MLST profiles [27].
The isolates from north west Germany investigated in the present study, showed a strong homogeneity of biochemical results and API 20E profiles with some variability in gelatine hydrolysis, VP reaction and Tween hydrolysis. Variation in some biochemical tests for Y. ruckeri has been reported in previous studies [27,30,31], but this variation could not be used as a distinguishing characteristic for phenotypical traits such as serotype or biotype [25,30,32]. While the majority of Y. ruckeri isolates from farmed rainbow trout in Peru [33] or Atlantic salmon in Chile [34] were motile, only 16 out of 82 isolates from the present study belonged to biotype 1 (motile and lipase positive). All other isolates could not hydrolyse Tween 80/20, were lipase-negative and lacked flagella.
The API 20E profiles 5,306,100, 5,107,100, 5,104,100 were previously obtained from Y. ruckeri isolates from south-west Germany [35], the profiles 5,307,100 and 5,304,100 were already reported from Spanish isolates [13]. While the profiles 5,106,100 and 5,307,500 were also previously reported from Y. ruckeri isolates, [36] there was no record about an isolate with the numeric profile 5,305,700. However, 16S rRNA sequencing confirmed this isolate as Y. ruckeri (identity = 100%, data not shown).
In addition to the biochemical characteristics, some genetic variability was observed in isolates using several repetitive sequence-based PCR techniques including BOX-PCR, ERIC-PCR, (GTG)5-PCR, REP-PCR as well as pulsed-field gel electrophoresis. An analysis of the isolates from north west Germany by repetitive sequence based PCRs gave 3 to 6 groups, with the majority of isolates in one group. Similar observations were already reported from Latin American isolates [33,34]. ERIC-PCR showed the highest discriminatory power. While in previous studies both ERIC-PCR and REP-PCR were considered of some value as tools to study ERM epidemiology [34], in our study, REP-PCR showed the lowest discriminatory power and could not differentiate the isolates very clearly. This could depend on the different origin of the isolates: our isolates originated mainly from rainbow trout, while Bastardo et al.[34] studied isolates from Atlantic salmon. When Y. ruckeri isolates from Peruvian rainbow trout were analysed by REP-PCR, more homogeneous amplicon patterns were observed as well [33].
PFGE has been frequently applied for detailed molecular typing of different bacteria [37-39], as well as for a differentiation among isolates of Y. ruckeri[15]. In the present study, the typing results by PFGE showed strong homogeneity among the isolates obtained from farmed fish species in north west Germany. More than half of the Y. ruckeri isolates belonged to the two PFGE types C1 and C10, with 36.1% of the isolates sharing the same PFGE type C1. These two PFGE types belong to the group C PFGE cluster, which comprises isolates from rainbow trout and represents the majority of the isolates. The isolates showing PFGE types A1/A2 or B1/B2 also differed from the other isolates in at least some of their repetitive sequences-based PCR patterns (Table 2). In the present study, no distinct differences were observed in PFGE types from biotype 1 and biotype 2 isolates from rainbow trout.
The data presented in this study may suggest that the isolates from north west Germany originated from the same ancestor and could indicate a clonal population structure. In a previous study on the genetic diversity of Y. ruckeri isolates, analysed by MLST, a large number of isolates also belonged to just two sequence types (ST), which formed one clonal complex with 21 out of 30 recognised STs [27]. However, the MLST analysis revealed that genetic recombination appeared to play a greater role than mutation for the generation and maintenance of genetic diversity within the population of Y. ruckeri[27]. This would be contradictory to the concept of clonal expansion of a population from a common ancestor that diversified mainly by mutation rather than recombination [40]. MLST results from Y. ruckeri rather suggest an epidemic population structure, as it was observed for instance for Vibrio parahaemolyticus[27,41]. The epidemiology of Yersinia ruckeri, which started as a geographically isolated disease and quickly became disseminated [31], also was considered to support the epidemic model [27].
