Abstract
Cells adjust to changes in environmental conditions using complex regulatory programs. These cellular programs are the result of an intricate interplay between gene expression, cellular growth rate, and protein degradation fluxes. New technologies that enable simultaneous and time-resolved measurements of these variables are necessary to dissect cellular homeostatic strategies. Here, we report the development of a novel automated flow-cytometry robotic setup that enables real-time measurement of precise and simultaneous relative growth and protein synthesis rates of multiplexed microbial populations across many conditions. These measurements generate quantitative profiles of dynamically-evolving protein synthesis and degradation rates. We demonstrate this setup in the context of gene regulation of the unfolded protein response (UPR) and uncover a dynamic and complex landscape of gene expression, growth dynamics, and proteolysis following perturbations.
Introduction
In response to perturbations in their environment, cells elicit physiological changes that involve intricate modulations of their growth programs and the composition of their proteome. It has long been appreciated that both these changes are individually necessary to restore cellular homeostasis1. Nonetheless, altered growth in response to perturbations and protein synthesis and degradation rates are themselves interlinked and their relationship is dynamic rather than static.
Although many quantitative approaches to measure gene expression levels exist, they are limited by their bulk nature, throughput, or time-resolution. Quantification of complex phenotypes in microbial cultures can be achieved using a combination of time-lapse fluorescence microscopy2, flow cytometry3, DNA microarrays4, next-generation RNA sequencing5 (RNA-seq) and DNA barcode arrays6. Microarray-based approaches and whole-transcriptome sequencing have excellent gene throughput and dynamical range but are inherently a bulk measurement and typically lack fine-time resolution. By contrast, time-lapse fluorescence measurement of cell populations inside microfluidic devices7 yields single cell information but is usually low throughput and subject to micro-environment inhomogeneity and light induced stress8. In microorganisms, such as Saccharomyces cerevisiae, flow cytometry allows easy measurement of phenotypic variables, like protein abundance and cell-to-cell variability across a population for about half the genome9. Nevertheless, widespread study of dynamics using flow cytometry is limited by the lack of hardware tools that allow for measurement of cell cultures in micro-well plates with low well-to-well variability, fine time resolution, and reliable reproducibility.
Here, we introduce an automated flow-cytometry system that achieves these properties and demonstrate its capabilities by determining quantitative instantaneous growth rates, protein synthesis rates and differential fluorescent protein degradation fluxes. This setup enables the facile measurement of protein synthesis rate for many genes or conditions at fine time resolution and allows for exploring the relationship between protein synthesis rate and growth changes at the single-cell level.
Results
High-throughput monitoring of microbial culture dynamics
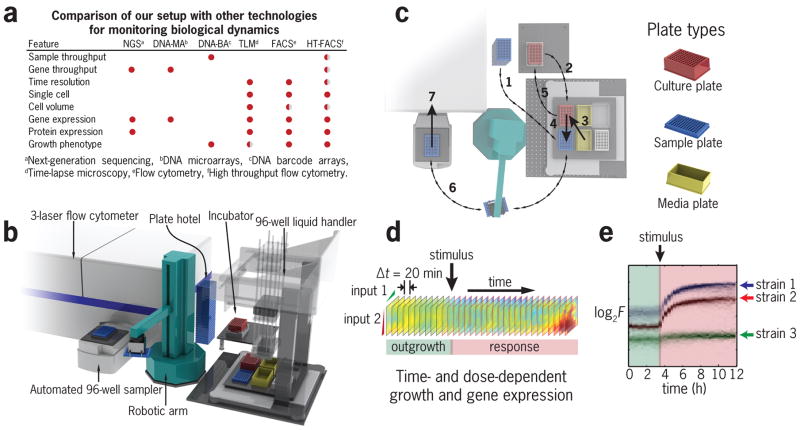
Simultaneous high-throughput measurement of growth and gene expression is challenging. For example, simple bulk growth and fluorescence measurements using plate readers suffer from poor reproducibility10,11. Although substantial progress has been made in time resolution using microfluidics12 and chemostats13,14, these technologies are limited in their ability to achieve simultaneous growth and gene expression measurements in high throughput at the single-cell level (Fig. 1a). To enable such measurements, we developed a measurement setup that integrates a flow cytometer, a liquid handler and a deep-well plate incubator using a robotic arm and custom control software (Fig. 1b, see Online Methods for implementation details). Briefly, using custom software, robotic and fluidic capabilities, samples of a culture are continuously transferred to a shallow 96-well plate, which is then moved by the robot to the flow cytometer for measurement (Fig. 1c). This capability enables us to repeat the above sequence of events to carry facile and reproducible stimulus-response experiments to explore phenotypes across time and stimulus dose (Fig. 1d). A typical experiment consists of two stages: an outgrowth phase where cells are brought to exponential growth, followed by a stimulus event where a treatment solution is added after the appropriate growth state has been achieved. Following the stimulus, we continuously monitor the culture evolution during the response period (Fig. 1e). Treatment and/or culture conditions are automatically maintained through the experiment (up to 24 hours) by adding the stimulus at its nominal concentration to compensate for dilution (Supplementary Fig. 1a). Different stimuli like pulses, nutrient depletions and ramps can also be easily implemented with a high degree of sample-to-sample and day-to-day reproducibility (Supplementary Fig. 1b–c). Additionally, several strains can be simultaneously cultured (multiplexed) in one well for internally controlled measurements of differential phenotypes under a large number of conditions.
Figure 1. Automated cell culture incubator with real-time flow cytometry readout.
