Abstract
Major depressive disorder (MDD) affects up to 17% of the population, causing profound personal suffering and economic loss (1). Clinical and pre-clinical studies have revealed that prolonged stress and MDD are associated with neuronal atrophy of cortical and limbic brain regions (2-9), but the molecular mechanisms underlying these morphological alterations have not yet been identified. Here, we show that stress increases levels of REDD1 (regulated in development and DNA damage responses 1), an inhibitor of mTORC1 (mammalian/mechanistic target of rapamycin complex 1) (10), in rat prefrontal cortex (PFC). This is concurrent with a decrease in phosphorylation of signaling targets of mTORC1, which is implicated in protein synthesis-dependent synaptic plasticity. We also found that REDD1 levels are increased in the postmortem PFC of human subjects with MDD relative to matched controls. Mutant mice with a deletion of REDD1 are resilient to the behavioral, synaptic, and mTORC1 signaling deficits caused by chronic unpredictable stress (CUS), while viral-mediated over expression of REDD1 in the rat PFC is sufficient to cause anxiety- and depressive-like behaviors and neuronal atrophy. Taken together, these postmortem and pre-clinical findings identify REDD1 as a critical mediator underlying the atrophy of neurons and depressive behavior caused by chronic stress exposure.
Structural alterations of the nervous system have been implicated in depression, including postmortem findings demonstrating atrophy of PFC in human subjects with MDD (2-4). Chronic stress decreases dendrite branching and spine density in rodents (5-9), which is associated with depressive behavior, notably anhedonia (11, 12). A recent postmortem study of MDD subjects reported decreases in mTORC1 signaling, which regulates synaptic protein synthesis (13). Conversely, rapid-acting antidepressants increase mTORC1 signaling and synaptogenesis in the rat PFC (14). These findings suggest that stress could decrease synaptogenesis via inhibition of mTORC1.
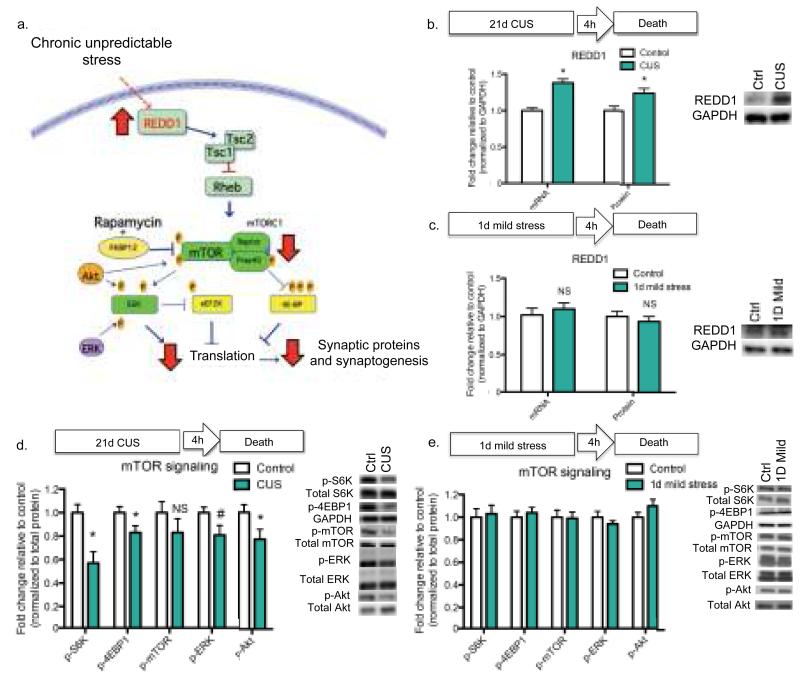
To address this issue, we examined the influence of chronic unpredictable stress (CUS) on REDD1 (DDIT4, RTP801), which stabilizes the TSC1/TSC2 complex, inhibiting mTORC1-dependent protein synthesis and cell growth (10) (Fig. 1a). Exposure to CUS for 21 d, which decreases the number and function of spine synapses (11), significantly increased REDD1 mRNA (p < 0.001) and protein (p < 0.05) in rat PFC (Fig. 1b). One day of mild stress had no effects of REDD1 (Fig. 1c), and levels of TSC2 protein were unaltered by CUS (Supplementary Fig. 1a-b). Consistent with our previous work (11), we found that PSD-95 in the PFC is decreased by CUS (p < 0.05) (Supplementary Fig. 1c). REDD1 levels were not altered by CUS in the hippocampus, another region in which atrophy is observed following chronic stress (Supplementary Fig. 1d).
Figure 1. Chronic unpredictable stress increases REDD1 and decreases mTORC1 signaling in rat PFC.
(a) Schematic showing how increased REDD1 could result in decreased mTORC1 signaling via Rheb and the TSC1/TSC2 complex (10), and subsequent decreased translation and synaptogenesis. (b) Rats received 21 d of CUS or regular handling (control) then sacrificed 4 h following the final stressor. Results are mean ± SEM fold change of PFC REDD1 mRNA [CUS n=8; Control n=8] and protein [CUS n=10; Control n=12] relative to control. (c) Rats received 1 d of mild stress or handling (control) then sacrificed 4 h following the final stressor. Results are mean ± SEM fold change PFC REDD1 mRNA [1 d mild stress n=6; control n=6] and protein [1 d mild stress n=6; control n=6] relative to control. For (b) and (c), protein levels were normalized to GAPDH, and representative blots are displayed to the right. (d) Rats received 21 d of CUS (n=10) or regular handling (control) (n=12), then sacrificed 4 h following the final stressor. (e) Rats received 1 d mild stress (n=6) or handling (control) (n=6), then sacrificed 4 h following the final stressor. For (d) and (e), phospho-protein levels in PFC were normalized to total protein levels; results are shown as mean (± SEM) fold change. Representative blots are displayed to the right. (*) p < 0.05 (#) p < 0.10 relative to control. Abbreviations: mTORC1, mammalian/mechanistic target of rapamycin complex 1; ERK, extracellular signal-regulated kinase; Akt, serine threonine kinase or protein kinase B; S6K, P70 ribosomal protein S6 kinase; 4E-BP1, eukaryotic translation initiation factor 4E-binding protein 1; TSC, tuberous sclerosis complex.