Pulsed-field gel electrophoretic analysis of Y. ruckeri isolates resulted in a discriminatory index (D) of 0.790, whereas repetitive sequence-based PCR approaches yielded low D values between 0.048 and 0.140. As the D value describes the probability that two randomly selected isolates show different profiles by the method applied, methods with low D values are considered not to be sufficiently discriminative for the detection of differences among Y. ruckeri isolates. In a previous analysis, “medium to high” D values ranging between 0.71–0.74 were obtained by the API 20E, lipopolysaccharide and outer membrane protein analysis [27], while ERIC-and REP-PCR were valuable in combination with other techniques. Similar results were reported from Y. ruckeri isolated in Turkey [42]. The combination of several typing methods, as performed by Bastardo et al. [27] showed that isolates with similar characteristics were associated with certain fish species and/or predominated in some geographical areas. In the present study, the combination of six typing methods, including API 20E, PFGE and four different repetitive sequence PCR techniques facilitated the identification of 27 different groups. With this set of methods, most of the isolates from rainbow trout (97.3%) could be differentiated from the isolates obtained from pike, koi and brown trout. Biotype 2 isolates were only detected in rainbow trout in this study. The genomic variation of Y. ruckeri isolates provides information for comparing their distribution and the relationships among individual isolates from different regions in north west Germany.
Conclusions
Most of the isolates from north west Germany belonged to biotype 2. The combination of different typing methods could well differentiate Y. ruckeri isolates. The results of this study showed the diversity of isolates present in north west Germany. Moreover, these data enable the identification and in-depth characterisation of pathogenic Y. ruckeri isolates. As a consequence of this detailed characterisation, these data can be used to examine the distribution of these isolates in the field and may be used as a basis for preventive disease monitoring plans.
Methods
Bacterial isolates and sample collection
In total, 83 Yersina ruckeri isolates were analysed, including 33 isolates from the strain collection of LAVES Niedersachsen, Food and Veterinary Institute Braunschweig/Hannover, Germany and the fish disease service at the Landesbetrieb Hessisches Landeslabor (LHL) Giessen, Germany. These bacteria were isolated from diseased rainbow trout suffering from ERM outbreaks. In addition, 48 isolates were sampled from rainbow trout (Oncorhynchus mykiss) in trout hatcheries at different sites in the German federal state NRW during the four seasons of 2011–2012. Sampling was done under the approval of Landesamt für Natur, Umwelt und Verbraucherschutz, NRW. Fish were randomly sampled when diseased fish were not present in the farm. The sampling was done following the guidelines for the diagnostic of fish diseases and regarded international and national guidelines for animal welfare. Of the remaining two isolates, one was the Y. ruckeri reference strain DSM 18,506 and the other a non-motile isolate also from NRW in 2008 and provided by Dr. Gould from MSD Animal Heath. All isolates were kept as glycerol stock cultures at−80°C. All isolates were cultured on trypticase soy agar (TSA; Sigma) at 25°C for 24–48 h and pure cultures were kept at−80°C using Cryobank beads (Copan Diagnostic INC., USA).
Identification of Y. ruckeri
All Y. ruckeri-suspect cultures were examined by Gram-staining and colony morphology. For species confirmation, the pure-cultured colonies were biochemically characterised by the API 20E system at 25°C for 48 h (bioMérieux, Craponne, France), as well as subjected to the following standard biochemical tests: cytochrome-oxidase production, catalase production, Methylred/Voges-Proskauer (VP), growth on McConkey agar, xylose utilisation, glucose utilisation/gas production, tween 80/20 hydrolysis, nitrate production, citrate utilisation, sorbitol fermentation, gelatine hydrolysis, and oxidative/fermentative glucose utilisation. Motility was checked by phase-contrast microscopy (1000×). To confirm the motility, bacteria were stab-inoculated with a needle into the bottom of API M medium (bioMérieux). In addition, silver staining [43] was used to investigate the isolates for the presence of flagella. After staining, the slides were microscopically inspected (Leica DFC320, Bensheim, Germany).
