Abstract
Plant growth is severely affected by toxic concentrations of the non-essential heavy metal cadmium (Cd). Comprehensive transcriptome analysis by RNA-Seq following cadmium exposure is required to further understand plant responses to Cd and facilitate future systems-based analyses of the underlying regulatory networks. In this study, rice plants were hydroponically treated with 50 µM Cd for 24 hours and ∼60,000 expressed transcripts, including transcripts that could not be characterized by microarray-based approaches, were evaluated. Upregulation of various ROS-scavenging enzymes, chelators and metal transporters demonstrated the appropriate expression profiles to Cd exposure. Gene Ontology enrichment analysis of the responsive transcripts indicated the upregulation of many drought stress-related genes under Cd exposure. Further investigation into the expression of drought stress marker genes such as DREB suggested that expression of genes in several drought stress signal pathways was activated under Cd exposure. Furthermore, qRT-PCR analyses of randomly selected Cd-responsive metal transporter transcripts under various metal ion stresses suggested that the expression of Cd-responsive transcripts might be easily affected by other ions. Our transcriptome analysis demonstrated a new transcriptional network linking Cd and drought stresses in rice. Considering our data and that Cd is a non-essential metal, the network underlying Cd stress responses and tolerance, which plants have developed to adapt to other stresses, could help to acclimate to Cd exposure. Our examination of this transcriptional network provides useful information for further studies of the molecular mechanisms of plant adaptation to Cd exposure and the improvement of tolerance in crop species.
Introduction
Heavy metal ions are highly reactive and toxic to living cells, and their accumulation in plants is a major agricultural problem. Heavy metals can be classified into two categories based on their influence on plant growth. The first category includes essential mineral elements, trace amounts of which may be required for adequate growth and development, and an excess of which is toxic as soon as the concentration exceeds the threshold that plants can endure. The second category includes non-essential metals with recognized toxicity including cadmium (Cd) and lead (Pb). In particular, Cd is absorbed by the roots from the soil and transported to the shoot, negatively affecting nutrient uptake and homeostasis in plants, even at low concentrations. It is also known to adversely impact various biochemical and physiological processes including changes in the transcriptome and proteome of plants, resulting in inhibited root and shoot growth and, ultimately, reduced yield [1], [2], [3]. Cd pollution in arable soil has dramatically increased worldwide over the last several decades through the use of phosphate fertilizers, sludge, and irrigation water containing Cd. Furthermore, accumulation of Cd in the edible parts of plants such as seed grains places humans at a risk when ingesting them because of its highly toxic effects on human health. Thus, it is important to study the mechanisms of plant responses and defenses to Cd exposure to overcome this problem.
Cd causes oxidative stress and generates reactive oxygen species (ROS) such as superoxide radicals (O2 −) and hydrogen peroxide (H2O2), which can cause damage in various ways such as reacting with DNA causing mutation, modifying protein side chains and destroying phospholipids [4]. Many genes that respond to oxidative stress in plants, as well as genes associated with defense systems, have been studied, including induction detoxification enzymes such as glutathione S-transferase (GST), peroxidase (Prx), thioredoxins (Trx), peroxiredoxin (PrxR) and catalase (Cat) under Cd exposure, which confer Cd tolerance in plants [5], [6], [7], [8]. For defense against Cd toxicity, chelation of metal ions by ligands such as cysteine-rich metallothioneins (MT) and phytochelatins (PC) is also induced under Cd exposure. Such molecules bind Cd2+ ions through S-containing amino acid ligands and sequestrate the complexes to reduce cellular metal toxicity [1], [9], [10]. However, the manner in which the genes in these multigenic families respond to Cd has not been well investigated in rice.