We also examined the effects of adrenal glucocorticoids, a key hypothalamic-pituitary-adrenal axis endocrine response to stress. We found that REDD1 mRNA (p < 0.0001) and protein (p < 0.001) were significantly increased in PFC by administration of the synthetic glucocorticoid, dexamethasone (Supplementary Fig. 2a), consistent with reports in muscle (15) and hippocampus (16). Physiological induction of corticosterone by immobilization stress (17) (Supplementary Fig. 2e) also significantly increased REDD1, and this effect was inhibited by glucocorticoid blockade with RU-486 (p < 0.05) (Supplementary Fig. 2d). These findings indicate that activation of hypothalamic-pituitary-adrenal axis underlies increased REDD1 expression in response to CUS. Consistent with previous work in the hippocampus (16), we also found that REDD2 in the PFC is decreased by glucocorticoid treatment (p < 0.05) (Supplementary Fig. 2b). REDD2 is also a negative regulator of mTORC1 (10) and may act with REDD1 to fine-tune mTORC1 signaling.
We also found that CUS (21 d), but not mild stress (1 d), decreases phosphorylation of the mTORC1 signaling targets p70 ribosomal S6 kinase (S6K) (p < 0.01) and 4EBP1 (p < 0.05) in the rat PFC (Fig. 1d-e). Further, we observed a significant decrease in phospho-Akt (p < 0.05) and a trend for phospho-ERK (p < 0.06), kinases linked to activation of mTORC1 signaling. There was a non-significant trend for decreased levels of phospho-mTOR. Dexamethasone treatment similarly decreased mTORC1 signaling, as measured by phospho-S6K (p < 0.001) (Supplementary Fig. 2c).
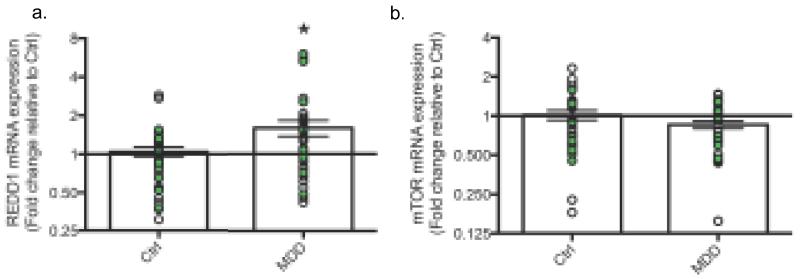
To assess the relevance of REDD1 to MDD, we analyzed REDD1 expression in postmortem dorsolateral PFC (dlPFC) of two separate cohorts of depressed subjects. In the first cohort, there was a 1.22-fold increase of REDD1 mRNA in the dlPFC of MDD subjects relative to controls (p = 0.18). This effect was corroborated in the second cohort, where there was a significant 2.57-fold increase (p = 0.025). Combined analysis of the two cohorts revealed a significant overall 1.60-fold increase of REDD1 in MDD patients relative to psychiatrically healthy controls (p = 0.012) (Fig. 2a). There was also a trend for decreased levels of mTOR mRNA in the combined samples (p = 0.098) (Fig. 2b). ANCOVA revealed that neither sex nor medication status was a significant covariate for REDD1 or mTOR expression. Sustained activation of the HPA axis, consistently observed in MDD patients (18), could underlie this observed increase of REDD1.
Figure 2. REDD1 mRNA is increased in the dlPFC of MDD patients.
(a) Results are the mean ± SEM fold change relative to control of REDD1 mRNA in dlPFC of MDD subjects or psychiatrically-healthy controls (Control n=36; MDD n=36). (b) Results are the mean ± SEM fold change relative to control of mTOR mRNA in dlPFC of MDD subjects or psychiatrically-healthy controls (Control n=35; MDD n=37). White data points indicate subjects from cohort 1; green data points indicate subjects from cohort 2. Target mRNA levels have been normalized to GAPDH mRNA. (*) p < 0.05 control vs. MDD.
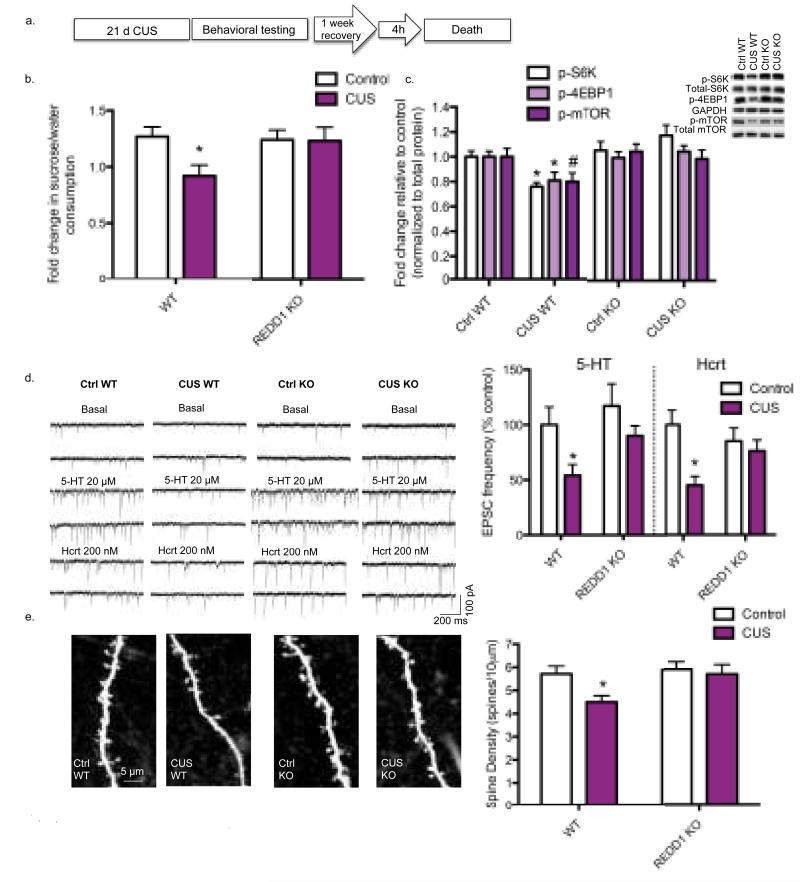
To directly test the role of REDD1 in the effects of chronic stress, we used REDD1 constitutive knock out (KO) mice. REDD1 deletion in PFC was confirmed (p < 0.0001) (Supplementary Fig. 3a). KO mice did not display any apparent physical or behavioral abnormalities despite a previous report suggesting a role for REDD1 in embryonic development (19) (Supplementary Fig. 3b–d). This lack of baseline behavioral differences was not surprising given the low level of REDD1 expression in brain (20). However, CUS exposure (21 d), which causes anhedonic behavior (i.e., decreased sucrose consumption) in WT mice, had no effect in REDD1 KO mice, indicating resilience to CUS (Fig. 3a-b). Sucrose consumption was significantly decreased in the WT CUS group relative to all other groups (p < 0.01). CUS also decreased phospho-S6K and phospho-4EBP levels in the PFC of WT, but not REDD1 KO mice (Fig. 3c). There was a non-significant trend for decreased levels of phospho-mTOR in the WT CUS group.