(a) Comparison of our HT-FACS robotic setup with other technologies for monitoring cell physiology. (b) Layout showing hardware arrangement for our system. (c) Sequence of main events that occur in a typical experiment. These include liquid transfers and plate transport that take place during the acquisition of one measurement. Steps 1–7 repeat every 20 minutes and involve (1) loading a fresh sample plate into the liquid handler, (2) removal of the culture from the incubator, (3) addition of fresh media and stimulus to the culture, (4) removal of a sample from the culture, (5) returning the culture to the incubator, (6) transport of the sample plate to the high throughput sampler (HTS) and (7) measurement of the sample plate in the flow cytometer. (d) Time and dose-dependent dynamical portraits can be acquired with our system to characterize regulatory networks. (e) Example of one of 96 possible conditions in which the fluorescence of 3 strains is monitored over time after a perturbation.
Quantification of protein expression rates
A confounding factor in interpreting protein synthesis rates using fluorescent reporters is the interdependence between the measured fluorescence and growth dynamics when perturbations impact growth. Here, we define growth slowdown as the decrease in the rate of cell division as measured by the time dependent accrual of the number of cells. Since the concentration of any cellular moiety can be affected by its turnover dynamics15,16 and the dynamics of cell growth, slowdown after stress may cause fluorescent proteins to accumulate17–19, resulting in inflated estimates of the extent of gene expression or rate of protein synthesis. In particular, we studied the instantaneous growth phenotype associated with increasing doses of tunicamycin-induced endoplasmic reticulum (ER) stress in the yeast S. cerevisiae. When cells are exposed to tunicamycin, they turn on the expression of about 200 genes in what is known as the unfolded protein response20–23 or UPR. Concomitantly, cells slow down growth and reduce the translational flux of proteins targeted to the secretory pathway, presumably as a way to avoid the accumulation of misfolded proteins24,25. To quantify the cell’s response to this ER stress, we used a transcriptional reporter consisting of a synthetic promoter that contains four copies of the UPRE-1 motif fused to a GFP coding sequence26.
The rate of change of fluorescence in the culture is the result of production and disappearance of the fluorophore:
| (1) |
FGFP,total is the total instantaneous fluorescence in the culture, α4XUPRE,total(t) is the protein synthesis rate associated with the p4XUPRE-GFP reporter and βGFP,total(F,t) is the degradation flux of the fluorescent protein. Since FGFP,total = N(t) × 〈FGFP,cell〉, where N(t) is the number of cells in the culture and 〈FGFP,cell〉 is the instantaneous average fluorescence per cell in the population, we write a general expression for the average protein synthesis rate per cell in terms of the measured fluorescence and cell number (See Supplementary Note for detailed derivation):
| (2) |
In this equation, the impact of growth rate on the change of GFP per cell is captured by the cell division rate γ(t), defined as the time derivative of the logarithm of the measured number of cells, dlogN/dt (Fig. 2a). A similar equation can be derived for the change of GFP concentration (GFP per unit volume), in which the change of cell volume also contributes to the dilution (see Online Methods). This contribution can be quantified by the cell volume accumulation rate θ(t), defined as dlog〈V〉/dt (where 〈V〉 is the average cell volume), which can be a significant contribution to biomass accrual rate when cells abruptly stop dividing (Fig. 2b). The average cell volume can be estimated using the cell’s light scattering (see Online Methods, Supplementary Fig. 2). However, the relationship between volume27–29 or cell cycle30 and scatter parameters can be complex and should be calibrated for each cell type and experimental conditions.
Figure 2. The automated FACS technology allows for facile and reproducible growth rate-corrected determination of reporter protein synthesis rate, and degradation fluxes as a function of time.
(a) Instantaneous cell division rate of cells stressed with tunicamycin (Tm). Cell division rates decrease in a dose-dependent way but (b) the average volume increases. (c) Average fluorescence measurement of the p4XUPRE-GFP transcriptional reporter, (d) Growth corrected expression rate of p4XUPRE-GFP. (e) Average fluorescence of pTDH3-mKate2. TDH3 is a housekeeping gene (f) Growth corrected expression rate of pTDH3. (g) Distributions of raw fluorescence across time for a mixture of two strains in a competition experiment where they are distinguished by the presence or absence of a pTDH3-mKate2 fluorescent tag (left). Distributions of green fluorescence in the two strains, containing GFP (red population) or a short-lived allele Ub-Tyr-GFP (blue population), both driven by the UPR synthetic promoter 4XUPRE (right). (h) Raw average fluorescence for the unstable GFP allele for different doses of ER stress induced by addition to tunicamycin. (i) Differential degradation flux of the unstable allele Ub-Tyr-GFP as a function of time. Experiment was replicated twice in the laboratory.
Equation (2) indicates that the instantaneous rate of change of fluorescence, d〈FGFP,cell〉/dt (Fig. 2c,e), is equal to the balance of the instantaneous protein synthesis rate α4XUPRE,cell(t) (Fig. 2d), its decrease due to cell division −γ(t)×〈FGFP,cell〉 and its disappearance by degradation βGFP,cell(F,t). If the fluorescent reporter is stable, its “degradation flux” βGFP,cell(F,t) is negligible. In this case, the instantaneous protein synthesis rate can be extracted from the fluorescence measurements and cell division rate, both measured in the same experiment. The estimated instantaneous protein synthesis rate α4XUPRE,cell(t) is a time-dependent, population-averaged effective rate of production of the fluorescent protein that can be interpreted as a lumped rate of transcription, translation and folding.
Our data clearly indicate that following perturbation by tunicamycin, the UPRE reporter undergoes a transient pulse of expression whose magnitude and duration depends on the extent of the stress (α4XUPRE,cell(t) in Fig. 2d). In particular, for low and medium doses of stress, these measurements trace the attenuation of UPR activation following homeostatic recovery. For high doses, some UPR attenuation still occurs. A colony counting assay reveals that cell death does not occur at these doses (Supplementary Fig. 3a,b). As a result, this UPR attenuation is not due to cell death but likely the consequence of global gene expression and growth arrest. The results of the colony-counting assay also quantitatively validate the growth measurements obtained by our setup (Supplementary Fig. 3c).
