Abstract
Acquisition of microbes by the neonate, which begins immediately during birth, is influenced by gestational age and mother’s microbiota and modified by exposure to antibiotics1. In neonates, prolonged duration of antibiotic therapy is associated with increased risk of sepsis after 4 days of life, known as late-onset sepsis (LOS)2, a disorder critically controlled by neutrophils3, but a role for the microbiota in regulating neutrophil behavior in the neonate has not been described. We exposed pregnant mouse dams to antibiotics in drinking water to limit transfer of maternal microbes to the neonates. Antibiotic exposure of dams decreased the total number of microbes in the intestine, altered the structure of intestinal microbiota and changed the pattern of microbial colonization. These changes were associated with decreased numbers of circulating and bone marrow neutrophils and granulocyte/macrophage restricted progenitor cells in the bone marrow. Antibiotic-exposure of dams attenuated the postnatal granulocytosis by reducing the number of interleukin (IL) 17-producing cells in intestine and consequent production of granulocyte colony stimulating factor (G-CSF). Relative granulocytopenia contributed to increased susceptibility of antibiotic-exposed neonatal mice to Escherichia coli K1 and Klebsiella pneumoniae sepsis, which could be partially reversed by administration of G-CSF. Restoration of normal microbiota, through TLR4- and MYD88-dependent mechanism, induced accumulation of IL17-producing type 3 innate lymphoid cells (ILC) in the intestine, promoted granulocytosis, and restored the IL17-dependent resistance to sepsis. Specific depletion of ILCs prevented the IL17- and G-CSF-dependent granulocytosis and resistance to sepsis. These data support a role for the intestinal microbiota in regulation of granulocytosis and host resistance to sepsis in the neonates.
Antibiotic exposure reduces the diversity of intestinal microbiota and delays the appearance of beneficial bacteria in children4; such alteration is associated with development of rheumatoid arthritis, inflammatory bowel disease and obesity5. In neonates, prolonged duration of antibiotic therapy is associated with increased risk of neonatal LOS2. While a role for the microbiota in neonatal LOS has been proposed6, the mechanisms involved are not understood. The intestinal microbiome undergoes dynamic changes during the neonatal period7, and is temporally associated with functional development of the immune system8. To ascertain the role of microbiota in susceptibility of neonates to LOS, we exposed pregnant dams to ampicillin, gentamicin, vancomycin, metronidazole and neomycin in their drinking water beginning 5 days before delivery. The dams and the neonatal mice continued to receive antibiotics for the duration of experiment. Thus neonatal antibiotic exposure refers to antibiotic exposure both in utero and after birth. Antibiotic exposure not only reduced the total number of intestinal microbes, but also modified to composition of intestinal microbiota in neonatal mice (Fig. 1a-b). Gammaproteobacteria transiently dominated the intestinal microbiota in postnatal day 3 mice. Bacilli and Clostridia were the predominant classes in postnatal day 5-14 mice, similar to patterns seen in human neonates9, while Bacteroidia were more prominent by day 14. Perinatal antibiotic exposure not only abolished the appearance of Gammaproteobacteria on day 3, but also prevented development of Bacteroidia on day 14 (Fig. S1a-b). These findings were associated with simplification of intestinal microbiota in antibiotic-exposed neonatal mice (Fig. S1c-f), consistent with observations that antibiotics decrease diversity of intestinal microbiota in human neonates10. Ampicillin, gentamicin, and vancomycin are the most commonly used antibiotics in the pregnant mothers and neonates11. Therefore, we confirmed these observations by exposing pregnant dams to the clinically relevant combination of ampicillin, gentamicin and vancomycin in their drinking water. This 3 antibiotic regimen similarly reduced the total number of intestinal microbes in the neonatal mice (Fig. 1a) and decreased the abundance of Gammaproteobacteria on day 3 and Bacteroidia on day 14, recapitulating the altered microbial composition in neonatal mice exposed to 5 antibiotics (Fig. S1a-b).
Figure 1. Perinatal antibiotic exposure alters the pattern of microbial colonization in the intestine and attenuates the postnatal granulocytosis.
(a) 16S rDNA copy numbers from the intestinal contents of neonatal mice exposed to combination of 3 (3ABX) or 5 (5ABX) antibiotics or no antibiotics (No ABX) was determined using real-time PCR. (b) Relative abundance of phylum and class level commensal bacteria obtained from 16S rDNA pyrosequencing of the intestinal contents of age- and sex-matched neonatal mice exposed to combination of 5 antibiotics (ABX) or no antibiotics (No ABX). Each bar represents the pooled intestinal contents from > 8 age-defined neonatal mice from more than 3 different litters. (c) Age- and sex-matched neonatal mice exposed to combination of 3 (3ABX) or 5 (5ABX) antibiotics or no antibiotics (No ABX) were examined for number of circulating or (d) bone marrow neutrophils. (e-f) Bone marrows from age- and sex-matched neonatal mice exposed to combination of 3 antibiotics (ABX) or no antibiotics (No ABX) were examined for number of hematopoietic stem cells or lineage committed progenitor cells. Flow cytometry plots are gated on live cells. Representative histograms from 3 separate experiments. (g) Age- and sex-matched neonatal mice exposed to combination of 3 (3ABX) or 5 (5ABX) antibiotics or no antibiotics (No ABX) were examined for plasma G-CSF levels. (h) Age-matched germ free (GF) or conventionalized (CNV) mice or neonatal mice exposed to combination of 5 antibiotics (ABX) were examined for number of circulating or (i) bone marrow neutrophils or (j) plasma G-CSF levels. Data are representative of three independent experiments containing 10-12 mice per group. Results are shown as the means ± s.e.m.