Transporters with heavy metal binding domains are key factors for root uptake of Cd from soil and efflux pumping of Cd at the plasma membrane. The roles of transporters such as PDR9 (pleiotropic drug resistance-type ATP-binding protein 9) [11], LCT1 (low-affinity cation transporter 1) [12], and HMA3 (heavy metal ATPase3) [13] have been the focus of studies aimed at elucidating the mechanisms of Cd transport in rice. HMA3, which is located in the tonoplast, is well known to detoxify excess Cd by selectively sequestrating Cd into root vacuoles to decrease the concentration in the cell, so that translocation of Cd from the roots to the shoot is limited [13]. Cd transport is often caused by Fe or Zn transporters, such as IRT1 (iron-regulated transporter 1) and ZIP1 [Zrt (zinc-regulated transporter)/IRT-like protein 1], because of their low substrate specificity [14], [15], [16], [17]. HMA2 and MTP1 belong to the CDF (cation diffusion facilitator) protein family and are transporters of Zn and Cd [18]. Nramp5, a natural resistance-associated macrophage protein (NRAMP) family transporter, has been shown to transport Cd and Mn [19]. Although the functions of a few transporters for Cd transition and the effects of other ions on their expression have been reported in rice, a comprehensive overview of heavy metal transporters that change in expression under Cd exposure remains to be elucidated.
The recent elucidation of scaffolding mechanisms for Cd signaling pathways has begun to solve the puzzle of the complex Cd defense system in plants. Other signaling pathways contributing to the Cd stress response have not been well investigated but some important candidate genes, such as drought responsive element binding protein (DREB)/C-repeat binding factor (CBF), were identified to be involved by microarray-based approaches in rice [20]. However, a detailed view of the transcriptomic changes triggered by Cd exposure cannot be obtained with the Rice 44K Microarray (G2519F#15241, Agilent Technologies, Palo Alto, CA, USA) platform because it contains a probe set representing approximately 30,000 genes (the array probes were designed based on full-length cDNA structures), allowing for the detection of only 55% of the genes annotated in the rice genome [21].
Recently, the RNA-Seq strategy using next generation sequencers has become a useful tool for analyzing genome-wide gene expression to accurately quantify and catalogue all transcripts, including mRNAs and non-coding RNAs. With high resolution and sensitivity, RNA-Seq can provide detailed information on the transcriptional structure of genes [22]. The molecular mechanisms by which plants respond to changes in Cd stress are complex but of great importance, and could be useful in developing strategies for elucidating the gene networks involved in plant responses to various kinds of stress. Thus, we performed transcriptome analysis using RNA-Seq to compare gene expression profiles in seedlings of Cd-exposed and control (unexposed) rice plants (Oryza sativa L. cv. Nipponbare). We identified many novel responsive transcripts involved in signal transduction, antioxidation, detoxification and metal transport that might confer to tolerance to Cd exposure in rice. Moreover, we demonstrated that the overall gene expression of the Cd stress signaling network as an acute toxic response is involved in controlling drought stress signaling pathways by RNA-Seq analysis. This study and our previous study on Pi-stress [22] will contribute in understanding the genome-wide gene expression network of basal response to the stress in rice.
Materials and Methods
Sample preparation
Rice (Oryza sativa ssp. japonica cv. Nipponbare) seeds were germinated and grown by hydroponic culture in nutrient media [1.425 mM NH4NO3, 0.323 mM NaH2PO4, 0.513 mM K2SO4, 0.998 mM CaCl2, 1.643 mM MgSO4, 0.009 mM MnCl2, 0.075 mM (NH4)6 Mo7O24, 0.019 mM H3BO3, 0.155 mM CuSO4, 0.036 mM FeCl3, 0.070 mM citric acid, and 0.152 mM ZnSO4] [23] in a growth chamber at 28°C and 70–80% humidity in a 16h light/8h dark cycle. After 10 days, the seedlings of uniform size and growth were subjected to Cd stress treatment by transferring them to a similar medium with 50 µM CdSO4. The plants were maintained under Cd stress conditions for 120 h and then details of plant growth were recorded and sampling was performed as described previously [21]. Total RNA was extracted from all tissue samples using an RNeasy Plant Kit (Qiagen, Hilden, Germany) according to manufacturer's instructions. Construction of 13 cDNA libraries (2 tissues, 3 conditions, and 2–3 replicates) from total RNA using a TruSeq RNA sample preparation kit and sequencing with the Illumina Genome Analyzer IIx (Illumina Inc., San Diego, CA, USA) was performed according to the manufacturer's protocols.