Raw fluorescence measurements of a transcriptional reporter for TDH3, a housekeeping gene31, show a stress-induced dose-dependent increase (Fig. 2e). By contrast, the growth-corrected instantaneous protein synthesis rate of pTDH3-mKate2 shows no change at early times for any dose (Fig. 2f) and data becomes noisy at later time-points for high doses of stress due to low cell counts (see Supplementary Fig. 4 for replicate data with higher starting OD and cell count). Importantly, this instantaneous protein synthesis rate measurement does not depend on any normalization, in contrast to previous measures of gene expression that need to be normalized by the total RNA abundance or a control that is assumed to be constitutively expressed. Taken together, these novel data highlight the fact that complex growth dynamics during perturbation experiments need to be integrated into analyses for the quantitative determination of gene expression profiles.
Quantification of relative protein degradation rates
Control of protein degradation is a crucial layer of regulation determining effective gene dosage for many genes32–35. Our analysis can be easily extended to calculate relative degradation flux for unstable proteins.
We co-cultured two strains containing the synthetic UPR promoter. In the first strain, the promoter is driving a long-lived GFP. In the second strain, the same promoter is driving an unstable GFP allele, Ub-Tyr-GFP36 containing a destabilizing Tyr residue37 that is unmasked by UBP138,39. We distinguished the two strains using an mKate240 fluorescent label: the stable GFP strain expresses mKate2 constitutively from a TDH3 promoter while the strain containing the Ub-Tyr-GFP allele lacks mKate2 expression (Fig. 2g). The basal fluorescence intensity of the unstable allele was lower than that of the stable allele and, following induction by of tunicamycin, it decreased after peaking. Both strains have the same genetic background and are in the same culture. Furthermore, reporters are under control of the same promoter, (see Supplementary Table 2, Supplementary Fig. 5a) and are subject to the same environmental conditions. As a result, we assume that the decrease in fluorescence is reflective of the faster degradation of the unstable allele (Fig. 2h). In this scenario, we can use a differential form of the protein turnover model (1) to compute a relative dose-dependent degradation flux Δβ per cell for the unstable allele (Fig. 2i, also see Supplementary Note):
| (3) |
Where Δβ(t) = βUb-Tyr-GFP,cell(F,t)-βGFP,cell(F,t), Δ(d〈F〉/dt) = d〈FUb-Tyr-GFP,cell〉/dt - d〈FGFP,cell〉/dt and Δ〈F〉 = 〈FUb-Tyr-GFP,cell〉-〈FGFP,cell〉. For a stable reference protein, this expression implies that βUb-Tyr-GFP,cell ≈ γ×Δ〈F〉 at steady state. Using the expression in Equation (3), the calculated degradation flux increases under ER stress in a dose- and time-dependent fashion (Fig. 2i). Such regulation of the degradation flux has been observed during ER stress22,41–44. Our measurements provide a quantitative and dynamic window into the regulation of this ER-associated degradation (ERAD)42,45 and its relationship with cytosolic protein degradation. Furthermore, since degradation fluxes are a function of substrate concentration 〈FUb-Tyr-GFP,cell〉, these measurement enable us to establish the regime in which the degradation of a given substrate has first-order kinetics (Supplementary Fig. 5b). This general approach, as opposed to methods based on bleaching and recovery46,47, enables the quantification of protein degradation in vivo, quantitatively and in real-time. Unlike traditional pulse-chase approaches36, our approach is not destructive48 and since it does not have the dead-time limitation of bleaching-based microscopy approaches47, it allows for transient measurements of degradation flux.
Automated measurement of multidimensional dose responses
Measurement of dose-responses is a powerful tool to establish the input-output mapping of biological modules, such as gene promoters49,50 or stress pathways1, and for the efficient constraining of complex dynamical models51,52. Furthermore, combinatorial stimulation is emerging powerful approach for interrogating the logic of cellular pathways53. As a first proof of concept, we measured the dose-response dependency of the UPR reporter protein synthesis rate for many combinations of ER stress induced by addition of tunicamycin (Tm) and synthetic transcriptional activation of the system (Fig. 3a). We achieved synthetic activation using an estradiol-inducible system54–56 where the addition of the drug estradiol (E2) at different doses induces the expression of an active allele of HAC1, HAC1i, the main transcription factor controlling the UPR. In this experiment, we multiplexed two strains, both of which contain an estradiol-inducible system driving expression of the HAC1i allele. In addition, one of the strains contains a stable allele of GFP driven by a synthetic UPR reporter, while a second strain contains a de-stabilized allele of GFP. This second strain is labeled by pTDH3-mKate2. For these two strains, multiplexed in the same well, we measured time-dependent growth, reporter protein synthesis rate and degradation flux for 96 combinations of the two inputs, E2 and Tm (Fig. 3b). For easy visualization, we summarized these quantities by time-averaging for three hours after induction (averaging window is depicted in gray in Fig. 2, panels c,d,e,f,h and i) in order to establish the 2D dose responses of four different phenotypic variables αTDH3, α4XUPRE, γ and Δβ (Fig. 3c).
Figure 3. Automated FACS technology allows for facile and reproducible determination of growth rate, gene expression rate, degradation fluxes, and cell-to-cell variability as a function of time and dosage.