Temporally associated with exposure to microbes, human neonates demonstrate increased circulating neutrophils 24-72 h after birth12. We observed a marked increase in circulating neutrophils in neonatal mice (postnatal day 1-3) approaching adult values by postnatal day 14. Perinatal exposure to either combination of the ampicillin, gentamicin, vancomycin, metronidazole and neomycin or the more clinically relevant combination of ampicillin, gentamicin and vancomycin abrogated this postnatal granulocytosis in the early neonatal period (Fig. 1c) as compared to age-matched controls. Premature birth and low birth weight strongly correlate with neutropenia in neonates13, therefore we assessed the effect of perinatal antibiotics on gestational age and birth weight. Antibiotic exposure did not significantly change the gestational age or birth weights or postnatal growth in neonatal mice (Fig. S1g-h).
The failure of postnatal granulocytosis in early neonatal period in antibiotic-exposed mice was accompanied by decreased numbers of bone marrow neutrophils (Ly6G+ cells) (Fig. 1d), decreased numbers of granulocyte/macrophage restricted progenitor (Lin−Sca-1−c-kit+CD16/32−CD34+) cells (Fig. 1e-f, S1i) and decreased levels of plasma G-CSF (Fig. 1g) suggesting impaired granulopoesis in antibiotic-exposed neonatal mice. To determine whether lack of postnatal granulocytosis might reflect antibiotic toxicity, we used germ-free (GF) mice, which have minimal microbiome but no antibiotic exposure. GF mice had lower circulating and bone marrow neutrophils and plasma GCSF (Fig. 1h-j) as compared to age-matched conventionalized (CNV) mice. These data, taken together, suggest that microbiota control postnatal granulocytosis in neonates.
It is unclear if neutrophils from antibiotic-exposed neonatal mice have distinct phenotypic and functional profiles as compared to the control mice. Chemokine receptor (CXCR) 2 and CXCR4 control the release of neutrophils from the bone marrow and their subsequent homing14. Surface expression of CXCR2, was lower in neutrophils from antibiotic-exposed neonatal mice whereas expression of CD11b and CD54, cell surface proteins involved in surface adhesion and transmigration (Fig. S1j-k) was unchanged. Phagocytic ability or ROS production was not significantly different in neutrophils from antibiotic-exposed neonatal mice as compared to control mice (Fig. S1l)
Neutrophils are essential in controlling infection due to E. coli serotype K1, an important cause of LOS and meningitis in human neonates15 and the leading cause of death in preterm infants16. Neutropenia is an important risk factor in fatal neonatal sepsis3. In order to test whether antibiotic exposure modified host defense against infection, neonatal mice (postnatal day 3 or 5 or 7 or 14 old) were exposed to the combination of 5 antibiotics or the more clinically relevant combination of ampicillin, gentamicin and vancomycin through their Dams beginning 5 days before birth and continuing until day 14. Antibiotic-exposed or age-matched (no antibiotic-exposed) controls, were inoculated intraperitoneally with 104 CFU g−1 of E. coli K1 or 106 CFU g−1 of Klebsiella pneumoniae, another important gram negative pathogen in neonates. Neonatal mice exposed to the 5-antibiotic combination or to the 3-antibiotic combination demonstrated marked susceptibility to E. coli K1 as compared to controls (median survival 8 h and 10 h respectively vs. >72 h) (Fig. 2a, Fig. S2a-b), with increased bacteria in blood, spleen and peritoneal fluid, indicating bacteremia and decreased neutrophil recruitment in the peritoneal fluid as compared to age-matched controls (Fig. S2c-e). Similarly, antibiotic-exposed postnatal day 5 old neonatal mice demonstrated marked susceptibility to K. pneumoniae as compared to controls (median survival 14 h vs. >72 h) (Fig. S2f).
Figure 2. Microbiota regulates postnatal granulocytosis and controls host resistance to E. coli.
(a) 5 day-old sex-matched neonatal mice exposed to combination of 3 (3ABX) or 5 antibiotics (5ABX) or no antibiotics (No ABX) were examined for susceptibility after inoculation with E. coli via intraperitoneal route. (b) 5 day-old sex-matched neonatal mice exposed to combination of 3 (3ABX) or (c) 5 antibiotics (5ABX) or no antibiotics (No ABX) were examined for circulating neutrophils or (d) plasma G-CSF levels 4h after inoculation with E. coli. (e) 5 day-old sex-matched neonatal mice exposed to combination of 3 (3ABX) or (f) 5 antibiotics (5ABX) or no antibiotics (No ABX) were treated with G-CSF and examined for susceptibility after inoculation with E. coli via intraperitoneal route. * Significantly different from neonatal mice not exposed to antibiotics (No ABX), ** significantly different from ABX-exposed neonatal mice. (g) Transfer of intestinal contents from postnatal day 3 old control (no antibiotic-exposed) mice to sex-matched neonatal mice exposed to combination of 5 antibiotics (5ABX) or (h) 3 antibiotics (3ABX) via oral gavage on postnatal day 3 and assessment of susceptibility to E. coli 48 h following transfer (postnatal day 5). (i) Transfer of intestinal contents from postnatal day 3 old control (no antibiotic-exposed) mice to sex-matched neonatal mice exposed to combination of 5 antibiotics (ABX) via oral gavage on postnatal day 3 and assessment of circulating or (j) bone marrow neutrophils 48 h following transfer (postnatal day 5). Data are representative of three independent experiments with 12 mice per group. Results are shown as the means ± s.e.m. * Significantly different from control (No ABX) neonatal mice, ** significantly different from ABX-exposed neonatal mice.