Sequencing and mapping of short reads onto the rice genome
More than two biological replicates for each set of conditions were highly correlated (coefficient > 0.92), and reads from the same treatment were merged for subsequent analysis. Trimming of Illumina adaptor sequences and low-quality bases (Q < 20) at the 5′ and 3′ ends of each read was performed by Cutadapt [24] (http://code.google.com/p/cutadapt/) and a custom-made program. These pre-processed reads were mapped to the IRGSP-1.0 genome assembly (http://rapdb.dna.affrc.go.jp/) to reconstruct the transcript structures by a series of programs; Bowtie for short-read mapping [25], TopHat for defining exon–intron junctions [26], and Cufflinks for gene structure predictions [27]. To estimate the expression levels of each transcript, all pre-processed reads were mapped to the Os-Nipponbare-Reference-IRGSP-1.0 genome assembly (http://rapdb.dna.affrc.go.jp/) by Bowtie with default parameters [28]. The expression level for each transcript was calculated as RPKM (Reads Per Kilobase exon Model per Million mapped reads)-derived read counts [29] based on the number of uniquely mapped reads that overlapped with exonic regions. The resulting RNA-Seq data were deposited in the DDBJ Sequence Read Archive (Accession No. DRA001092).
Analysis of the responsive transcripts and alternative splicing patterns
We performed G-test to detect differentially expressed transcripts in control and Cd treatments based on the statistical null hypothesis that the proportions of mapped reads to the transcripts are the same between the two conditions. The frequency distribution of transcripts was determined by constructing 2×2 contingency tables with variables corresponding to the number of mapped and unmapped reads on a given transcript in control and each Cd treatment, respectively. A false discovery rate (FDR<0.01) was used in multiple hypothesis testing to correct for multiple comparisons. When calculating fold changes, 1 was added to avoid division by 0. Gene Ontology (GO) terms were assigned to each transcript from RAP-DB for each GO category. Enrichment of GO terms in the biological process category was evaluated by Fisher's exact test with a FDR threshold of 5% for responsive transcripts in major clusters at 1 h and 24 h after Cd treatment. The results were plotted as −log10 of FDR values in a heatmap. Microarray data (GSE6901, http://www.ncbi.nlm.nih.gov/geo/) were used to compare the responsive transcripts between Cd exposure (24 h) and other abiotic stresses (drought, salt and cold). The responsive transcripts (≥2-fold or ≤ 0.5-fold) in each treatment were used for Venn diagram analysis using the “venn” function of the R base package gplot version 2.10.1. For analysis of alternative splicing patterns in Cd-responsive genes, RPM (Reads per Million mapped reads) values of the splice sites in 5,222 representative loci were calculated using reads mapped on the sites. The splicing patterns in sites with RPM values > 1 were compared between control and Cd exposure samples.
qRT-PCR
The expression of Cd-upregulated metal ion transporter genes in root and shoot samples were confirmed by quantitative RT-PCR analysis. Rice seeds were germinated and grown in water in a growth chamber. After 10 days, the seedlings were subjected to different stress treatments by transferring them to water containing different reagents (50 µM CdSO4 for Cd exposure, 50 µM AlCl3 for Al exposure, 50 µM CuSO4·5H2O for Cu exposure, 50 µM FeSO4·7H2O for Fe exposure condition, 50 µM HgCl for Hg exposure, 50 µM MgCl2·6H2O for Mg exposure, 50 µM MnCl2·4H2O for Mn exposure, 50 µM NaCl for Cl exposure, 50 µM NiCl2 for Ni exposure, 50 µM RbCl for Rb exposure or 50 µM ZnCl2 for Zn exposure). Ten-day-old rice seedlings transferred to water were used as a control for the stress treatment. Total RNA was extracted from samples collected after 24 h of each treatment. After DNase I (Takara, Shiga, Japan) treatment, first-strand cDNA was synthesized using the Transcriptor First Strand cDNA synthesis kit (Roche, Basel, Switzerland) according to the manufacturer's protocol. The resulting cDNA was used for PCR amplification in the LightCycler 480 system (Roche, Basel, Switzerland) with each primer set. The detection threshold cycle for each reaction was normalized using Ubiquitin1 with 5′-CCAGGACAAGATGATCTGCC-3′ and 5′-AAGAAGCTGAAGCATCCAGC-3′ as primers. Three technical replicates for each treatment were used for analysis.