(a) Perturbation strategy involving the perturbation the UPR pathway using an environment input (tunicamycin) and a synthetic input (an inducible spliced allele of HAC1). (b) Experimental design making use of the construction of a full two-dimensional dose response for 96 combinations of these two inputs. (c) Protein synthesis rate of the 4XUPRE (α4XUPRE), protein synthesis rate of pTDH3 (αTDH3), growth phenotype (γ) and differential degradation flux (Δβ) for every dose of tunicamycin and estradiol (E2). The data are reported as an average over a time window of 3 hours. (d) Time-dependent distributions of single-cell green fluorescence, red fluorescence, forward-scatter and side-scatter for no treatment (left), 80 nM estradiol (center) and 5 μg/mL tunicamycin (right).
As expected, the rate of protein synthesis from the UPR reporter (α4XUPRE) increases both with expression of HAC1i and tunicamycin-induced ER stress (Fig. 3c, top left). Nonetheless, synthetic activation with the inducible system generates a slightly lower UPR reporter protein synthesis rate than stress-induced activation. Simultaneous activation with both ER stress and HAC1i expression yields a similar protein synthesis rate as activation with ER stress alone. This is consistent with a model in which both ER stress and the dose of HAC1i expression modulate the UPR independently even in the presence of different growth phenotypes. Here again, the growth-corrected TDH3 rate of protein synthesis is constant after stress or UPR over-activation (Fig. 3c, top right). This, together with the observation that cells are viable after removal of stress in our plating assay (Supplementary Fig. 3b), reveals that the cell’s protein synthesis capacity is not saturated in these regimes and suggests that the observed growth phenotype results from arrest in the cell cycle and not a failure to accumulate biomass.
Moreover, while ER stress causes a major growth defect (Fig. 3c, bottom left), HAC1i overexpression caused only a transient pause in cell division rate γ(t) (Supplementary Fig. 3d, top) that is offset by cell volume growth θ(t) (Supplementary Fig. 3d, middle, bottom). Degradation flux was also dependent on both Tm and E2 dosage consistent with a role for the UPR in modulating the degradation flux across all doses of stress (Fig. 3c, bottom right).
Quantification of cell-to-cell variability
The regulatory response of a population of cells to environmental changes is often as accompanied by a change in population structure that reflects changes in cell-cycle and cellular growth. A unique strength of our flow-cytometry setup is that it allows for quantifying both cell-to-cell variability in a population and changes in population structure as a function of time. For example, examination of p4XUPRE and pTDH3 fluorescence as a function of tunicamycin and estradiol doses shows the temporal emergence and disappearance of multimodal distributions (Fig. 3d, Supplementary Fig. 7 and 8). At the same time, measurement of forward scatter (FSC) and side-scatter signals (SSC) reveals complex dynamics resembling the reporter protein synthesis rate of the constitutive gene (Supplementary Fig. 7c–d/8c–d). As a result, correction by SSC for example, removes the multimodality and points to its root in the interplay of stress and growth (Supplementary Fig. 7e–f). Furthermore, our measurements indicate that while the same steady-state mean fluorescence of p4XUPRE can be achieved by different inputs, the variability around this mean can be different based on the nature of the stimulus (estradiol or tunicamycin, Fig. 3d and Supplementary Fig. 11a). Quantification of cell-to-cell variability by the coefficient of variation (CV) as a function of mean for different doses of tunicamycin (Supplementary Fig. 9a) and estradiol (Supplementary Fig. 9b) reveals that even for the same input, fluorescence trajectories can traverse the same mean value more than once, but that the population distributions of these transient states can be different (Supplementary Fig. 10b).
More quantitatively and using single cell data for all conditions and timepoints, we observe that on average the CV2 of the UPRE reporter fluorescence decreases as an inverse function of its mean at low levels of expression and reaches a noise floor at high expression levels as expected from dominance of extrinsic cellular noise at these values (Supplementary Fig. 11). Dissection of this phenomenon, however, uncovers a fine microstructure in which the variability has different dependence on the physical population parameters, such as side- and forward-scatter (Supplementary Fig. 12) based on the mode of stimulation in the system. Specifically, we found that systematic over-dispersion of the variability over the Poisson limit is present at smaller cell sizes when the system is stimulated by tunicamycin, but not by estradiol (Supplementary Fig. 13). These data point to an increase in global cellular variability under stressful conditions.
Discussion
In this work, we developed a novel technology to simultaneously measure the quantitative temporal profiles of molecular phenotypes and growth dynamics in stimulus-response experiments. These datasets can be exploited by different means to study important layers of cellular regulation. By combining several subpopulations in the same well by mixing strains with different genetic backgrounds (or when subpopulations arise as a consequence of cell-to-cell variability), we were able to dissect and compare phenotypes like relative protein stability and growth rates. We also leveraged the high throughput nature of our measurements to establish entire time-dependent output surfaces for a system as a function of dual-perturbations. An important advantage of our technology is that it documents phenotypic variables in single cells, and hence provides their distribution across a population. For example, we show that the cell-to-cell variability can be quantified across cell size to identify different physiological regimes. Future exploitation of these dynamically evolving multivariate distributions will help to uncover the quantitative features of the underlying regulatory processes57,58, including the cell-cycle dependencies of gene expression and protein degradation59–62. Finally, we were able to monitor cell growth in terms of both division rates and a surrogate of cell volume, enabling us to quantify different contributions to cell growth fluctuations. We expect that this technology will be instrumental for in vivo and dynamic studies of protein turnover, multi-species ecology, and dynamic mapping of genetic interactions.