Figure 3. REDD1 knock out mice are resilient to CUS-induced alterations in the PFC.
(a) REDD1 KO mice and their wild-type littermates received 21 d of CUS or regular handling (Ctrl), then were behaviorally tested. After testing, mice continued to receive CUS or regular handling for 7 d, then sacrificed 4 h after the final stressor. (b) Ratio of sucrose to water consumption (± SEM) over a 1 h test is depicted for WT Ctrl (n=13), WT CUS (n=14), KO Ctrl (n=16), and KO CUS (n=17) groups. ANOVA revealed a significant effect of genotype [F(1,56) = 4.47; p < 0.05], stress [F(1,56) = 5.809; p < 0.03], and genotype x stress interaction [F(1,56) = 4.157; p < 0.05]. (c) Phospho-protein levels in PFC were analyzed in WT Ctrl (n=13), WT CUS (n=14), KO Ctrl (n=16), and KO CUS (n=17) groups. Results are the mean ± SEM fold change. Phospho-protein levels were normalized to total protein levels and representative blots are displayed. ANOVA for phospho-S6K revealed a significant genotype x stress interaction [F(1, 56) = 7.632, p < 0.01] and a main effect of genotype [F(1, 56) = 12.08, p < 0.01], but no main effect of stress. ANOVA for phospho-4EBP1 showed a significant genotype x stress interaction [F(1, 56) = 4.939, p < 0.05] and a main effect of genotype [F(1, 56) = 4.077, p < 0.05], but no main effect of stress. (d) Left. Sample whole-cell voltage-clamp traces of serotonin (5-HT) and hypocretin (Hcrt)-induced EPSCs in layer V pyramidal cells. Scale is depicted on the lower right. Right. Mean ± SEM frequency of 5-HT and Hcrt-induced EPSCs from WT Ctrl (5-HT n=16; Hcrt n=13), WT CUS (5-HT n=16; Hcrt n=16), KO Ctrl (5-HT n=16; Hcrt n=15), and KO CUS (5-HT n=16; Hcrt n=16) groups is depicted. ANOVA for 5-HT-induced EPSCs in layer V pyramidal neurons of the mPFC showed a significant main effect of stress [F(1,60) = 6.131, p < 0.05] and a trend for a main effect of genotype [F(1,60) = 3.124, p < 0.10], but no stress x genotype interaction. ANOVA of Hcrt-induced EPSCs revealed a significant main effect of stress [F(1,56)=7.809, p < 0.01] and a significant stress x genotype interaction [F(1,56)=4.046, p < 0.05], but no main effect of genotype. (e) Left. Representative images are shown of high magnification Z-stack projections of segments of the layer V pyramidal cell apical tuft dendrites (scale: 5 μm). Right. Mean ± SEM of spine density from WT Ctrl (n=30), WT CUS (n=41), KO Ctrl (n=27), and KO CUS (n=23) groups is depicted. ANOVA for spine density in the recorded cells revealed a significant main effect of stress [F(1,117) = 4.487, p < 0.05] and genotype [F(1,117) = 4.183, p < 0.05], but no stress x genotype interaction (*) p < 0.05 relative to WT Ctrl mice. (#) p < 0.10 relative to WT Ctrl mice.
CUS decreases serotonin (5-HT) and hypocretin (Hcrt)-induced EPSCs (excitatory postsynaptic potentials) and the number of spine synapses in the distal tufts of layer V pyramidal neurons in the medial PFC (mPFC) (11). Here, we show that REDD1 KO mice are resilient to these CUS-induced cellular deficits. First, we found that EPSC responses to both 5-HT and Hcrt, which are stress-sensitive probes targeting the apical dendrite (21), were significantly decreased in the WT CUS group compared to the unstressed groups (p < 0.05) and there was a trend for a decrease relative to the KO CUS group (p < 0.10) (Fig. 3d). There were no differences in basal responses between groups. Spine density analysis using two-photon laser imaging in the recorded cells revealed a decrease in spine density in WT CUS mice as compared to all other groups (p < 0.02) (Fig. 3e). The results demonstrate that REDD1 KO mice display resilience to the effects of chronic stress on anhedonic behavior, mTORC1 signaling, and number and function of spine synapses in PFC.