Results and Discussion
Changes in plant morphology under Cd exposure
In the hydroponically cultured rice, growth was greatly affected by Cd exposure (Figure 1). In Cd-exposed rice, growth retardation of the shoot was apparent after 24 h, many dark spots appeared on all leaves after 48 h, the leaves turned yellow and the leaf tips of the seedlings began to wilt after 72 h, and after 120 h of 50 µM cadmium exposure all leaf blades were curled completely and the seedlings were shrunken and wilting, which is attributable to supply of Cd (Figure 1). This concentration or higher concentrations of Cd have been previously shown to elicit robust physiological responses and gene expression as acute toxic responses in rice seedlings [3], [30], [31]. The wilting occurred gradually compared with drought treatment for 24 h, in which symptoms started to appear after 2 h treatment and plants were completely dried up after 24 h, in the same growth chamber (Figure S1). The detoxification processes of the plant are insufficient to cope with the toxic metal beyond a 10 µM dose, and wilting and reduction of plant fresh weight are accompanied by decreased leaf conductance and increased stomatal closing [32]. It has been suggested that part of the fatal damage to plants from Cd exposure occurs through drought stress.
Figure 1. Rice phenotypes under Cd exposure.
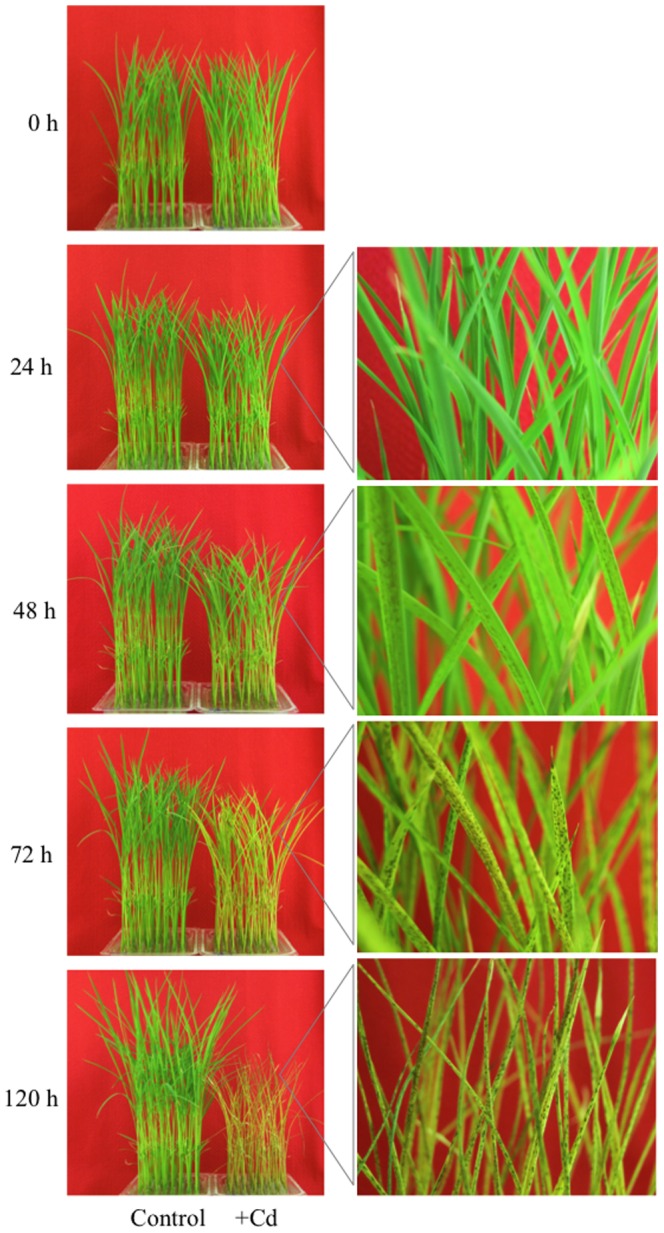
Phenotypic changes in rice plants grown from 0 to 120 h in culture medium with 50 µM CdSO4 for Cd stress. The shoots showed growth retardation. Black spots on the leaves gradually increased and leaves curled up slowly during Cd exposure. RPKM fold changes at 1 h and 24 h were calculated for Cd treated samples compared with non-treated samples (0 h).