Online Methods
Automated Flow Cytometry Hardware Setup
The automated flow cytometry hardware setup consists of a 3-laser flow cytometer (LSR II, Beckton-Dickinson Co.), a liquid handler (Multimek 96, Beckman Coulter) and an open deep well plate magnetic heater/shaker (Variomag Teleshake, Inheco GmbH). Plates are transferred between these three devices with a robotic arm (Plate Crane XL, Hudson Robotics Inc.). All hardware was secured on a steel breadboard and partially enclosed with an aluminum frame to decrease temperature fluctuations and reduce contamination. Cell cultures are grown in a 1mL 96-well polystyrene plate (Riplate®, Ritter GmbH, Schwabmünchen, Germany) that is agitated at 900 rpm and kept at 30°C in heater/shaker.
In our setup, the liquid handler takes a 10–100 μL sample (sample volume, Vs) of the 400–600 μL cell culture (culture volume, Vc) every 10–20 minutes (sampling frequency, ts). A volume of fresh media equal to the sample volume is added at every time-point to maintain the culture volume constant. The sample is then placed in a second shallow 96-well microplate (Model 3795, Corning Inc.) containing 70 uL of Tris-EDTA buffer (pH = 7.4). The diluted samples are then measured using a high throughput sampler (BD® High Throughput Sampler 338301, Beckton-Dickinson Co.) to inject samples into the flow cytometer. The parameters ts, Vs and Vc deter mine an effective culture dilution rate (d2) that can be arbitrarily and dynamically set for each experiment so as to achieve the desired concentration of cells and to accommodate different growth rates (Supplementary Fig.1). The values used in our experiments are Vc = 500 μL, Vs = 30 μL and ts = 20 min, resulting in d2 = 0.94. This equals a mean residence time of a volume element in the culture (t½) of approximately 4 hours, which means that the observed number of cells of a strain growing with a doubling time of 4 hours will remain constant, while strains that are faster or slower either accumulate or are washed away.
The stimulus event, which takes place at t0, consists of the media plate being instantaneously swapped for a media plate containing the stimulus at a titer that brings the effective concentration of stressor to 1X in the culture. This titer equals the dilution factor of the media (d1) multiplied by the desired concentration of stressor (d1X). This is followed for later dilution events with a media plate with 1X titer for the response phase. A typical experiment consists of an outgrowth phase of 3 hours followed by response phase lasting 8 hours.
Pipette tips are reused through the experiment and washed with 50% ethanol and water between samples.
Reproducibility experiments show no carry over and measurements of cell division rate and fluorescence are reproducible to better than a few percent with no internal controls (Supplementary Figure 1b–d). Although we do not exploit this in the data presented here, the use of internal controls and other experimental designs decreases error substantially. Since these figures are dependent on the particular experimental design and data analysis strategy we report reproducibility data only on the most basic measurements. Although there is no appreciable bias or unevenness in the heating of individual wells as measured using a thermocouple sensor, there is a reproducible vertical temperature gradient of about 1 degree in each well (data not shown).
Software and data processing
We used a custom-written software for data acquisition and control of each piece of hardware independently and their coordination (Labview®, National Instruments Corp.). A personal computer runs concurrent threads that control the flow cytometer, its high throughput sampler, the liquid handler, the robotic arm and the heater/shaker where cells grow. Sensors embedded in each piece of hardware report on their individual state and allow for a master thread to coordinate the sampling, dilution and measurement of cell density and fluorescence using the flow cytometer. An Ethernet connection to the flow cytometer, set up to run continuously, provides access of the stream of data events that make up the raw flow cytometry data set. The continuous data stream is displayed in real time as it is acquired and simultaneously stored as individual binary data files for offline analysis. Offline data analysis is performed using custom scripts that histogram all various parameters and computes summary statistics like fluorescence distributions and equivalent cell densities (Matlab R2012b, Mathworks). These data were first processed to remove outliers by removing events with no fluorescence, events with forward scatter values of less than 5000 and events out of a four standard deviation of the joint side and forward scatter. Fluorescence was corrected by cell size utilizing the SSC values for each event1. Using cell sorting and automated microscopy, we found that the SSC parameter is a good surrogate of cell volume (Supplementary Fig. 2) and thus is a good surrogate for cell size. Cell densities are estimated from the (Poisson) rate at which cells enter the flow cytometer. We do this by fitting an exponential distribution to inter-arrival times (τi) for each well to deter mine the mean <τ>. Cell density N is then calculated as (<τ>*L)−1, where L is the sample injection flow rate (usually 1 μL/s).
Plasmids and strains
All plasmids used in this study are listed in Supplementary Table 1. The Ub-Tyr-GFP2 construct is driven by a crippled CYC1 promoter containing 4 cis-acting UPRE motifs. The 4XUPRE synthetic promoter contains four copies of the UPRE motif CAGCGTG3, which is a known binding target for the transcription factor Hac1p. All plasmids used in this work are single genomic integration plasmids.
The promoter of CYC1 was amplified from S. cerevisiae (W303a strain) genomic DNA with Elongase Enzyme Mix (Life Technologies) and cloned between PspOMI and XhoI sites in a pNH605 plasmid. GFP was then cloned between XhoI and BamHI sites. The region containing the UPRE elements was amplified with Elongase Enzyme Mix (Life Technologies) from the Ub-Tyr-GFP plasmid and was cloned into KpnI and PspOMI restriction sites, resulting in plasmid pAAD53. Stress reporters were constructed by amplifying the promoters of SSA1, HSP12, HSP82, HOR2, GPD1 and ERO1 from genomic DNA and cloned between PspOMI and XhoI sites in a plasmid containing the GFP ORF, resulting in plasmids pAAD7-12. The Splicing Reporter is a modified HAC1 construct, in which the first exon has been replaced by GFP4. The barcoding construct was assembled by cloning the promoter of TDH3 between PspOMI and XhoI restriction sites followed by mKate2 between XhoI and BamHI sites into a plasmid containing the HIS3 coding sequence from C. albicans. This plasmid was amplified with primers containing homology regions to the CAN1 locus and then transformed into yeast to get the barcoded strain. The rest of the plasmids were sequentially transformed into the W303a strain by linearizing them and making the yeast cells competent with a standard lithium acetate method. All the resulting strains are listed in Supplementary Table 2.