Rapidly-responding ‘emergency’ granulocytosis is necessary for sustained output of circulating neutrophils during infection17. To determine if antibiotic exposure alters granulocytosis in response to infection, we measured the number of circulating and bone marrow neutrophils in neonatal mice exposed to either the 5- or 3-antibiotic protocol and age-matched (no antibiotic-exposed) controls 4 h after E. coli K1 challenge. The number of circulating neutrophils representing recruitment from bone marrow (Fig. 2b-c) and plasma G-CSF concentration (Fig. 2d) were significantly lower in neonatal mice exposed to combination of 5 or 3 different antibiotics, suggesting impaired granulocytic response to E. coli K1 sepsis.
The extent to which relative granulocytopenia accounted for impaired host defense was determined by antibody-mediated depletion of neutrophils in neonatal mice. Neonatal mice treated with anti-Ly6G antibody (clone 1A8 or RB6-8C5) were more susceptible to E. coli K1 as compared to age-matched isotype antibody-treated mice (Fig. S2g). To test if restoring number of neutrophils in antibiotic-exposed neonatal mice could alter susceptibility to infection, we treated the 3-day-old antibiotic-exposed mice with G-CSF. G-CSF treatment increased the number of circulating neutrophils (Fig. S2h) and improved resistance to E. coli K1 sepsis in antibiotic-exposed neonatal mice (median survival 36 h) (Fig. 2e-f). Taken together, these data confirm an important role for microbiota-stimulated postnatal granulocytosis in susceptibility of antibiotic-exposed neonates to sepsis.
To determine whether reconstitution of antibiotic-exposed mice with normal microbiota could restore neutrophil number and host defense, we transferred intestinal contents from postnatal 1 or 3 or 5 or 7 day old neonatal mice into sex-matched antibiotic-exposed neonatal mice of same age by gavage (Fig. S2i). Transfer of normal intestinal microbiota was able to partially restore resistance to E. coli K1 in neonatal mice exposed to the 5-antibiotic protocol (mean survival 38h) or more the clinically relevant 3-antibiotic protocol (mean survival 46h) (Fig. 2g-h, Fig. S2j-k). Additionally, the number of circulating and bone marrow neutrophils (Fig. 2i-j) and plasma G-CSF (Fig. S2l) in antibiotic-exposed neonatal mice given intestinal contents of the control mice were higher than those administered PBS. This is consistent with previous observations that the intestinal microbiota plays a key role in affecting the outcome of infection18,19. It is difficult to ascertain if specific microbes or rather microbial ecology and diversity regulate postnatal granulocytosis. Nevertheless, these data, taken together, suggest that intestinal microbiota controls postnatal granulocytosis and modulates host resistance to infection in neonates.
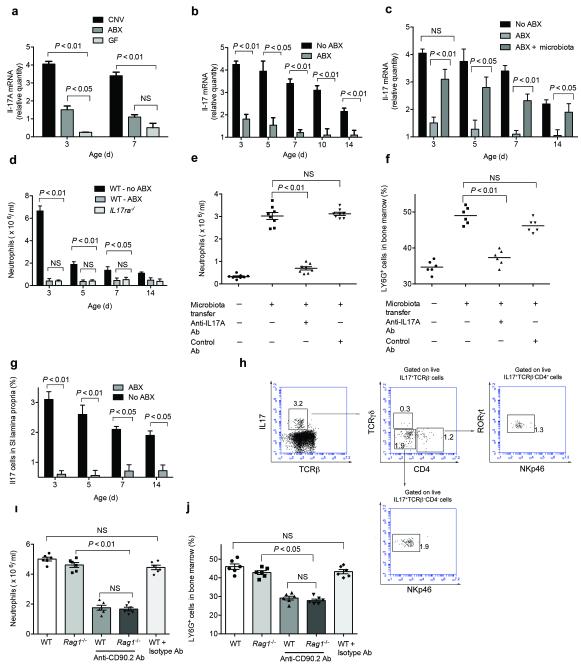
Regulation of G-CSF by IL17A is a key mechanism controlling granulocytosis20 and neutrophil recruitment during bacterial infection21. Transcript levels of IL17A and were significantly lower in intestine but not lung of GF mice (Fig. 3a, S3a) and antibiotic-exposed neonatal mice (Fig. 3b) suggesting a role for IL17A in regulation of postnatal granulocytosis. Furthermore, transfer of normal microbiota restored IL17A levels in the intestine of antibiotic-exposed neonatal mice, though not to the same extent as age-matched controls (Fig. 3c). To determine the importance of IL17 signaling in postnatal granulocytosis, we utilized mice deficient in IL17A receptor (Il17ra−/−) or blocked IL17A signaling with a neutralizing antibody. The number of circulating neutrophils was significantly lower in Il17ra−/− neonatal mice (Fig. 3d). Interestingly, the number of circulating neutrophils in neonatal Il17ra−/− mice was no higher than antibiotic-exposed age- and strain-matched controls. Treatment of neonatal mice with anti-IL17A blocking antibody resulted in lower neutrophil numbers than neonatal mice treated with isotype-control antibody (Fig. S3b), and in addition, blocked the increase in circulating and bone marrow neutrophils (Fig. 3e-f) and plasma G-CSF (Fig. S3c) in antibiotic-exposed neonatal mice after transfer of normal intestinal microbiota. These data confirm that intestinal microbiota regulates postnatal granulocytosis via IL17A.
Figure 3. Microbiota-derived signals regulate postnatal granulocytosis via IL17 and G-CSF dependent pathway.