DNA extraction
Bacterial cells were lysed and genomic DNA was extracted, following the manufacturers protocol provided in the innuPREP Bacteria DNA Kit (Analytik Jena, Jena, Germany). The purity of DNA was confirmed using a NanoDrop spectrophotometer (NanoDrop, Delaware, USA).
Repetitive sequence-based PCR approaches
For genotyping, four different repetitive sequence-based PCRs were performed including BOX-A1R-based repetitive extragenic palindromic-PCR (BOX-PCR), (GTG)5-PCR, enterobacterial repetitive intergenic consensus (ERIC-PCR) and repetitive extragenic palindromic (REP-PCR). These PCR assays were performed for all Y. ruckeri isolates [44], using primers listed in Table 3. PCRs were performed in 25 μl volumes containing 2 μl of DNA template, 0.2 mM concentrations of deoxynucleoside triphosphates, 2.5 μl of 10 × PCR buffer (Qiagen, Hilden, Germany), 2 mM MgCl2, 8 μM, 1 μM and 2 μM of each forward and reverse primer for BOX-/(GTG)5-, ERIC-and REP-PCR, respectively, and 5 U of Qiagen HotstarTaq DNA Polymerase (Qiagen). The reaction was performed in a Biometra T3000 thermocycler (Analytik Jena, Germany) with an initial denaturation cycle at 95°C for 7 min, followed by 30 cycles of amplification (denaturation for 1 min at 94°C, annealing for 1 min at 53°C, 52°C and 44°C for BOX, ERIC/(GTG)5, and REP-PCR, respectively, and extension for 8 min at 65°C), and a final elongation for 16 min at 65°C. The PCR products were detected by electrophoresis by using 2.5% agarose gels. The images obtained were analysed by the UPGMA method using GelCompar II (Applied Maths, Belgium).
Table 3.
Primers used in this study
| PCR | Name | Sequence (5′ → 3′) | References |
|---|---|---|---|
| Specific PCR of Y. ruckeri |
YER8 |
GCGAGGAGGAAGGGTTAAGTG |
[45] |
| YER10 |
GAAGGCACCAAGGCATCTCTG |
[45] |
|
| (GTG)5-PCR |
(GTG)5 |
GTGGTGGTGGTGGTG |
[44] |
| BOX-PCR |
BOXA1R |
CTACGGCAAGGCGACGCTGACG |
[44] |
| ERIC-PCR |
ERIC2 |
AAGTAAGTGACTGGGGTGAGCG |
[44] |
| ERIC1R |
ATGTAAGCTCCTGGGGATTCAC |
[44] |
|
| REP-PCR | REP2-I |
ICGICTTATCIGGCCTAC |
[44] |
| REP1R-I | IIIICGICGICATCIGGC | [44] |
16S rDNA gene sequencing
A specific PCR for the identification of Y. ruckeri was applied to five isolates which showed patterns different from most of the isolates in repetitive sequence-based PCR. For this PCR, the primers YER8 and YER10 (listed in Table 3) were used according to Gibello et al.[45]. The PCR was performed using the Titanium Taq PCR Kit according to the manufacturer’s protocol (Clontech, Saint-Germain-en-Laye, France). Briefly, the PCR was performed in 20 μl volumes containing 1 μl of DNA template, 0.2 mM deoxynucleoside triphosphates, 2 μl of 10× Titanium Taq buffer (Clontech, USA), 10 μM of each forward and reverse primer, and 0.1 μl of 50 × Titanium Taq DNA Polymerase (Clontech). The reaction was performed in a BIORON thermocycler (BIORON, Germany) with an initial denaturation cycle for 3 min at 95°C, followed by 40 cycles of amplification (denaturation for 30 s at 95°C, annealing for 30 s at 60°C, and extension for 1 min at 72°C), and a final elongation for 7 min at 72°C. PCR products were sent to LGC Genomics (Berlin, Germany) for sequencing. The resulting 16S rDNA sequences were compared in a BLAST search with the sequences previously deposited in the NCBI database (http://www.ncbi.nlm.nih.gov).