Cd treatment induced overall changes in gene expression in rice
To ascertain our hypothesis at the transcriptional level, we generated transcriptome profiles of the early response to Cd exposure using RNA-Seq during plant growth, particularly at 1 h and 24 h after 50 µM Cd treatment and before treatment (0 h). For each set of conditions, an average of approximately 48.3 million (90.7%) quality-evaluated reads were mapped to the rice genome sequence and used for further analysis (Table S1). Among the 60,163 transcripts detected in total (RPKM > 0), 36,222 were identified to be responsive to Cd exposure and were used to dissect the transcriptional responses associated with tissue (root and shoot) and time of collection (1 h and 24 h). The number of responsive transcripts was 16,814 in roots and 14,264 in shoots at 1 h, whereas it was 26,098 in roots and 23,924 in shoots at 24 h (Figure 2, Table S2), suggesting the effect of Cd exposure was enhanced gradually up to 24 h. Several MTs, Prxs and heat shock proteins (Hsps) were strongly upregulated among the 20 genes with the greatest relative expression at 24 h. Table 1 shows strongly upregulated (top 20) transcripts without probes in the Rice 44K Microarray in roots and shoots at 24 h. We also found transcription factors (e.g. AP2-EREBP, NAC) that may function as regulatory factors under Cd exposure among these novel Cd-responsive transcripts, most of which had uncharacterized functions. Furthermore, the responsive transcripts included 1.84–7.98% unannotated transcripts predicted by the Cufflinks program in each particular tissue/time. A validation experiment on the responsive unannotated transcripts was performed by qRT-PCR analysis [21]. These novel transcripts may represent interesting novel targets to understand Cd regulatory pathways in rice. Moreover, among 44,519 representative loci on the rice genome (IRGSP-1.0), alternative splicing isoforms were confirmed in 5,222 (11.7%) representative loci. Changes in the alternative splicing patterns of these loci were examined against Cd exposure and 2,873 loci (6.5%) showed different patterns. RNA-Seq is a promising tool for analyzing alternative patterns that microarrays are unable to, and approximately ∼48% of rice genes showed alternative splicing patterns [33]. This suggests that Cd responsive expression is also regulated by alternative splicing mechanisms with respect to the timing of the response or tissue specificity. In conclusion, RNA-Seq data were far superior to data derived from the microarray and that Cd treatment affected many genes and caused drastic changes in gene expression in rice.
Figure 2. Distribution of upregulated and downregulated transcripts in roots and shoots responsive to Cd treatment.