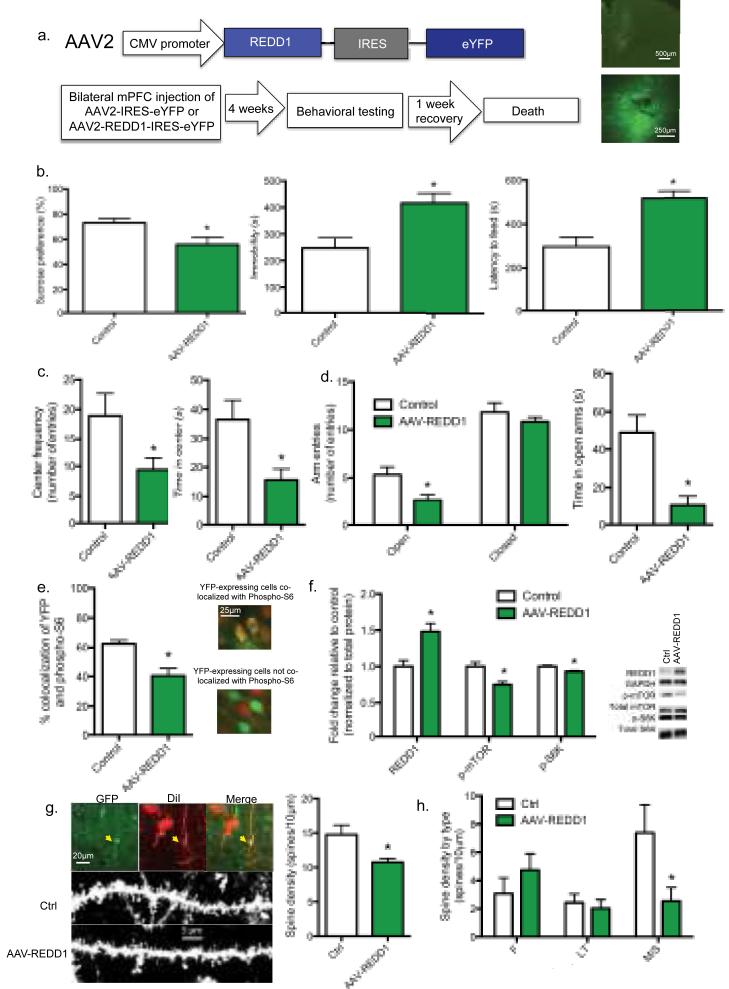
We next tested whether REDD1 expression in the mPFC is responsible for the behavioral effects of CUS using a viral expression approach (Fig. 4a). We found that rats given a bilateral mPFC infusion of a REDD1-expressing viral vector (AAV-REDD1) showed behaviors similar to those observed in rats exposed to CUS, including a significant decrease in sucrose preference (p < 0.05), increased immobility in the forced swim test (p < 0.05), and increased latency to feed in the novelty suppressed feeding test (p < 0.01) (Fig. 4b). Further, AAV-REDD1 infused rats showed an anxiogenic phenotype, spending significantly less time in the center of the open field relative to controls (p < 0.05) (Fig. 4c) and less time and decreased entries in the open arms in the elevated plus maze (p < 0.01 and p < 0.05, respectively) (Fig. 4d). There was no effect on velocity or distance traveled, indicating that locomotor activity was unaffected.
Figure 4. REDD1 over-expression in the rat mPFC increases depressive and anxiety-related behaviors and decreases mTORC1 signaling.
(a) Top. Human REDD1 was subcloned into an AAV2 plasmid under transcriptional regulation of the CMV promoter (AAV2-REDD1-IRES-eYFP). AAV2-IRES-eYFP plasmid without the human REDD1 gene was used as the control. Bottom. Schematic of experiment. Representative photomicrographs of injection sites in the mPFC are shown to the right. (b) Left. Mean sucrose preference (± SEM) for control (n=7) and AAV-REDD1 infused animals (n=7). Center. Mean immobility time (± SEM) for control (n=6) and AAV-REDD1 injected (n=7) animals. Right. Mean latency to feed (± SEM) for control (n=6) and AAV-REDD1 injected (n=7) animals. (c) Left. Mean entries to the center in the open field (+/− SEM) for control (n=7) and AAV-REDD1 injected (n=7) animals. Right. Mean center duration (± SEM) in the open field for control (n=7) and AAV-REDD1 injected (n=7) animals. (d) Left. Mean number (± SEM) of open arm entries in the elevated plus maze for control (n=6) and AAV-REDD1 injected (n=7) animals. Center. Mean open arm duration (± SEM) in the elevated plus maze for control (n=6) and AAV-REDD1 injected (n=7) animals. Right. Mean number (± SEM) of closed arm entries in the elevated plus maze for control (n=6) and AAV-REDD1 injected (n=7) animals. (e) Mean (± SEM) percent co-localization of eYFP and phospho-S6 Ribosomal protein in control (n=5) and REDD1 virus-injected (n=6) rats. Right. Representative co-localization images. (f) Mean (± SEM) fold change of protein from mPFC microdissections from control (n=10) or AAV-REDD1 injected (n=10) rats. Representative blots are shown to the right. (g) Left Top. Representative images of a co-localized mPFC layer V pyramidal neurons positive for GFP and DiI. Left Bottom. Representative images of high magnification Z-stack projections of segments of layer V pyramidal cell primary apical dendrites. Right. Mean ± SEM of spine density from control (n=12) and AAV-REDD1 (n=12) groups. (h) Mean ± SEM of spine density of Filopodia (F), Long-Thin (LT), and Mushroom/Stubby (M/S) spines from control and AAV-REDD1 groups is depicted (F Ctrl n=11; F REDD1 n=12; LT Ctrl n=11; LT REDD1 n=11; M/S Ctrl n=12; M/S REDD1 n=11). (*) p < 0.05 relative to control.
To verify the expression and function of AAV-REDD1, we performed fluorescent double labeling for eYFP, to visualize virus-infected neurons, and phospho-S6, a downstream substrate of S6K (Fig. 4e). We found that AAV-REDD1 expression significantly decreased levels of phospho-S6 when co-localized with eYFP (p < 0.05). This was confirmed in a separate cohort by immunoblot analysis of microdissected mPFC. AAV-REDD1 infusion significantly increased REDD1 levels (p < 0.01) and decreased levels of phospho-mTOR and phospho-S6K compared to control virus (p < 0.01), demonstrating that REDD1 over-expression causes functional inhibition of mTORC1 signaling in the mPFC (Fig. 4f).