Pulsed-field gel electrophoresis (PFGE)
Isolates to be examined by pulsed-field gel electrophoresis were grown on TSA plates for 48 h at 25°C. Agarose plugs containing Yersinia ruckeri genomic DNA were prepared according to the method of Wagley at el[39]. Slices of the agarose plugs that contain Y. ruckeri genomic DNA were digested with the restriction endonuclease NotI (100 U) for 4 h at 37°C. A Salmonella Braenderup H9812 molecular standard was prepared by the same method and restricted with XbaI [46]. NotI digested agarose plugs were loaded into a 1% (w/v) PFGE-grade agarose gel (Roth, Germany) and subjected to electrophoresis in 0.5 × TBE buffer using a Bio-Rad CHEF-DRIII system. PFGE conditions were 5.6 V/cm with switch times of 1 to 15 s at 14°C for 20 h. Images were captured under ultraviolet light. The images obtained were analysed by the UPGMA method using GelCompar II (Applied Maths).
Discriminatory power of typing methods
Simpson’s index of diversity was applied to calculate the discriminatory power of the different typing methods: API 20E, REP-PCR, BOX-PCR, (GTG)5-PCR, ERIC-PCR and PFGE according to Hunter and Gaston [47].
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YH, MR, SS and DS designed the study. YH carried out the biochemical identification and repetitive sequence-based PCRs according to the suggestions from MR and AJ. GBM and YH performed the PFGE and analysed the results under the supervision of SS and DS. YH drafted the manuscript and all authors critically revised the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Different origins of Yersinia ruckeri.
Contributor Information
Yidan Huang, Email: danni-hyd@hotmail.com.
Martin Runge, Email: martin.runge@laves.niedersachsen.de.
Geovana Brenner Michael, Email: geovana.michaelbrenner@fli.bund.de.
Stefan Schwarz, Email: stefan.schwarz@fli.bund.de.
Arne Jung, Email: arne.jung@tiho-hannover.de.
Dieter Steinhagen, Email: dieter.steinhagen@tiho-hannover.de.
Acknowledgements
We thank Dr. S. Braune (LAVES, Hannover), Dr. C. Gould (MSD Animal Health) and Dr. A. Nilz (LHL Hessen) for the generous provision of isolates that were used in this study. Dr. W. Schäfer and D. Mock (LANUV, Albaum) gave support during the field sampling, and R. Becker (FLI, Mariensee) provided valuable technical assistance. This work was supported by Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen (LANUV).
References
- Noga EJ. Fish disease: diagnosis and treatment. 2. Iowa: Wiley-Blackwell; 2010. [Google Scholar]
- Valtonen ET, Rintamaki P, Koskivaara M. Occurrence and pathogenicity of Yersinia ruckeri at fish farms in northern and central Finland. J Fish Dis. 1992;9:163–171. doi: 10.1111/j.1365-2761.1992.tb00650.x. [DOI] [Google Scholar]
- Oraić D, Zrnčić S, Šoštarić B, Bažulić D, Lipej Z. Occurrence of enteric redmouth disease in rainbow trout(Oncorhynchus Mykiss) on farms in Croatia. Acta Vet Hung. 2002;9(3):283–291. doi: 10.1556/AVet.50.2002.3.4. [DOI] [PubMed] [Google Scholar]
- Lesel R, Lesel M. Outbreak of enteric redmouth disease in rainbow trout, Salmo gairdneri Richardson, in France. J Fish Dis. 1983;9:385–387. doi: 10.1111/j.1365-2761.1983.tb00091.x. [DOI] [Google Scholar]
- Fuhrmann H, Boehm KH. An outbreak of enteric redmouth disease in West Germany. J Fish Dis. 1983;9:309–311. doi: 10.1111/j.1365-2761.1983.tb00080.x. [DOI] [Google Scholar]
- Meier W. Enteric redmouth disease: outbreak in rainbow trout in Switzerland. Dis Aquat Organ. 1986;9:81–82. [Google Scholar]
- Ross AJ, Rucker RR, Ewing WH. Description of a bacterium associated with redmouth disease of rainbow trout (Salmo Gairdneri) Can J Microbiol. 1966;9(4):763–770. doi: 10.1139/m66-103. [DOI] [PubMed] [Google Scholar]
- Tebbit GL, Erickson JD, Vande Water RB. Development and use of Yersinia ruckeri bacterins to control enteric redmouth disease. International symposium on fish biologies: serodiagnostics and vaccines. vol. 49: Developments in biological standardization. 1981. pp. 395–401.