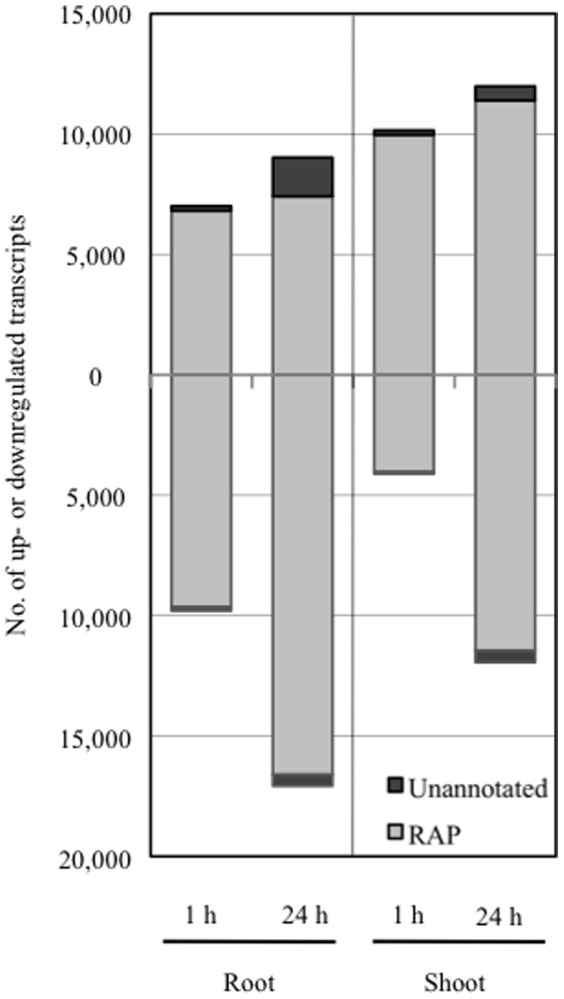
The total numbers of upregulated (upper) or downregulated (lower) transcripts in roots (left) and shoots (right) identified by RNA-Seq were determined by G-tests (FDR < 0.01) at each stress timepoint (1, 24 h) during Cd exposure in comparison with non-treatment (0 h). Each bar shows the distribution of transcripts with matching RAP-DB annotations (grey) and unannotated transcripts (black). The x-axis shows the time course and the y-axis shows number of transcripts.
Table 1. Cadmium-upregulated transcripts identified in roots and shoots by RNA-Seq analysis.
| Root 24 h | Description | Fold change |
| Os12t0568166-01 | Conserved hypothetical protein | 341.43 |
| Os05t0211700-00 | - | 210.16 |
| Os08t0404900-00 | Conserved hypothetical protein | 170.60 |
| Os09t0492900-00 | Conserved hypothetical protein | 170.46 |
| Os01t0653300-00 | VQ domain containing protein | 151.31 |
| Os01t0661750-00 | Conserved hypothetical protein | 142.92 |
| Os06t0133500-00 | Conserved hypothetical protein | 129.63 |
| Os06t0662550-01 | Conserved hypothetical protein | 108.59 |
| Os07t0154201-00 | Hypothetical gene | 103.63 |
| Os06t0147250-00 | Conserved hypothetical protein | 102.14 |
| Os02t0464550-01 | Conserved hypothetical protein | 101.50 |
| Os11t0495400-00 | PHF5-like protein | 100.00 |
| Os04t0429050-00 | AP2-EREBP | 97.98 |
| Os10t0525200-01 | Cytochrome P450 family protein | 93.75 |
| Os01t0498802-01 | Non-protein coding transcript | 93.09 |
| Os12t0418600-01 | Hypothetical conserved gene | 82.22 |
| Os07t0162000-00 | - | 82.14 |
| Os06t0146650-00 | Conserved hypothetical protein | 70.88 |
| Os08t0336200-01 | Hypothetical gene | 70.61 |
| Os10t0525301-00 | Hypothetical gene | 68.00 |
Reads were mapped to the rice genome and responsive genes were identified by G-tests. The top 20 upregulated transcripts in roots and shoots that are not represented on the Rice 44K microarray platform are shown.
Functional characterization of Cd-responsive transcripts
To investigate the functions of cadmium stress-responsive transcripts, we performed Gene Ontology (GO) analysis and found an overrepresentation of specific GO keywords that denote involvement with processes triggered by stress, including metal ion transport (GO:0030001) (upregulated, root), response to stress (GO:0006950) (upregulated, root and shoot), trehalose biosynthetic process (GO:0005992) (upregulated, shoot), DNA replication (GO:0006260) (downregulated, root), DNA repair (GO:0006281) (downregulated, root), translation (GO:0006412) (downregulated, root and shoot), and photosynthesis (GO:00015979) (downregulated, shoot) (Figure S2). These enriched GO terms among the upregulated transcripts indicate that RNA-Seq was successful in identifying Cd-responsive genes. The enriched GO terms among downregulated transcripts result in growth retardation (Figure 1). These results imply that Cd controls a significant part of the defense to stress and plant growth.