In a separate cohort of AAV-REDD1 injected animals, we used the Enhanced Spine Platform (ESP) (Afraxis, Inc.) to perform dendritic spine analysis by assessing eYFP-positive layer V neurons co-localized with DiOlistic labeling. We found that AAV-REDD1 led to atrophy of primary apical dendrites of layer V neurons (p < 0.05), as measured by spine density (Fig. 4g). Moreover, the spine density decrease in AAV-REDD1 injected animals was observed specifically in functionally mature mushroom/stubby spines (p < 0.05), but not immature filopodia or long-thin spines (Fig. 4h).
Taken together, the results demonstrate that REDD1 is increased in the PFC following CUS, and is necessary and sufficient for the synaptic deficits and depressive behaviors caused by CUS (Fig. 1c). The synaptic alterations due to REDD1 inhibition of mTORC1 are also consistent with regulation of mTORC1 via other convergent pathways. For example, stress decreases the expression of brain derived neurotrophic factor (BDNF) (22), which increases mTORC1 signaling via activation of Akt and inhibition of the TSC1/2 complex (23, 24). Consistent with this, mice lacking BDNF show reduced dendrite arbor and spine number, and occlusion of hippocampal neuronal atrophy caused by chronic stress (25, 26). The BDNF Val66Met polymorphism, which blocks the processing and release of BDNF, is associated with decreased hippocampal volume in humans and atrophy of neurons in the PFC of Met knock-in mice (27-29). We have also reported that expression of GATA1, a transcriptional repressor, is increased in CUS-exposed rats and in postmortem dlPFC of MDD patients and is sufficient to decrease expression of synapse-related genes (4). Together, these findings demonstrate that chronic stress decreases the expression of synapse-related genes and synaptic protein synthesis by regulation of both transcriptional and translational mechanisms. These findings also identify points for novel therapeutic interventions to block or reverse the effects of chronic stress on mTORC1 signaling, synaptogenesis, and depressive behaviors.
Materials & Methods
Male Sprague-Dawley rats with initial weights of 175–250g were pair-housed and maintained in standard conditions with a 12-hour light/dark cycle. Male REDD1 −/− (KO) mice or their wild-type littermates (22–30 g) were single-housed in plastic cages and maintained on a 12-hr light/dark cycle. Rats and mice were handled before use, and food and water were provided ad libitum throughout the experiments except when noted. Animal use and procedures were in accordance with the National Institutes of Health guidelines and approved by the Yale University Animal Care and Use Committee.
Drugs
Dexamethasone (Sigma) was administered I.P. at a concentration of 10mg kg−1. RU-486 (Sigma) was administered I.P. at a concentration of 30mg kg−1.
CUS Procedures
All animals were age- and weight-matched prior to commencement of CUS. Rats were subjected to a sequence of 11 stressors over the course of 21 d. These stressors included cold (4 °C for 1 h), cage rotation (1 h), isolation overnight, food and water deprivation overnight, light on overnight, light off during day (3 h), odor overnight, stroboscope overnight, crowding overnight, tilted cage overnight (45 °). Mice were subjected to a sequence of ten stressors over the course of 21 d. These stressors included cold (4 °C for 1 h), swim stress (18 °C for 10 min), cage rotation (1 h), restraint (1 h), food and water deprivation overnight, light on overnight, light off during day (3 h), odor overnight, stroboscope overnight, wet bedding overnight, and tilted cage overnight (45 °). All stressors were randomly interspersed throughout the stress period. For the 1 d mild stress group, rats were subjected to only the final day of stressors used in the 21 d CUS protocol.
Sucrose Preference Test
Rats were habituated to 1% sucrose for 48 h, and the side of the bottle was counterbalanced across animals and days. On testing day, rats were water-deprived for 6 h, then presented with pre-weighed, identical bottles of 1% sucrose and water. One hour later, the bottles were removed and weighed to determine consumption of each fluid.
Sucrose Consumption Test
Mice were habituated to 1% sucrose for 48 h. After overnight fluid deprivation, sucrose intake was measured the following morning by a 1 h test. The amount of sucrose intake was normalized to the amount of water intake (calculated from a 1 h consumption test following overnight fluid deprivation) for each animal.
Forced Swim Test
Rats were subjected to one 15 min session of swimming in water. Sessions were scored off-line by a blind observer. Mice were subjected to one 10 min session of swimming in water. Minutes two through six were scored off-line by a blind observer. Immobility was defined as the least amount of movement possible to stay afloat.
Novelty Suppressed Feeding Test
Rats were food-deprived overnight, then placed in an open field in the dark with pellets of food in the center. Time elapsed until the first bite of food was recorded. Immediately after the novelty test, home cage feeding was tested to verify motivation for food.
Elevated Plus Maze
Activity in an elevated plus maze apparatus with two open and two closed arms was measured over a 5 min period. For mice, frequency and time spent in open and closed arms were measured using the AnyMaze tracking system, while for rats, this was measured by a blind observer.
Open Field
Activity in an open field was measured over a 10 min period with AnyMaze (mice) or Ethovision (rats). Time spent in margin and center, total distance, and mean speed were measured during the session.
Western Blotting
Rodents were rapidly decapitated, and synaptoneurosome-enriched fractions were prepared from prefrontal cortex tissue samples and sonicated in lysis buffer. Protein was electrophoretically separated on a 7.5% SDS PAGE gel and transferred to a PVDF membrane. Blots were incubated in the appropriate primary antibody [phospho-S6 kinase (Thr 389; Cell Signaling 9234; 1:1,000); S6 kinase (Cell Signaling 2708; 1:1,000); phospho-mTOR XP (Ser 2448; Cell Signaling 5536; 1:500); mTOR (Cell Signaling 2983; 1:1,000); phospho-4EBP1 (Thr 37/46; Cell Signaling 2855; 1:500); phospho-ERK (Thr 202/Tyr204; Cell Signaling 4370; 1:1,000); ERK (Cell Signaling 4695; 1:1,000); phospho-Akt (Ser 473; Cell Signaling 4058; 1:1,000); Akt (Cell Signaling 9272; 1:1,000); GAPDH (Advanced Immunochemical 2-RGM2; 1:20,000); REDD1 (Proteintech 10638-1-AP; 1:500); TSC2 (Santa Cruz sc-893; 1:1,000); PSD-95 (Invitrogen 51-6900; 1:1,000] and developed using enhanced chemiluminescence (GE Biosciences). Optical densities of the bands were analyzed using NIH Image J software. For analysis, protein levels were normalized to total protein levels then expressed as a percentage of that in control animals. For figure panels, contrasts have been adjusted linearly for easier viewing of bands.