- Davies RL. O-Serotyping of Yersinia ruckeri with special emphasis on European isolates. Vet Microbiol. 1990;9:299–307. doi: 10.1016/0378-1135(90)90016-O. [DOI] [PubMed] [Google Scholar]
- Davies RL. Clonal analysis of Yersinia ruckeri based on biotypes, serotypes and outer membrane protein-types. J Fish Dis. 1991;9:221–228. doi: 10.1111/j.1365-2761.1991.tb00591.x. [DOI] [Google Scholar]
- Romalde JL, Margarinos B, Barja JL, Toranzo AE. Antigenic and molecular characterization of Yersinia ruckeri: proposal for a new intraspecies classification. Syst Appl Microbiol. 1993;9:411–419. doi: 10.1016/S0723-2020(11)80274-2. [DOI] [Google Scholar]
- Arias CR, Olivares-Fuster O, Hayden K, Shoemaker CA, Grizzle JM, Klesius PH. First report of Yersinia ruckeri biotype 2 in the USA. J Aquat Anim Health. 2007;9:35–40. doi: 10.1577/H06-011.1. [DOI] [PubMed] [Google Scholar]
- Fouz B, Zarza C, Amaro C. First description of non-motile Yersinia ruckeri serovar I strains causing disease in rainbow trout, Oncorhynchus mykiss (Walbaum), cultured in Spain. J Fish Dis. 2006;9:339–346. doi: 10.1111/j.1365-2761.2006.00723.x. [DOI] [PubMed] [Google Scholar]
- Austin DA, Robertson PAW, Austin B. Recovery of a new biogroup of Yersinia ruckeri from diseased rainbow trout (Oncorhynchus mykiss, Walbaum) Syst Appl Microbiol. 2003;9:127–131. doi: 10.1078/072320203322337416. [DOI] [PubMed] [Google Scholar]
- Wheeler RW, Davies RL, Dalsgaard I, Garcia J, Welch TJ, Wagley S, Bateman KS, Verner-Jeffreys DW. Yersinia ruckeri biotype 2 isolates from mainland Europe and the UK represent different clonal groups. Dis Aquat Organ. 2009;9:25–33. doi: 10.3354/dao02039. [DOI] [PubMed] [Google Scholar]
- Welch TJ, Verner-Jeffreys DW, Dalsgaard I, Wiklund T, Evenhuis JP, Cabrera JAG, Hinshaw JM, Drennan JD, LaPatra SE. Independent emergence of Yersinia ruckeri biotype 2 in th United States and Europe. Appl Environ Microbiol. 2011;9(10):3493–3499. doi: 10.1128/AEM.02997-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein BU, Kleingeld DW, Bohm KH. First isolation of a non-motile/tween 80 negative Yersinia ruckeri strain in Germany. Bull Eur Assoc Fish Pathol. 1994;9(5):165–166. [Google Scholar]
- Karatas S, Candan A, Demircan D. Enteric red mouth disease in cultured rainbow trout (Oncorhynchus Mykiss) on the black sea coast of Turkey. Isr J Aquacult-Bamid. 2004;9(3):226–231. [Google Scholar]
- Wang KY, Fan FL, Huang XL, Geng Y, Chen DF, Huang JL. Occurrence and treatment of a channel catfish(Ictalurus punctatus(Rafinesque)) disease, caused by Yersinia ruckeri. Sci Fish Farming. 2009;9:50–51. [Google Scholar]
- Coquet L, Cosette P, Quillet L, Petit F, Junter GA, Jouenne T. Occurrence and phenotypic charaterization of Yersinia ruckeri strains with biofilm-forming capacity in a rainbow trout Farm. Appl Environ Microbiol. 2002;9(2):470–475. doi: 10.1128/AEM.68.2.470-475.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch RA. Enteric redmouth disease(Hagerman strain) Mar Fish Rev. 1978;9:42–51. [Google Scholar]