Among the GO keywords identified, response to stress (GO:0006950) included many drought stress responsive genes such as the Arabidopsis cor47 homolog Dip1 [34], Rab 21 ( = Rab16A) (responsive to abscisic acid 21) genes [35], and other Rab 16 genes [36] in roots and shoots at 24 h under Cd exposure (Table S3). The signaling pathways in drought, high-salinity and low temperature stress show high levels of cross-talk among abiotic stresses in Arabidopsis [37]. Our results suggested that expression of these genes responsive to drought stress (also implying a relationship to high-salinity and low temperature stresses) was affected by Cd exposure, but a relationship between Cd and drought stresses at the transcription level has not been reported as far as we know.
Cd exposure triggered upregulation of drought stress responsive genes
We investigated the expression of well-characterized drought stress-related transcription factors (TFs) and their downstream genes under Cd exposure to ascertain the relationship between Cd and drought stresses.
i) DREB/CBF TFs
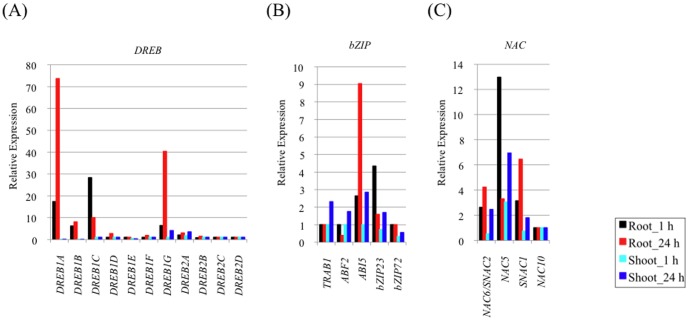
Among the DREB/CBF TFs, which contain drought-responsive elements (DRE), DREB1A/CBF3 and DREB1G were drastically upregulated in roots after 24 h of Cd exposure and DREB1C/CBF2 was drastically upregulated in roots after 1 h (Figure 3A). Over-expression of DREB1A/CBF3 improved tolerance to drought, high-salinity and low temperature, and resulted in growth retardation under non-stress conditions, suggesting functional similarity to Arabidopsis [38], [39]. Over-expression of DREB1G significantly elevated tolerance to drought [40]. As the transcriptional networks mediated by DREB1 are conserved in plants and DREB1 regulates many downstream target genes under abiotic stress [37], part of the signaling pathway related to DREB1 might be enhanced under Cd exposure in rice.
Figure 3. Effects of Cd exposure on rice growth and the expression of abiotic stress-related TF genes.
The relative expression of (A) DREBs, (B) bZIPs and (C) NACs in rice is shown. The x-axis shows transcripts and the y-axis shows relative expression. The black bar shows the relative expression in roots at 1 h, the red bar roots at 24 h, the light blue bar shoots at 1 h and the blue bar shoots at 24 h.
ii) bZIP TFs
Several bZIP domain TFs were characterized that regulated expression of ABA-responsive genes in rice. The abiotic stress-inducible genes bZIP23 [41] and ABI5 [42] were upregulated in roots at 1 h and 24 h during Cd exposure, respectively (Figure 3B). bZIP23 and ABI5 are thought to function as key components in ABA-dependent transcriptional networks in rice, suggesting the existence of gene expression regulation through ABA-dependent pathways under Cd exposure. The existence of an ABA-dependent pathway was supported by strong upregulation of NCED3 and NCED9 in roots (Table S3). In our BLASTP search, the Arabidopsis proteins AtNCED3 [43] and AtNCED9 [44], key enzymes of ABA synthesis during drought stress and seed development, respectively, were the top hits to NCED3 and NCED9 in rice. ABA is known to accumulate rapidly and confer stress tolerance by inducing many genes under drought [43], [44], [45], [46]. Synthesis of ABA following Cd stress can lead to decreased leaf conductance as Cd2+ toxicity perturbs the plant–water relationship [32] (Figure 1). LEA3-1 was upregulated in bZIP23 overexpressors [41], and here it was also upregulated in roots and shoots (Table S3). LEA genes often appear to be ABA-dependent [47], and their proteins may function in protecting macromolecules such as enzymes, protein complexes and membranes [48]. Overexpression of LEA3-1 in rice can significantly improve relative yield under drought stress [49].