Quantitative real-time PCR
RNA was purified using RNAqueous (Ambion), and 500 ng of total RNA was used for cDNA synthesis (Genisphere). Gene-specific primers for REDD1 were designed using Primer 3 software. PCR was run on the Eppendorf Realplex Mastercycler using SYBR Green (Qiagen). Ct values of genes of interest were normalized to that of housekeeping genes (HMBS or GAPDH).
Enzyme-linked Immunosorbent Assay (ELISA)
Trunk blood was collected, then allowed to sit overnight at 4 °C. The following day, samples were centrifuged at 3,000 rcf, then the supernatant was collected and stored at −80 °C until use. All samples were run in duplicate using the Corticosterone ELISA (Enzo) and concentration was determined as percentage bound using a standard curve ranging from 32 – 20,000 pg mL−1.
Electrophysiological recordings and neurobiotin-labeled spine density analysis
Brain slices were prepared as previously described (14, 21). Briefly, mice were anesthetized (chloral hydrate, 400 mg kg−1, i.p.) and brains were removed. Coronal slices 400 μm thick were cut from a block of tissue containing the mPFC in ice-cold sucrose-ACSF with an oscillating-blade tissue slicer (Leica VT 1000S; GMI.Inc). Slices were placed in a submerged recording chamber; bath temperature was then raised slowly to 32 °C. The standard ACSF (pH 7.35), equilibrated with 95% O2/5% CO2, contained 128 mM NaCl, 3 mM KCl, 2 mM CaCl2, 2 mM MgSO4, 24 mM NaHCO3, 1.25 mM NaH2PO4, and 10 mM, d-glucose. There was recovery period of 1–2 h before recording.
Pyramidal neurons in layer V were patched under visual control using a microscope (60x IR lens; Olympus, Center Valley Pennsylvania) with infrared differential interference contrast microscopy (IR/DIC). Patch pipettes (3–5 MΩ) were pulled from glass tubing by using a Flaming-Brown Horizontal Puller (Sutter, Novato, California). The pipette solution contained the following: 115 mM K gluconate, 5 mM KCl, 2 mM MgCl2, 2 mM Mg-ATP, 2 mM Na2ATP, 10 mM Na2-phosphocreatine, 0.4 mM Na2GTP, and 10 mM Hepes, pH 7.33. Neurobiotin (0.3%) was added to the pipette solution to mark cells for later processing and imaging.
Whole-cell recordings were made with an Axoclamp-2B amplifier (Molecular Devices, Sunnyvale, California). The output signal was low-pass-filtered at 3 KHz and digitized at 15 kHz; data were acquired by pClamp 9.2/Digidata 1320 software (Molecular Devices). Postsynaptic currents were studied in the continuous single-electrode voltage-clamp mode (3,000 Hz low-pass filter) clamped near resting potential (75 mV ± 5 mV).
After completion of recording, slices were transferred to 4% paraformaldehyde (0.1 M phosphate buffer) then processed with streptavidin conjugated to Alexa 594 (1:1,000) for visualization of labeled cells. Labeled neurons within layer V of prelimbic mPFC were imaged with a two-photon Ti:sapphire laser scanning system (810 nanometers; Mai Tai, Spectra Physics, Mountain View, California) coupled to direct detection Radiance 2000 BioRad laser scanner (Zeiss Microimaging, Thornwood, New York) mounted on a Olympus BX50WI microscope, using a 60x (0.9 numerical aperture) water-immersion objective. For spine density analysis, Z-stacks usually consisted of 2–5 scans at high zoom at 1-μm steps in the z-axis. Spine density was sampled in apical tufts.
Ballistic dye labeling and microscopy and Afraxis ESP dendritic spine analysis
Ballistic delivery of DiI (48 h diffusion) to 300 μm thick sections was used to individually label neurons. Dendritic segments of 50 μm in length were scanned (0.1 × 0.1 0.33 μm) using laser□scanning confocal microscopy (Olympus FV1000, 63X, 1.42 NA). GFP-positive layer V pyramidal neurons were identified for scanning within mPFC, blind to experimental condition.
Blind deconvolution (AutoQuant) was applied to raw three-dimensional digital images, which were then analyzed for spine density and morphology by trained analysts. Individual spines were measured manually for (a) head diameter, (b) length, and (c) neck diameter from image Z-stacks using custom-built Afraxis ESP software (C++, Java). Each dendrite was analyzed by 2–3 independent analysts. Analysts were blinded to all experimental conditions (including treatment, brain region, cell type, and branch order). Data between analysts was averaged to report data for each cell.
Immunohistochemistry
Rats were rapidly and deeply anesthetized with chloral hydrate (400 mg kg−1, i.p.) and perfused through the heart with ice-cold phosphate buffered saline (PBS), followed by ice-cold 4% paraformaldehyde (PFA) in 0.1M phosphate buffer (PB). Free-floating sections (60 μm) containing the PFC were cut using a vibratome. Sections were stained with anti-GFP antibody (AbCam ab13970) and anti-phospho-S6-ribosomal protein antibody (Cell Signaling 5364) then visualized on a fluorescent microscope (Olympus). Co-localization was measured by blind selection of 10 GFP-labeled cells followed by assessment of co-localization with phospho-S6 Ribosomal Protein. Any fluorescent expression of phospho-S6 Ribosomal Protein was counted as positive labeling. Cell counts are expressed as a percentage of those GFP-labeled cells also expressing phospho-S6 Ribosomal Protein.