- Roberts MS. A report of an epizootic in hatchery-reared rainbow trout, salmo gairdneri Richardson, at an English trout farm, caused by Yersinia ruckeri. J Fish Dis. 1983;9:551–552. doi: 10.1111/j.1365-2761.1983.tb00111.x. [DOI] [Google Scholar]
- Noga EJ. Enteric redmouth disease (ERM, redmouth, yersiniosis, blood spot, yersinia ruckeri infection) Ames: Iowa State University Press; 2000. [Google Scholar]
- Danley ML, Goodwin AE, Killian HS. Epizootics in farm-raised channel catfish, Ictalurus punctatus(Rafinesque), caused by the enteric redmouth bacterium Yersinia ruckeri. J Fish Dis. 1999;9:451–456. doi: 10.1046/j.1365-2761.1999.00196.x. [DOI] [Google Scholar]
- Stevenson RMW, Airdrie DE. Serological variation among Yersinia ruckeri strains. J Fish Dis. 1984;9:247–254. doi: 10.1111/j.1365-2761.1984.tb00930.x. [DOI] [Google Scholar]
- Davies RL, Frerichs GN. Morphological and biochemical differences among isolates of Yersinia ruckeri obtained from wide geographical areas. J Fish Dis. 1989;9:357–365. doi: 10.1111/j.1365-2761.1989.tb00324.x. [DOI] [Google Scholar]
- Bastardo A, Ravelo C, Romalde JL. Multilocus sequence typing reveals high genetic diversity and epidemic population structure for the fish pathogen Yersinia ruckeri. Environ Microbiol. 2012;9(8):1888–1897. doi: 10.1111/j.1462-2920.2012.02735.x. [DOI] [PubMed] [Google Scholar]
- Schill WB, Phelps SR, Pyle SW. Multilocus electrophoretic assessment of the genetic structure and diversity of Yersinia ruckeri. Appl Environ Microbiol. 1984;9(5):975–979. doi: 10.1128/aem.48.5.975-979.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucangeli C, Morabito S, Caprioli A, Achene L, Busani L, Mazzolini E, Fabris A, Macri A. Molecular fingerprinting of strains of Yersinia ruckeri serovar O1 and Photobacterium damsela subsp piscicida isolated in Italy. Vet Microbiol. 2000;9(3):273–281. doi: 10.1016/S0378-1135(00)00241-8. [DOI] [PubMed] [Google Scholar]
- Bastardo A, Ravelo C, Romalde JL. A polyphasic approach to study the intraspecific diversity of Yersinia ruckeri strains isolated from recent outbreaks in salmonid culture. Vet Microbiol. 2012;9(1–2):176–182. doi: 10.1016/j.vetmic.2012.05.024. [DOI] [PubMed] [Google Scholar]
- Austin B, Austin DA. Bacterial fish pathogens: diseases of farmed and wild fish. Chichester, UK: Praxis Publishing; 2007. [Google Scholar]
- Sousa JA, Magarinos B, Eiras JC, Toranzo AE, Romalde JL. Molacular characterization of Portuguese strains of Yersinia ruckeri isolated from fish culture systems. J Fish Dis. 2001;9:151–159. doi: 10.1046/j.1365-2761.2001.00281.x. [DOI] [Google Scholar]
- Bastardo A, Sierralta V, Leon J, Ravelo C, Romalde JL. Phenotypical and genetic characterization of Yersinia ruckeri strains isolated from recent outbreaks in farmed rainbow trout Oncorhynchus mykiss (Walbaum) in Peru. Aquaculture. 2011;9:229–232. doi: 10.1016/j.aquaculture.2011.03.040. [DOI] [Google Scholar]
- Bastardo A, Bohle H, Ravelo C, Toranzo AE, Romalde JL. Serological and molecular heterogenetity among Yersinia ruckeri strains isolated from farmed Atlantic salmon Salmo salar in Chile. Dis Aquat Organ. 2011;9:207–214. doi: 10.3354/dao02296. [DOI] [PubMed] [Google Scholar]