Growth conditions
Starting from single colonies picked from YPD (yeast extract, peptone, 2% w/v glucose) agar plates, yeast cultures were inoculated and grown for 24 hours in exponential phase at 30 °C in YPD liquid media. Before the multiplexed experiment, the two strains were combined at an approximate ratio of 1:1 and diluted to a total of a density of 106 cells/mL. 500 μL of the mix were transferred into a 96 deep-well polycarbonate plate.
Before the start of the experiment, cells are continuously grown while diluting them with fresh YPD media that does not contain any stimulus. Stimuli are applied by subjecting to combinations of an exogenous inducer (β-estradiol, Sigma-Aldrich) and an ER-stressor (tunicamycin, Calbiochem). Serial dilutions of a 33.3X solution of either estradiol or tunicamycin were combined in equal amounts into a 96-well 2 mL block, resulting in a 16.6X set of solutions. These solutions replaced the fresh media in the liquid handler in the stimulus event. A further 1:16.6 dilution was made to obtain the 1X solution that was used to keep the titer constant during the response phase.
To confirm that cells do not undergo stress by exposure to the liquid handling shear forces associated with constant dilution, we measured the fluorescence of several stress-responsive transcriptional reporters that reflect the activity of various stress-responsive transcription factors for ER stress (hac1 splicing rate4, pERO1 and p4XUPRE transcription), general stress (pHSP12), heat-shock (pSSA1 and pHSP82) and high-osmolality (pGPD1, pHOR2). Supplementary Figure 6 shows time-course data with no treatment (control, left), UPR activation (80 nM estradiol, center) and ER stress (5 μg/mL Tunicamycin, right). These data demonstrate that liquid handling and the various manipulations in our setup do not elicit stress responses, as sensed by these various pathways.
Inducible heterologous gene expression system
The inducible system consists of a chimeric transcription factor construct (plasmid pPW2078) and a gene expression construct (pPW2085). Both of which are integrated into the genome sequentially and in different loci.
The chimeric transcription factor (GERM construct) is a fusion of the Gal4p DNA binding domain (GAL4[DBD]), the human estradiol receptor lipid binding domain (ER[LBD]) and the MSN2 activation domain (MSN2[AD]) (GERM). The expression of the GERM construct is driven by the ADH1 promoter1. The gene expression system consists of the intron-less allele of HAC1 (HAC1i, an active mRNA form of HAC1 not subject to Ire1 regulation5,6) coding sequence driven by the GAL1 promoter. Upon estradiol addition, the GERM transcription factor localizes into the nucleus and activates the transcription of genes driven by Gal4p binding site-containing promoters, including the HAC1i construct.
Cell Size Measurements
Forward (FSC) and side scatter (SSC) were measured and used as a proxy for cell volume (Supplementary Fig.2a). To establish the correspondence between flow cytometry parameters and cellular parameters, an exponentially growing cell population was sorted and fixed using 3% formaldehyde. Cells were sorted based on their forward scatter (FSC-A) value and their volume was estimated using bright field microscopy with a 40X objective. Cells were automatically located in the image and their volume was estimated by revolving their contour along their longest axis of symmetry. The plot in (Supplementary Fig.2b) shows the estimated robust mean of the cell volume as a function of the centroid of the FSC-A gate (blue trace). For reference purposes, the population histogram is shown (green).
Cell viability assay
Five TE-diluted samples from our setup were plated into YPD agar plates 50uL. These corresponded to the cells exposed to the individual stimuli and combinations of them, as well as a control. Plates were left in an incubator at 30ºC and after two days images were taken. Colony forming units were counted from the images with an automated image analysis taking an equivalent squared-area from each of the pictures (Supplementary Fig.3).
Supplementary Material
Acknowledgments
We would like to thank J. DeRisi (UCSF) for useful conversations, engineering advice as well as access to equipment. We thank J. Stewart-Ornstein (UCSF) for the use of the estradiol-inducible system, D. Pincus (UCSF) and the Walter lab for the HAC1i construct. V. Chubukov and Chen-Shan Chin provided early insight on reactor design and flow cytometry interfacing. This work was funded by the NIGMS system biology center (P50 GM081879), the David and Lucille Packard Foundation (HES and HL) and NIH grants R01-GM070808 (HL).
Footnotes
Author contributions
IZ, HES and HL conceived the hardware setup. IZ designed, implemented and characterized the hardware setup, control software and mathematical framework. IZ and AAD designed and carried over the experiments. IZ, AAD, HL and HES analyzed and interpreted the data. AAD cloned the strains necessary for the experiments. IZ, AAD, HL and HES prepared the manuscript.
The authors declare no competing financial interests.