Construction, preparation, and infusion of recombinant adeno-associated virus
Human REDD1 was subcloned (Quark Pharmaceuticals) into an AAV2 plasmid under transcriptional regulation of CMV promoter (AAV2-REDD1-IRES-eYFP). The same backbone without the REDD1 gene was used as control plasmid. Human embryonic kidney 293 (HEK293) cells were transfected with AAV2, RC, and helper plasmids using a CaCl2 method. Seventy-two h after transfection, cells were harvested and lysed, then purified using HiTrap heparin HP affinity columns (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). Viral titers were determined in HT-1080 cells.
Under a mixture of ketamine (80 mg kg−1) and xylazine (6 mg kg−1) anesthesia, 1 μL of virus per side was delivered bilaterally to rat mPFC at a rate of 0.1 μL min−1, followed by 3 min of rest to allow diffusion. The coordinates for the mPFC were relative to Bregma (+3.5mm, ±0.5mm, −4.0mm). Rats were given carprofen (5 mg kg−1) as an analgesic and 5 mL of 0.9% saline to prevent fluid loss. Behavioral testing commenced four weeks following viral injection.
Human subjects
Brain tissue was collected as previously described from subjects diagnosed with MDD and psychiatrically-healthy age-matched subjects (30, 31). Briefly, the right hemisphere of the brain was blocked coronally, then immediately frozen and stored at −80 °C. Twenty μm sections were cut serially then collected in tubes with Trizol reagent and purified by RNeasy columns (Qiagen, USA) and RNA integrity was measured using RIN.
Statistical Analysis
For rodent studies, data was analyzed using two-tailed Student’s t-tests (for two-group comparisons) or ANOVAs (for multiple-group comparisons) (SPSS, Inc.), and differences were considered significant if p < 0.05. Post hoc analyses were carried out using Fischer’s LSD test. For postmortem studies, ANCOVA was used to compare gene expression levels in MDD and psychiatrically healthy controls, and differences were considered significant if p < 0.05. In the initial analysis, diagnostic status was the fixed factor, and age, PMI, pH, sex, and medication were included as covariates. In the reported analysis, we again used diagnostic group as the main effect, and included only those covariates that were significant for each gene in the analysis. ANCOVA revealed significant effects of age, PMI, and pH, but not sex or medication on REDD1 expression. ANCOVA revealed significant effects of age and PMI but not of pH, sex, or medication on mTOR expression. Outliers that were two standard deviations outside of the mean were excluded for all analyses.
Supplementary Material
Acknowledgements
This work is supported by NIMH R37MH45481 (RSD), NIMH R01MH93897 (RSD), NIMH F32MH98513 (KTO), NIGMS P30GM103328 (CAS), the State of Connecticut, and Yale University. We are grateful to the families consenting to donate brain tissue and be interviewed for the human tissue samples, and to the Cuyahoga County Medical Examiner’s Office for their assistance. We thank G. Rajkowska for identification of anatomically comparable regions of dlPFC. We thank Quark Pharmaceuticals for kindly providing the REDD1 plasmid and REDD1 KO mice. We thank M. Banasr for helpful discussions on behavioral experiments, X.-Y. Li for assistance in breeding and genotyping REDD1 KO mice, and A. Lepack and W. Andres for technical assistance. We are grateful for critical reading of the manuscript by J. Taylor, M. Picciotto, and A. Nairn.
Footnotes
Competing Financial Interests: The authors declare no competing financial interests.
Author Contributions: K.T.O. prepared the original draft of the manuscript and was involved in all aspects of the experimental design and research, including execution and analysis of all behavioral, biochemical, and molecular experiments, rodent surgeries and dissections, and design, construction, and preparation of recombinant AAVs. R.-J.L. performed all electrophysiological recordings and neurobiotin spine density analyses. B.V. assisted with optimization and preparation of the recombinant AAVs and with rat surgeries. J.G.M. and R.J.D. assisted in design and construction of the overexpression construct. V.D. assisted with qPCR execution and analysis. M.I. assisted with rodent behavioral testing and sample preparation. S.D. and C.D. assisted with sample preparation. S.B. and C.R. were responsible for ESP spine density imaging and analysis. D.A.L. and C.A.S. were responsible for human tissue generation and preparation of relevant human subjects’ information. G.K.A. was involved in the analysis and interpretation of the electrophysiological and spine density experiments. R.S.D. was involved in all aspects of study design, data analysis, interpretation of results, and preparation of the manuscript and figures. All authors discussed the results presented in the manuscript.
References
- 1.Kessler RC, Chiu WT, Demler O, Merikangas KR, Walters EE. Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62:617–627. doi: 10.1001/archpsyc.62.6.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drevets WC, Price JL, Simpson JR, Todd RD, Reich T, et al. Subgenual prefrontal cortex abnormalities in mood disorders. Nature. 1997;386:824–827. doi: 10.1038/386824a0. [DOI] [PubMed] [Google Scholar]
- 3.Rajkowska G, Miguel-Hidalgo JJ, Wei J, Dilley G, Pittman SD, et al. Morphometric evidence for neuronal and glial prefrontal cell pathology in major depression. Biol Psychiatry. 1999;45:1085–1098. doi: 10.1016/s0006-3223(99)00041-4. [DOI] [PubMed] [Google Scholar]
- 4.Kang HJ, Voleti B, Hajszan T, Rajkowska G, Stockmeier CA, et al. Decreased expression of synapse-related genes and loss of synapses in major depressive disorder. Nat Med. 2012;18:1413–1417. doi: 10.1038/nm.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radley JJ, Rocher AB, Miller M, Janssen WG, Liston C, et al. Repeated stress induces dendritic spine loss in the rat medial prefrontal cortex. Cereb Cortex. 2006;16:313–320. doi: 10.1093/cercor/bhi104. [DOI] [PubMed] [Google Scholar]
- 6.Radley JJ, Sisti HM, Hao J, Rocher AB, McCall T, et al. Chronic behavioral stress induces apical dendritic reorganization in pyramidal neurons of the medial prefrontal cortex. Neuroscience. 2004;125:1–6. doi: 10.1016/j.neuroscience.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Willner P. Chronic mild stress (CMS) revisited: consistency and behavioural-neurobiological concordance in the effects of CMS. Neuropsychobiology. 2005;52:90–110. doi: 10.1159/000087097. [DOI] [PubMed] [Google Scholar]