(a) Age-matched germ free (GF) or conventionalized (CNV) mice or neonatal mice exposed to combination of 5 antibiotics (ABX) or (b) age- and sex-matched neonatal mice exposed to combination of 5 antibiotics (ABX) or no antibiotics (No ABX) were examined for IL17A transcripts in the intestine. (c) Transfer of intestinal contents from age- and sex-matched control (no antibiotic-exposed) mice to neonatal mice exposed to combination of 5 antibiotics (ABX) via oral gavage and assessment of IL17A transcripts 48 h following transfer. (d) Neonatal mice deficient in receptor for IL17A (Il17ra−/−) and age- and sex-matched wild type (WT) littermates exposed or not exposed to combination of 5 antibiotics (ABX) were examined for number of circulating neutrophils. (e) Treatment with neutralizing antibody against IL17A daily on postnatal day 0-3 and transfer of intestinal contents from postnatal day 3 control (no antibiotic-exposed) mice to neonatal mice exposed to combination of 5 antibiotics (ABX) via oral gavage on postnatal day 3 and assessment of circulating or (f) bone marrow neutrophils 48 h following transfer (postnatal day 5). (g) Age- and sex-matched neonatal mice exposed to combination of 5 antibiotics (ABX) or no antibiotics (No ABX) were examined for frequency of IL17+ cells in small intestine lamina propria. (h) 3 day-old neonatal mice were examined for IL17+ cells in small intestine lamina propria. Flow cytometry plots are gated on live IL17+ cells and frequency of each distinct population of cells is indicated. Representative histograms from 5 separate experiments. (i) Treatment of neonatal Rag1−/− mice with anti-CD90.2 antibody or isotype control antibody (5 μg g-1 body weight) daily on postnatal day 0-3 and assessment of circulating and (j) bone marrow neutrophils on postnatal day 3. Data are representative of three independent experiments with 8-12 mice per group. Results are shown as the means ± s.e.m.
IL17 is produced by various lymphocyte subtypes22,23 as well as other cells residing in the intestinal lamina propria24, suggesting that intestine is a key source of IL17 production in basal conditions. The number of IL17 producing cells was lower in the intestines of antibiotic-exposed neonatal mice as compared to age and sex-matched control mice (Fig. 3g, S3d). The exact identity of IL17-producing cells in neonatal intestine is not known. CD4+ T cells are important sources of IL17 in adult mice22, but circulating and bone marrow neutrophil numbers were not diminished in neonatal Rag1−/− mice (Fig. S3e-f), which lack mature T and B cells25 suggesting that mature T cells are dispensable in regulation of postnatal granulocytosis in neonatal mice. IL17-producing cells in lamina propria of neonatal mice were either CD4−TCRβ−TCRγδ+(10%) or CD4− TCRβ−TCRγδ−RORγt+NKp46 (55%) or CD4+TCRβ−TCRγδ−RORγt+NKp46− (35%) (Fig. 3h), suggesting that majority of IL17-producing cells belong to recently described group 3 innate lymphoid cells (ILC)26, which includes lymphoid tissue inducer cells and natural cytotoxicity receptor (NCR) negative ILC27. ILCs are present in increased numbers at mucosal sites and play an important role in maintaining intestinal homeostasis28. To confirm the role of ILCs, we administered anti-CD90.2 antibody to neonatal Rag1−/− mice to deplete the ILC populations. The number of circulating and bone marrow neutrophils were lower in Rag1−/− mice treated with anti-CD90.2 (Thy1.2) antibody as compared to mice treated with isotype-control antibody (Fig. 3i-j). These data taken together suggest that intestinal microbiota controls postnatal granulocytosis by inducing a population of ILCs.
IL23, secreted by macrophages and dendritic cells in intestine in response to microbial products, has been shown to induce IL17 production by CD4−TRCβ− TCRγδ+ cells and innate lymphoid cells28 in mouse models of experimental colitis, but the number of circulating and bone marrow neutrophils (Fig. S3g) were not diminished in neonatal (Il23−/−) mice, suggesting that IL23 is dispensable in regulation of postnatal granulocytosis in neonatal mice.
Since both commensal and pathogenic bacteria can be recognized by a number of pattern recognition receptors (PRR)29, we hypothesized that signals derived from intestinal microbiota could regulate postnatal granulocytosis through signaling via TLR and NOD family members. We examined the postnatal granulocytic response of mice deficient in TLR2 (Tlr2−/−), TLR4 (Tlr4−/−), NOD2 (Nod2−/−) and MYD88 (MyD88−/−), the latter of which encodes a critical adaptor molecule, which regulates signaling through multiple TLRs30. The number of circulating and bone marrow neutrophils in neonatal Tlr4−/− and MyD88−/− mice (Fig. 4a-d), but not Tlr2−/− and Nod2−/− mice (Fig. S4a-d) was lower as compared to age- and strain-matched controls, consistent with previous observations that commensal detection by PRR and subsequent signaling via MYD88 promotes systemic immune responses31. Tlr4−/− mice have distinctly different microbiota from WT mice32, therefore to control for potential differences in the microbiota, we blocked TLR4 signaling in WT neonatal mice with a neutralizing antibody. Treatment of neonatal mice with anti-TLR4 neutralizing antibody resulted in decreased number of circulating and bone marrow neutrophils compared to neonatal mice treated with isotype-control antibody (Fig. S4e-f). Nevertheless, Tlr4−/− mice or mice treated with anti-TLR4 neutralizing antibody or MyD88−/− mice failed to completely recapitulate the phenotype observed in antibiotic-treated or Il17ra−/− neonatal mice, suggesting that TLR4 and MYD88 signaling pathways do not completely account for postnatal granulocytosis and alternate PRR pathways are involved in regulating postnatal granulocytosis. TIR-domain-containing adapter-inducing interferon-β (TRIF), which relays signals from TLR4 independent of MYD8833 is critical in neutrophil homeostasis34. Therefore TRIF, or members of the NOD like receptor (NLR) family, such as NLR635 which recognize microbial patterns and mediate intestinal homeostasis36 could be additional microbial sensor in regulating postnatal granulocytosis. Furthermore, levels of short-chain fatty acids (SCFA), gut microbiota-derived fermentation products, which have a critical role in intestinal inflammation and host resistance are decreased in the intestinal contents of antibiotic-exposed mice37. Therefore SCFA could play a potential role in regulating postnatal granulocytosis.