Bibliography
- 1.Zhang Q, Andersen ME. Dose response relationship in anti-stress gene regulatory networks. PLoS Comput Biol. 2007;3:e24. doi: 10.1371/journal.pcbi.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muzzey D, van Oudenaarden A. Quantitative time-lapse fluorescence microscopy in single cells. Annu Rev Cell Dev Biol. 2009;25:301–27. doi: 10.1146/annurev.cellbio.042308.113408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calvert MEK, Lannigan Ja, Pemberton LF. Optimization of yeast cell cycle analysis and morphological characterization by multispectral imaging flow cytometry. Cytometry A. 2008;73:825–33. doi: 10.1002/cyto.a.20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–6. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 5.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–9. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pierce SE, et al. A unique and universal molecular barcode array. Nat Methods. 2006;3:601–3. doi: 10.1038/nmeth905. [DOI] [PubMed] [Google Scholar]
- 7.Cookson S, Ostroff N, Pang WL, Volfson D, Hasty J. Monitoring dynamics of single-cell gene expression over multiple cell cycles. Mol Syst Biol. 2005;1:2005.0024. doi: 10.1038/msb4100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacquet M, Renault G, Lallet S, De Mey J, Goldbeter A. Oscillatory nucleocytoplasmic shuttling of the general stress response transcriptional activators Msn2 and Msn4 in Saccharomyces cerevisiae. J Cell Biol. 2003;161:497–505. doi: 10.1083/jcb.200303030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newman JRS, et al. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–6. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- 10.Blomberg A. Measuring growth rate in high-throughput growth phenotyping. Curr Opin Biotechnol. 2011;22:94–102. doi: 10.1016/j.copbio.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 11.Zaslaver A, et al. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods. 2006;3:623–8. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- 12.McKenna BK, Evans JG, Cheung MC, Ehrlich DJ. A parallel microfluidic flow cytometer for high-content screening. Nat Methods. 2011;8:401–3. doi: 10.1038/nmeth.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chin CS, Chubukov V, Jolly ER, DeRisi J, Li H. Dynamics and design principles of a basic regulatory architecture controlling metabolic pathways. PLoS Biol. 2008;6:e146. doi: 10.1371/journal.pbio.0060146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levy S, et al. Strategy of transcription regulation in the budding yeast. PLoS One. 2007;2:e250. doi: 10.1371/journal.pone.0000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schimke RT, Doyle D. Control of enzyme levels in animal tissues. Annu Rev Biochem. 1970;39:929–76. doi: 10.1146/annurev.bi.39.070170.004433. [DOI] [PubMed] [Google Scholar]
- 16.Belle A, Tanay A, Bitincka L, Shamir R, O’Shea EK. Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci U S A. 2006;103:13004–9. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leveau JH, Lindow SE. Predictive and interpretive simulation of green fluorescent protein expression in reporter bacteria. J Bacteriol. 2001;183:6752–62. doi: 10.1128/JB.183.23.6752-6762.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Subramanian S, Srienc F. Quantitative analysis of transient gene expression in mammalian cells using the green fluorescent protein. J Biotechnol. 1996;49:137–51. doi: 10.1016/0168-1656(96)01536-2. [DOI] [PubMed] [Google Scholar]
- 19.Warner JB, Lolkema JS. LacZ-promoter fusions: the effect of growth. Microbiology. 2002;148:1241–3. doi: 10.1099/00221287-148-5-1241. [DOI] [PubMed] [Google Scholar]
- 20.Kawahara T, Yanagi H, Yura T, Mori K. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol Biol Cell. 1997;8:1845–62. doi: 10.1091/mbc.8.10.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell. 2010;40:238–52. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 22.Haynes CM, Titus Ea, Cooper Aa. Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol Cell. 2004;15:767–76. doi: 10.1016/j.molcel.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 23.Travers KJ, et al. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–58. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 24.Steffen KK, et al. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics. 2012;191:107–18. doi: 10.1534/genetics.111.136549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delic M, et al. Oxidative protein folding and unfolded protein response elicit differing redox regulation in endoplasmic reticulum and cytosol of yeast. Free Radic Biol Med. 2012;52:2000–12. doi: 10.1016/j.freeradbiomed.2012.02.048. [DOI] [PubMed] [Google Scholar]
- 26.Pollard MG, Travers KJ, Weissman JS. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol Cell. 1998;1:171–82. doi: 10.1016/s1097-2765(00)80018-0. [DOI] [PubMed] [Google Scholar]
- 27.Latimer P. Light scattering vs. microscopy for measuring average cell size and shape. Biophys J. 1979;27:117–26. doi: 10.1016/S0006-3495(79)85206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzur A, Moore JK, Jorgensen P, Shapiro HM, Kirschner MW. Optimizing optical flow cytometry for cell volume-based sorting and analysis. PLoS One. 2011;6:e16053. doi: 10.1371/journal.pone.0016053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mullaney PF, Dean PN. The small angle light scattering of biological cells. Theoretical considerations. Biophys J. 1970;10:764–72. doi: 10.1016/S0006-3495(70)86334-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Münch T, Sonnleitner B, Fiechter A. The decisive role of the Saccharomyces cerevisiae cell cycle behaviour for dynamic growth characterization. J Biotechnol. 1992;22:329–51. doi: 10.1016/0168-1656(92)90150-8. [DOI] [PubMed] [Google Scholar]
- 31.Delgado ML, et al. The glyceraldehyde-3-phosphate dehydrogenase polypeptides encoded by the Saccharomyces cerevisiae TDH1, TDH2 and TDH3 genes are also cell wall proteins. Microbiology. 2001;147:411–7. doi: 10.1099/00221287-147-2-411. [DOI] [PubMed] [Google Scholar]
- 32.Pratt JM, et al. Dynamics of protein turnover, a missing dimension in proteomics. Mol Cell Proteomics. 2002;1:579–91. doi: 10.1074/mcp.m200046-mcp200. [DOI] [PubMed] [Google Scholar]