- 8.Izquierdo A, Wellman CL, Holmes A. Brief uncontrollable stress causes dendritic retraction in infralimbic cortex and resistance to fear extinction in mice. J Neurosci. 2006;26:5733–5738. doi: 10.1523/JNEUROSCI.0474-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shansky RM, Hamo C, Hof PR, McEwen BS, Morrison JH. Stress-induced dendritic remodeling in the prefrontal cortex is circuit specific. Cereb Cortex. 2009;19:2479–2484. doi: 10.1093/cercor/bhp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corradetti MN. The Stress-inducted Proteins RTP801 and RTP801L Are Negative Regulators of the Mammalian Target of Rapamycin Pathway. Journal of Biological Chemistry. 2005;280:9769–9772. doi: 10.1074/jbc.C400557200. [DOI] [PubMed] [Google Scholar]
- 11.Li N, Liu R-J, Dwyer JM, Banasr M, Lee B, et al. Glutamate N-methyl-D-aspartate receptor antagonists rapidly reverse behavioral and synaptic deficits caused by chronic stress exposure. Biol Psychiatry. 2011;69:754–761. doi: 10.1016/j.biopsych.2010.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banasr M, Valentine GW, Li X-Y, Gourley SL, Taylor JR, et al. Chronic unpredictable stress decreases cell proliferation in the cerebral cortex of the adult rat. Biol Psychiatry. 2007;62:496–504. doi: 10.1016/j.biopsych.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Jernigan CS, Goswami DB, Austin MC, Iyo AH, Chandran A, et al. The mTOR signaling pathway in the prefrontal cortex is compromised in major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35:1774–1779. doi: 10.1016/j.pnpbp.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li N, Lee B, Liu R-J, Banasr M, Dwyer JM, et al. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science. 2010;329:959–964. doi: 10.1126/science.1190287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Kubica N, Ellisen LW, Jefferson LS, Kimball SR. Dexamethasone represses signaling through the Mammalian Target of Rapamycin in muscle cells by enhancing expression of REDD1. Journal of Biological Chemistry. 2006;281:39128–39134. doi: 10.1074/jbc.M610023200. [DOI] [PubMed] [Google Scholar]
- 16.Polman JA, Hunter RG, Speksnijder N, van den Oever JM, Korobko OB, et al. Glucocorticoids Modulate the mTOR Pathway in the Hippocampus: Differential Effects Depending on Stress History. Endocrinology. 2012;153:4317–4327. doi: 10.1210/en.2012-1255. [DOI] [PubMed] [Google Scholar]
- 17.Boersma G, Benthem L, van Dijk G, Steimer TJ, Scheurink AJ. Pharmacological treatment of hyperinsulineamia in rats depends on coping style. Eur J Pharmacol. 2011;654:122–127. doi: 10.1016/j.ejphar.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 18.Stetler C, Miller GE. Depression and hypothalamic-pituitary-adrenal activation: a quantitative summary of four decades of research. Psychosom Med. 2011;73:114–126. doi: 10.1097/PSY.0b013e31820ad12b. [DOI] [PubMed] [Google Scholar]
- 19.Feng Q, Zou X, Lu L, Li Y, Liu Y, et al. The stress-response gene redd1 regulates dorsoventral patterning by antagonizing Wnt/beta-catenin activity in zebrafish. PLoS One. 2012;7:e52674. doi: 10.1371/journal.pone.0052674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shoshani T, Faerman A, Mett I, Zelin E, Tenne T, et al. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol Cell Biol. 2002;22:2283–2293. doi: 10.1128/MCB.22.7.2283-2293.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu RJ, Aghajanian GK. Stress blunts serotonin- and hypocretin-evoked EPSCs in prefrontal cortex: role of corticosterone-mediated apical dendritic atrophy. Proc Natl Acad Sci U S A. 2008;105:359–364. doi: 10.1073/pnas.0706679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duman RS. Role of neurotrophic factors in the etiology and treatment of mood disorders. Neuromolecular Med. 2004;5:11–25. doi: 10.1385/NMM:5:1:011. [DOI] [PubMed] [Google Scholar]
- 23.Hoeffer CA, Klann E. mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci. 2010;33:67–75. doi: 10.1016/j.tins.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takei N, Inamura N, Kawamura M, Namba H, Hara K, et al. Brain-derived neurotrophic factor induces mammalian target of rapamycin-dependent local activation of translation machinery and protein synthesis in neuronal dendrites. J Neurosci. 2004;24:9760–9769. doi: 10.1523/JNEUROSCI.1427-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magariños AM, Li CJ, Gal Toth J, Bath KG, Jing D, et al. Effect of brain-derived neurotrophic factor haploinsufficiency on stress-induced remodeling of hippocampal neurons. Hippocampus. 2011;21:253–264. doi: 10.1002/hipo.20744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Z-Y, Jing D, Bath KG, Ieraci A, Khan T, et al. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006;314:140–143. doi: 10.1126/science.1129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bueller JA, Aftab M, Sen S, Gomez-Hassan D, Burmeister M, et al. BDNF Val66Met allele is associated with reduced hippocampal volume in healthy subjects. Biol Psychiatry. 2006;59:812–815. doi: 10.1016/j.biopsych.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 28.Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, et al. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004;24:10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szeszko PR, Lipsky R, Mentschel C, Robinson D, Gunduz-Bruce H, et al. Brain-derived neurotrophic factor val66met polymorphism and volume of the hippocampal formation. Mol Psychiatry. 2005;10:631–636. doi: 10.1038/sj.mp.4001656. [DOI] [PubMed] [Google Scholar]
- 30.Kang HJ, Adams DH, Simen A, Simen BB, Rajkowska G, et al. Gene expression profiling in postmortem prefrontal cortex of major depressive disorder. J Neurosci. 2007;27:13329–13340. doi: 10.1523/JNEUROSCI.4083-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sibille E, Morris HM, Kota RS, Lewis DA. GABA-related transcripts in the dorsolateral prefrontal cortex in mood disorders. Int J Neuropsychopharmacol. 2011;14:721–734. doi: 10.1017/S1461145710001616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.