Figure 4. Microbiota-derived signals regulate postnatal granulocytosis via TLR4 and MyD88 dependent pathway.
(a and c) Neonatal mice deficient in MyD88 (Myd88−/−) or TLR4 (Tlr4−/−) and age and sex matched wild type (WT) littermates exposed or not exposed to combination of 5 antibiotics (ABX) were examined for number of circulating neutrophils or (b and d) bone marrow neutrophils. (e) Administration of LPS (10 ng) to neonatal mice exposed to combination of 5 antibiotics (ABX) or no antibiotics (No ABX) via oral gavage and assessment of IL17 transcripts in small intestine or (f) plasma G-CSF (g) circulating neutrophils or (h) bone marrow neutrophils 48 h following gavage. Data are representative of three independent experiments with 8-10 mice per group. Results are shown as the means ± s.e.m.
Components of intestinal microbiota have been shown to differentially regulate adaptive immune function22,38,. We tested the effect of LPS, a ligand of TLR4, which is upstream of MyD88, hypothesizing it could regulate postnatal granulocytosis in neonatal mice. LPS has previously been detected in the intestinal homogenate of newborn mice within 1 h after birth39 and fecal LPS levels were shown to be reduced in antibiotic-exposed mice40. Administration of LPS (25 ng) by gavage was previously shown to increase the macrophage inflammatory protein 2 transcripts in the intestine of neonatal mice delivered by caesarean section to equivalent levels seen in vaginally delivered neonatal mice39. Therefore, we administered LPS (10 ng by gavage) to antibiotic-exposed neonatal mice (postnatal day 3). Twenty-four hours later, the levels of IL17 transcripts in intestine, plasma GCSF levels and the number of circulating and bone marrow neutrophils in antibiotic-exposed neonatal mice given LPS were higher than those administered PBS (Fig. 4e-h). Treatment of neonatal mice with anti-IL17A antibody blocked the increase in circulating and bone marrow neutrophils and plasma G-CSF in antibiotic-exposed neonatal mice after LPS administration (Fig. S4g-i). These data suggest that LPS is one of the microbiota-derived signals regulating postnatal granulocytosis in neonates.
In conclusion, we identified a role for microbiota, signaling through TLR4- and MYD88-dependent mechanism to induce IL17A- and GCSF-dependent regulation of postnatal granulocytosis. Better understanding of how the intestinal microbiota affects postnatal development and function of neutrophils, could lead to therapeutically relevant strategies to restore granulocytosis. These results could form basis for future clinical studies for microbiota manipulation and transplantation to ameliorate antibiotic-induced microbiota dysbiosis and improve neonatal mortality.
METHODS
Animals and neonatal antibiotic-exposure
We bred C57/B6, mice deficient in TLR2, TLR4, RAG1, IL17, IL23 (subunit p19), MYD88, NOD2 and appropriate wild type controls at Children’s Hospital of Philadelphia animal facility. We maintained the germ free C57/B6 neonatal mice (gift from D. Artis, University of Pennsylvania) in plastic isolator cages with autoclaved feed and water in University of Pennsylvania Germ Free Core facility. We treated pregnant wild type or gene targeted female mice with sterile drinking water mixed with 5 different antibiotics (ampicillin, gentamicin, metronidazole, vancomycin and neomycin) (all 1 mg ml−1, from Sigma-Aldrich), or 3 different antibiotics (ampicillin, gentamicin and vancomycin) (all 1 mg ml−1) starting from day E15. After birth, neonatal mice from multiple litters were pooled and randomly distributed to control for founder effect and to minimize in-cage variations. The dams and the neonatal mice continued to receive antibiotic containing drinking water for the duration of experiment. Thus neonatal antibiotic exposure refers to antibiotic exposure both in utero and after birth. We used neonatal C57/B6, Rag1−/−, Tlr2−/−, Tlr4−/−, Nod2−/−, Il17ra−/−, Myd88−/−, Il23−/− mice and germ free C57/B6 mice between ages of 1-21 days and appropriate, age-, gender- and genetic strain-matched controls in subsequent experiments to account for any variations in data. The Children’s Hospital of Philadelphia Institutional Animal Care and Use Committee (IACUC) approved all protocols.
Neonatal late onset sepsis
We grew E. coli serotype K1 or K. pneumoniae (ATCC, 43816) (37°C, 200 rpm) in Luria Bertani (LB) broth containing 100 μg ml−1 rifampin (Sigma) or tryptic soy (TS) broth respectively to log-phase growth. To mimic E. coli or K. pneumoniae late onset neonatal sepsis, we inoculated neonatal mice (postnatal day 3-14) with either E. coli (104 CFU g−1) or K. pneumoniae (106 CFU g−1) respectively via intraperitoneal (i.p.) injection15 and euthanized 72 h later or earlier if moribund. To asses bacterial burden we homogenized the spleen and liver in sterile PBS. We plated serial dilutions of spleen or liver homogenates or blood or peritoneal fluid in LB agar plates with rifampin (100 μg ml−1) and incubated (37°C, overnight) to count the number of CFU of E.coli.