- Wortberg F, Nardy E, Contzen M, Rau J. Identification of Yersinia ruckeri from diseased salmonid fish by fourier transform infrared spectroscopy. J Fish Dis. 2012;9:1–10. doi: 10.1111/j.1365-2761.2011.01317.x. [DOI] [PubMed] [Google Scholar]
- Furones MD, Rodgers CJ, Munn CB. Yersina ruckeri, the causal agent of enteric redmouth disease(ERM) in fish. Annu Rev of Fish Dis. 1993;9:105–125. [Google Scholar]
- Aral H, Morita Y, Izumi S, Katagiri T, Kimura H. Molecular typing by pulsed-field gel electrophoresis of Flavobacterium psychrophilum isolates derived from Japanese fish. J Fish Dis. 2007;9:345–355. doi: 10.1111/j.1365-2761.2007.00809.x. [DOI] [PubMed] [Google Scholar]
- Soto E, Mauel M, Lawrence M. Improved pulsed-field gel electrophoresis procedure for the analysis of Flavobacterium columnare isolates previously affected by DNA degration. Vet Microbiol. 2008;9:207–212. doi: 10.1016/j.vetmic.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Wagley S, Koofhethile K, Wing JB, Rangdale R. Comparison of V parahaemolyticus isolated from seafoods and cases of gastrointestinal disease in the UK. Int J Environ Health Res. 2008;9(4):283–293. doi: 10.1080/09603120801911064. [DOI] [PubMed] [Google Scholar]
- Smith JM, Smith MH, O’Rourke M, Spratt BG. How clonal are bacteria? Proc Natl Acad Sci U S A. 1993;9(10):4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Escalona N, Martinez-Urtaza J, Romero J, Espejo RT, Jaykus LA, DePaola A. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J Bacteriol. 2008;9(8):2831–2840. doi: 10.1128/JB.01808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onuk EE, Ciftci A, Findik A, Ciftci G, Altun S, Balta F, Ozer S, Coban AY. Phenotypic and molecular characterization of Yersinia ruckeri isolates from rainbow trout (Oncorhynchus mykiss, Walbaum,1792) in Turkey. Berl Munch Tierarztl Wochenschr. 2011;9:320–328. [PubMed] [Google Scholar]
- Blenden DC, Goldberg HS. Silver impregnation stain for Leptospira and flagella. Am Soc Microbiol. 1965;9(3):899–900. doi: 10.1128/jb.89.3.899-900.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann S. Untersuchung zur Klassifizierung, identifizierung und differenzierung von Yersinia Arten . Wien: University Wien; 2007. [Google Scholar]
- Gibello A, Blanco MM, Moreno MA, Cutuli MT, Domenech A, Moniguez L, Fernandez-Garayzabal JF. Development of a PCR assay for detection of Yersinia ruckeri in tissues of inoculated and naturally infected trout. Appl EnvironMicrobiol. 1999;9(1):346–350. doi: 10.1128/aem.65.1.346-350.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter SB, Vauterin P, Lambert-Fair MA, VanDuyne MS. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field get electrophoresis protocols: converting the national databases to the new size standard. J Clin Microbiol. 2005;9:1045–1050. doi: 10.1128/JCM.43.3.1045-1050.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of simpson’s index of diversity. J Clinical Microbiol. 1988;9(11):2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Different origins of Yersinia ruckeri.