- 33.Bachmair A, Varshavsky A. The degradation signal in a short-lived protein. Cell. 1989;56:1019–32. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 34.Varshavsky A. The early history of the ubiquitin field. Protein Sci. 2006;15:647–54. doi: 10.1110/ps.052012306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lopez AD, et al. Proteasomal degradation of Sfp1 contributes to the repression of ribosome biogenesis during starvation and is mediated by the proteasome activator Blm10. Mol Biol Cell. 2011;22:528–40. doi: 10.1091/mbc.E10-04-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varshavsky A. Ubiquitin fusion technique and related methods. Methods Enzymol. 2005;399:777–99. doi: 10.1016/S0076-6879(05)99051-4. [DOI] [PubMed] [Google Scholar]
- 37.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–86. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 38.Bollenbach T, Quan S, Chait R, Kishony R. Nonoptimal microbial response to antibiotics underlies suppressive drug interactions. Cell. 2009;139:707–18. doi: 10.1016/j.cell.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker RT, Tobias JW, Varshavsky A. Ubiquitin-specific proteases of Saccharomyces cerevisiae. Cloning of UBP2 and UBP3, and functional analysis of the UBP gene family. J Biol Chem. 1992;267:23364–75. [PubMed] [Google Scholar]
- 40.Shcherbo D, et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem J. 2009;418:567–74. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–57. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen M, Gutierrez GJ, Ronai Za. Ubiquitin-recognition protein Ufd1 couples the endoplasmic reticulum (ER) stress response to cell cycle control. Proc Natl Acad Sci U S A. 2011;108:9119–24. doi: 10.1073/pnas.1100028108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanna J, Meides A, Zhang DP, Finley D. A ubiquitin stress response induces altered proteasome composition. Cell. 2007;129:747–59. doi: 10.1016/j.cell.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 44.Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280:31582–6. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- 45.Carvalho P, Stanley AM, Rapoport Ta. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143:579–91. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yewdell JW, Lacsina JR, Rechsteiner MC, Nicchitta CV. Out with the old, in with the new? Comparing methods for measuring protein degradation. Cell Biol Int. 2011;35:457–62. doi: 10.1042/CBI20110055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eden E, et al. Proteome half-life dynamics in living human cells. Science. 2011;331:764–8. doi: 10.1126/science.1199784. [DOI] [PubMed] [Google Scholar]
- 48.Lévy F, Johnsson N, Rümenapf T, Varshavsky A. Using ubiquitin to follow the metabolic fate of a protein. Proc Natl Acad Sci U S A. 1996;93:4907–12. doi: 10.1073/pnas.93.10.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson JC, Voigt Ca, Arkin AP. Environmental signal integration by a modular AND gate. Mol Syst Biol. 2007;3:133. doi: 10.1038/msb4100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Müller D, Stelling J. Precise regulation of gene expression dynamics favors complex promoter architectures. PLoS Comput Biol. 2009;5:e1000279. doi: 10.1371/journal.pcbi.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aldridge BB, Gaudet S, Lauffenburger Da, Sorger PK. Lyapunov exponents and phase diagrams reveal multi-factorial control over TRAIL-induced apoptosis. Mol Syst Biol. 2011;7:553. doi: 10.1038/msb.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Birtwistle MR, et al. Emergence of bimodal cell population responses from the interplay between analog single-cell signaling and protein expression noise. BMC Syst Biol. 2012;6:109. doi: 10.1186/1752-0509-6-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bollenbach T, Kishony R. Resolution of gene regulatory conflicts caused by combinations of antibiotics. Mol Cell. 2011;42:413–25. doi: 10.1016/j.molcel.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, O’Malley BW, Tsai SY. A regulatory system for use in gene transfer. Proc Natl Acad Sci U S A. 1994;91:8180–4. doi: 10.1073/pnas.91.17.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stewart-Ornstein J, Weissman JS, El-Samad H. Cellular noise regulons underlie fluctuations in Saccharomyces cerevisiae. Mol Cell. 2012;45:483–93. doi: 10.1016/j.molcel.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McIsaac RS, et al. Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Mol Biol Cell. 2011;22:4447–59. doi: 10.1091/mbc.E11-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neuert G, et al. Systematic identification of signal-activated stochastic gene regulation. Science. 2013;339:584–7. doi: 10.1126/science.1231456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Munsky B, Trinh B, Khammash M. Listening to the noise: random fluctuations reveal gene network parameters. Mol Syst Biol. 2009;5:318. doi: 10.1038/msb.2009.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Howlett NG, Avery SV. Flow cytometric investigation of heterogeneous copper-sensitivity in asynchronously grown Saccharomyces cerevisiae. FEMS Microbiol Lett. 1999;176:379–86. doi: 10.1111/j.1574-6968.1999.tb13687.x. [DOI] [PubMed] [Google Scholar]
- 60.Kafri R, et al. Dynamics extracted from fixed cells reveal feedback linking cell growth to cell cycle. Nature. 2013;494:480–3. doi: 10.1038/nature11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knijnenburg Ta, et al. A regression model approach to enable cell morphology correction in high-throughput flow cytometry. Mol Syst Biol. 2011;7:531. doi: 10.1038/msb.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown MR, et al. Flow-based cytometric analysis of cell cycle via simulated cell populations. PLoS Comput Biol. 2010;6:e1000741. doi: 10.1371/journal.pcbi.1000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
Bibliography
- 1.Stewart-Ornstein J, Weissman JS, El-Samad H. Cellular noise regulons underlie fluctuations in Saccharomyces cerevisiae. Mol Cell. 2012;45:483–93. doi: 10.1016/j.molcel.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varshavsky A. Ubiquitin fusion technique and related methods. Methods Enzymol. 2005;399:777–99. doi: 10.1016/S0076-6879(05)99051-4. [DOI] [PubMed] [Google Scholar]
- 3.Mori K, Ogawa N, Kawahara T, Yanagi H, Yura T. Palindrome with spacer of one nucleotide is characteristic of the cis-acting unfolded protein response element in Saccharomyces cerevisiae. J Biol Chem. 1998;273:9912–20. doi: 10.1074/jbc.273.16.9912. [DOI] [PubMed] [Google Scholar]
- 4.Pincus D, et al. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. 2010;8:e1000415. doi: 10.1371/journal.pbio.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 6.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–9. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.