Manipulation of intestinal microbiota in the antibiotic exposed neonatal mice
We collected intestinal contents from non-antibiotic exposed neonatal mice at various ages (postnatal day 1 or 3 or 5 or 7 or 10 or 21 days) and weighed and suspended in sterile water. We transferred intestinal contents (200 μg in 50 μl PBS) from postnatal 1 or 3 or 5 or 7 or 10 or 21 day old neonatal mice or vehicle (50 μl PBS) to sex-matched antibiotic-exposed postnatal 1 or 3 or 5 or 7 or 10 or 21 day neonatal mice respectively by a single oral gavage via fine polyethylene tubing. We pooled the neonatal mice in each experimental group and randomly redistributed to minimize in-cage variations. The dams and neonatal mice continued to receive antibiotic containing drinking water and after 48 h were examined for circulating and bone marrow neutrophils or plasma G-CSF or inoculated via i.p. route with E. coli.
Treatment with monoclonal depleting or neutralizing monoclonal antibodies, G-CSF and TLR agonist
We injected neonatal mice with anti-Ly6G antibody (Clone RB6-865, BD Biosciences or Clone 1A-8, Bioxcell) or anti-IL17A antibody (Clone 50104, R&D), anti-TLR4 antibody (Clone MTS510, eBiosciences), anti-CD90.2 antibody (Clone 30H12, Bioxcell), or anti-IgG2A (Clone 54447, R&D) (all 5 μg g−1 body weight) via i.p. route daily on postnatal day 0-3. We injected antibiotic-exposed neonatal mice with recombinant mouse GCSF (10 μg g−1 body weight) (Cat 414-CS, R&D) via i.p. route on postnatal day 3. We treated antibiotic-exposed neonatal mice (postnatal day 3) with LPS (Invitrogen) (10 ng in 50 μl PBS) or vehicle by oral gavage (50 μl endotoxin-free PBS) via fine polyethylene tubing and after 48 h inoculated them via i.p. route with E. coli.
Bone marrow isolation and flow cytometry
For characterization of bone marrow neutrophils, we pooled the femurs from 2-3 neonatal mice, flushed them with 5% FCS DMEM and incubated them (4°C, 5 min) with ACK lysis buffer. We then incubated (4°C, 30 min) the cells (2 × 106) with Live/Dead Blue viability dye (Invitrogen) and then stained them with allophycocyanin (APC) conjugated anti-mouse Ly6G antibody (Clone RB6-8C5, R&D), fluorescein isothiocyanate (FITC)-conjugated anti-mouse CXCR4 antibody (Clone 12G5, Biolegend), phycoerthrin (PE)-conjugated anti-mouse CXCR2 antibody (Clone 5E8, Biolegend), PE/Cy7-conjugated anti-mouse CD45 antibody (Clone 30F-11, Biolegend), washed (2x) and resuspended them in flow cytometry buffer (PBS, 0.5% FCS and 0.1% sodium azide). To determine the hematopoietic progenitor cells in the bone marrow, we incubated the cells (2 × 106) with Live/Dead Blue viability dye and then stained them with FITC-conjugated anti-mouse lineage marker antibodies (CD3, CD4, CD8a, TCRβ, TCRγδ, CD45R, CD11c, CD11b, CD19, TER119, Ly6G/Ly6C, NK1.1 and FceR1), Alexaflour647-conjugated anti-mouse CD34 antibody (Clone RAM34, BD Biosciences), PE/Cy7-conjugated anti-mouse c-kit antibody (Clone 2B8, Biolegend), Brilliant Violet (BV) 711-conjugated anti-mouse Sca-1 (Clone D7, Biolegend), PE-conjugated anti-mouse CD16/CD32 (Clone 93, Biolegend) washed (2x) and resuspended them in flow cytometry buffer. We used isotype-matched antibodies as negative controls. We collected the data with LSRII (BD Biosciences) and analyzed the data with Flowjo (Treestar).
Isolation of lamina propria lymphocytes and detection of IL-17 producing innate lymphoid cells (ILC17)
We pooled and cut the freshly resected terminal ilea from 3-4 neonatal mice into 2- to 5-mm pieces and incubated (37°C, 15 min) them in extraction buffer (HBSS, 15 mM HEPES and 1 mM EDTA) to remove the epithelial cells. We then incubated (37°C, 30 min) the cut tissues with shaking (150 rpm) in digestion buffer (RPMI 1640 with 10% FBS, 15 mM HEPES, 1% penicillin/streptomycin (wt/vol), and 300 U ml−1 collagenase VIII). We isolated the lamina propria lymphocytes from the resultant single cell suspension by discontinuous Percoll (GE Healthcare) gradient at 40-80% interface and incubated (37°C, 5 h, 5%CO2) the cells (4 × 107) in culture medium containing RPMI 1640 with 10 % FCS, 1X nonessential amino acids, 10 mM HEPES, 2 mM L-glutamine (all from Invitrogen) and 1% penicillin/streptomycin with 1:1000 Golgi Stop (554724, BD Biosciences), 10 ng/ml phorbol 12-myristate 13-acteate (PMA) and 500 ng/ml calcium ionophore A23187 (both from Sigma-Aldrich). We washed and incubated (4°C, 10 min) the cells (107) with anti-mouse CD16/CD32 (eBiosciences) and then reincubated (4°C, 30 min) with FITC-conjugated anti-mouse TCRβ antibody (Clone KJ16-133.18, eBiosciences), PE/Cy7-conjugated anti-mouse TCRγδ antibody (Clone GL3, Biolegend), APC-conjugated anti-mouse CD4 antibody (Clone RM4-5, Biolegend) and BV-conjugated anti-mouse NKp46 antibody (Clone lone 29A1.4, eBiosciences). For intracellular staining for IL-17, we washed and fixed (4°C, 60 min) the surface-stained cells in 1X Cytofix/Cytoperm buffer (BD Biosciences) and permeabilized them (4°C, overnight) using 1X Permeabilization Buffer (BD Biosciences) according to manufacturer instructions. We stained the cells intracellularly with PE-conjugated anti-mouse IL-17 antibody (Clone TC11-18H10.1, Biolegend) or PerCP-conjugated anti-mouse RORγ antibody (Clone B2D, eBiosciences) then washed (2x) and resuspended them in flow cytometry buffer. We collected the data with LSRII (BD Biosciences) and analyzed the data with Flowjo (Treestar).
Analysis of microbiota from intestinal contents and quantification of bacterial 16 S ribosomal (r) DNA
We collected intestinal contents from control (non-antibiotic exposed) neonatal mice and antibiotic exposed neonatal mice at various ages (1-21 days) and snap froze them (−80°C). As intestinal contents were not available in adequate amounts for individual sampling from neonatal mice, we pooled intestinal contents from 6-8 neonatal mice from 3 separate litters per treatment group so as to minimize cage effects. We extracted DNA from intestinal contents using QIAamp DNA Stool Mini Kit (Qiagen), quantified 16S rDNA by RT-PCR using degenerate41 or class specific bacterial 16S rDNA primers and probes for Gammaproteobacteria42 or Bacilli43. For 16S rDNA sequencing we amplified the V2 region of microbial 16S rRNA by high fidelity PCR with barcoded 8F and 338R universal primers with A and B sequencing adaptors respectively and bifido primers (Roche) and sequenced them with Genome Sequencer GS-FLX Titanium system (Roche) at University of North Carolina Microbiome core facility (Chapel Hill, NC). We decoded and processed the sequences using the QIIME software package (Version 1.7) and custom R package code44. We used phylogenetic diversity (PD) to compute and visualize α diversity and unweighted and weighted Unifrac for β diversity. We tested the observed differences in Unifrac distances between antibiotic treated groups and across different ages for significance using a t test, and we corrected the reported P values for multiple comparisons using a Monte Carlo permutation procedure with 10,000 iterations.
We determined the number of circulating and bone marrow neutrophils, calculated the phagocytosis index and determined the ROS production by the bone marrow neutrophils as described before45. We isolated the RNA and determined the transcript levels of IL17 in the neonatal small intestine and the lungs as described before45. We determined the levels of plasma G-CSF as described before45.
Statistical analysis
To determine group sizes necessary for adequate statistical power, power analysis was performed using preliminary data sets. The investigators were blinded to group allocation during collection and analysis of the data and all inclusion/exclusion criteria were pre-established. All data meet the assumptions of the statistical tests used. We compared differences between groups by the unpaired Student’s t test or Student Neumann Keul’s test (GraphPad Prism 4). We considered P values <0.05 significant. We used the Kaplan-Meier log-rank test to compare survival between groups.
Supplementary Material
ACKNOWLEDGEMENTS
We thank D. Artis (Department of Microbiology and Pathology, University of Pennsylvania) for providing germ free mice. We thank N. Butz (Microbiome Core Facility, University of North Carolina) for her assistance with microbial DNA isolation. We thank the Children’s Hospital of Philadelphia Research Institute Flow Cytometry and Cell Sorting Core Laboratory for technical advice and support. We thank S. Guttentag, K. Hudock and C. Hergott for their helpful comments. H.S.D. is supported by 5T32HD060556, P.M.O. is supported by 5R01AI093566, J.K.K. is supported by 5R01HL062052, 3R37HL079142 and 5P60AA009803, J.N.W. is supported by 1R01AI105168 and 5R01AI038446 and G.S.W. is supported by 1R01AI099479 and 5R01HL105834.
Footnotes
AUTHOR CONTRIBUTIONS:
H.S.D., G.N.W and G.S.W. conceived the study. H.S.D. and G.S.W. designed the experiments. P.M.O. and J.K.K. provided reagents. H.S.D., O.R.M., L.Y., D.N., M.J. and C.E.O. carried out experiments. H.S.D. and G.S.W. analyzed the data and wrote the manuscript.
REFERENCES
- 1.Penders J, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 2.Kuppala VS, Meinzen-Derr J, Morrow AL, Schibler KR. Prolonged initial empirical antibiotic treatment is associated with adverse outcomes in premature infants. The Journal of pediatrics. 2011;159:720–725. doi: 10.1016/j.jpeds.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sarkar S, Bhagat I, Hieber S, Donn SM. Can neutrophil responses in very low birth weight infants predict the organisms responsible for late-onset bacterial or fungal sepsis? Journal of perinatology: official journal of the California Perinatal Association. 2006;26:501–505. doi: 10.1038/sj.jp.7211554. [DOI] [PubMed] [Google Scholar]
- 4.Schwiertz A, et al. Development of the intestinal bacterial composition in hospitalized preterm infants in comparison with breast-fed, full-term infants. Pediatric research. 2003;54:393–399. doi: 10.1203/01.PDR.0000078274.74607.7A. [DOI] [PubMed] [Google Scholar]
- 5.Pflughoeft KJ, Versalovic J. Human microbiome in health and disease. Annual review of pathology. 2012;7:99–122. doi: 10.1146/annurev-pathol-011811-132421. [DOI] [PubMed] [Google Scholar]
- 6.Mai V, et al. Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PloS one. 2013;8:e52876. doi: 10.1371/journal.pone.0052876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS biology. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vael C, Desager K. The importance of the development of the intestinal microbiota in infancy. Current opinion in pediatrics. 2009;21:794–800. doi: 10.1097/MOP.0b013e328332351b. [DOI] [PubMed] [Google Scholar]
- 9.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alm B, et al. Neonatal antibiotic treatment is a risk factor for early wheezing. Pediatrics. 2008;121:697–702. doi: 10.1542/peds.2007-1232. [DOI] [PubMed] [Google Scholar]
- 11.Clark RH, Bloom BT, Spitzer AR, Gerstmann DR. Empiric use of ampicillin and cefotaxime, compared with ampicillin and gentamicin, for neonates at risk for sepsis is associated with an increased risk of neonatal death. Pediatrics. 2006;117:67–74. doi: 10.1542/peds.2005-0179. [DOI] [PubMed] [Google Scholar]
- 12.Manroe BL, Weinberg AG, Rosenfeld CR, Browne R. The neonatal blood count in health and disease. I. Reference values for neutrophilic cells. The Journal of pediatrics. 1979;95:89–98. doi: 10.1016/s0022-3476(79)80096-7. [DOI] [PubMed] [Google Scholar]
- 13.Gessler P, et al. Neonatal neutropenia in low birthweight premature infants. American journal of perinatology. 1995;12:34–38. doi: 10.1055/s-2007-994396. [DOI] [PubMed] [Google Scholar]
- 14.Martin C, et al. Chemokines acting via CXCR2 and CXCR4 control the release of neutrophils from the bone marrow and their return following senescence. Immunity. 2003;19:583–593. doi: 10.1016/s1074-7613(03)00263-2. [DOI] [PubMed] [Google Scholar]
- 15.Pluschke G, Pelkonen S. Host factors in the resistance of newborn mice to K1 Escherichia coli infection. Microbial pathogenesis. 1988;4:93–102. doi: 10.1016/0882-4010(88)90051-4. [DOI] [PubMed] [Google Scholar]
- 16.Cohen-Wolkowiez M, et al. Early and late onset sepsis in late preterm infants. The Pediatric infectious disease journal. 2009;28:1052–1056. doi: 10.1097/inf.0b013e3181acf6bd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieschke GJ, et al. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood. 1994;84:1737–1746. [PubMed] [Google Scholar]
- 18.Sekirov I, et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infection and immunity. 2008;76:4726–4736. doi: 10.1128/IAI.00319-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke TB, et al. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nature medicine. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarzenberger P, et al. Requirement of endogenous stem cell factor and granulocyte-colony-stimulating factor for IL-17-mediated granulopoiesis. Journal of immunology. 2000;164:4783–4789. doi: 10.4049/jimmunol.164.9.4783. [DOI] [PubMed] [Google Scholar]
- 21.Ye P, et al. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. The Journal of experimental medicine. 2001;194:519–527. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanov II, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 23.Ivanov II, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell host & microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nature immunology. 2007;8:1086–1094. doi: 10.1038/ni1511. [DOI] [PubMed] [Google Scholar]
- 25.Mombaerts P, et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 26.Spits H, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nature reviews. Immunology. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 27.Sawa S, et al. Lineage relationship analysis of RORgammat+ innate lymphoid cells. Science. 2010;330:665–669. doi: 10.1126/science.1194597. [DOI] [PubMed] [Google Scholar]
- 28.Buonocore S, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464:1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medzhitov R. TLR-mediated innate immune recognition. Seminars in immunology. 2007;19:1–2. doi: 10.1016/j.smim.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnare M, et al. Toll-like receptors control activation of adaptive immune responses. Nature immunology. 2001;2:947–950. doi: 10.1038/ni712. [DOI] [PubMed] [Google Scholar]
- 31.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ubeda C, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. The Journal of experimental medicine. 2012;209:1445–1456. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 34.Bugl S, et al. Steady-state neutrophil homeostasis is dependent on TLR4/TRIF signaling. Blood. 2013;121:723–733. doi: 10.1182/blood-2012-05-429589. [DOI] [PubMed] [Google Scholar]
- 35.Elinav E, et al. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nature reviews. Cancer. 2013;13:759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 36.Henao-Mejia J, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cho I, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 39.Lotz M, et al. Postnatal acquisition of endotoxin tolerance in intestinal epithelial cells. The Journal of experimental medicine. 2006;203:973–984. doi: 10.1084/jem.20050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daenen S, Goris H, de Boer F, Halie MR, van der Waaij D. Recovery of murine myelopoiesis after cytostatic reduction by Ara-C. Effect of bacitracin-induced changes in the intestinal microflora and influence of timing. Leukemia research. 1991;15:1013–1018. doi: 10.1016/0145-2126(91)90106-4. [DOI] [PubMed] [Google Scholar]
- 41.Hill DA, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal immunology. 2010;3:148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudi K, Tannaes T, Vatn M. Temporal and spatial diversity of the tap water microbiota in a Norwegian hospital. Applied and environmental microbiology. 2009;75:7855–7857. doi: 10.1128/AEM.01174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oliwa-Stasiak K, Kolaj-Robin O, Adley CC. Development of real-time PCR assays for detection and quantification of Bacillus cereus group species: differentiation of B. weihenstephanensis and rhizoid B. pseudomycoides isolates from milk. Applied and environmental microbiology. 2011;77:80–88. doi: 10.1128/AEM.01581-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mei J, et al. Cxcr2 and Cxcl5 regulate the IL-17/G-CSF axis and neutrophil homeostasis in mice. The Journal of clinical investigation. 2012;122:974–986. doi: 10.1172/JCI60588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.