iii) NAC TFs
Several NAC TFs, including NAC5 [50] and SNAC1 [51], are induced by drought, high-salinity and low-temperature. NAC5 was upregulated in roots at 1 h and shoots at 24 h, and SNAC1 was upregulated in roots at 24 h during Cd exposure (Figure 3C). NAC5 is regulated through an ABA-dependent pathway [52]. LEA3, which was identified as a NAC5 downstream gene [50], [53], and a rice ERD1 (early responsive to drought 1) homologue (OsERD1) were also upregulated in roots and shoots (Table S3). The expression of OsERD1 is probably regulated by SNAC1 through the NAC recognized sequence (NACRS) promoter [51], [54]. Over-expression of SNAC1 can significantly improve drought resistance and confer strong tolerance to high-salinity stress [51].
iv) Other TFs
Many other stress-related TFs, including AP2/ERFs (AP37, AP59), C2H2 zinc finger (ZFP252), TIFY (TIFY11) and MYB (Myb4), were also upregulated under Cd exposure (Table S3). Overexpression of AP37, AP59 [55] and ZFP252 [56] results in drought and high-salinity tolerance. Overexpression of ZFP252 in rice increased the amount of soluble sugars and free proline [56]. The accumulation of soluble raffinose and the upregulation of galactinol synthase genes, key enzymes of raffinose synthesis, have been reported in drought-, high salinity- and low temperature-treated Arabidopsis plants [57]. In our study, galactinol synthase and raffinose synthase genes were strongly upregulated in roots under Cd exposure (Table S3). Cd treatment has also been shown to increase the level of raffinose in Arabidopsis [58]. The AtGolS3 galactinol synthase gene is controlled by DREB1A through DRE and DRE-like cis-elements in Arabidopsis [57], [59]. These results suggest raffinose is a metabolite for adaptation to Cd and might be produced by enhancing a DREB-related pathway in rice. P5CS, a key enzyme of osmoprotectant proline synthesis, was upregulated in shoots under Cd exposure (Table S3). Accumulation of proline as an enzyme protectant has been reported in rice seedlings exposed to Cd [60], [61], suggesting that proline is also a metabolite for adaptation to Cd. Overexpression of TIFY11a [62] resulted in high-salinity and drought tolerance, and overexpression of Myb4 [63] resulted in tolerance to low temperature, so upregulation of TIFY11 and Myb4 may also function in tolerance to Cd exposure (Table S3). Thus, many reports suggest that overexpression of stress-inducible TFs can increase abiotic stress tolerance to drought, high-salinity, or low temperature in plants, so upregulation of such TFs could be useful for developing transgenic crops with enhanced tolerance to Cd stress.
Some (DREB, bZIPs and NCED) were more upregulated in roots than in shoots under Cd exposure up to 24 h and the tendency of their expression patterns was confirmed by qRT-PCR analysis, which was different to the tendency under drought stress for 3 h (Figure S3, Table S4). As OsDREB1A and OsDREB1B were included in the 50 genes with the greatest relative expression in roots exposed to 10 µM Cd-treatment for 3 h [20], roots might be affected more directly and earlier by Cd exposure. In conclusion, our data clearly indicated that Cd affects the expression of genes in the drought-related signaling pathway, especially in roots, because the Cd exposure treatment was performed in hydroponic culture.
We also confirmed the upregulation of Jacalin1, LOX (lipoxygenase), BBTI 1 (Bowman Birk trypsin inhibitor 1), Receptor kinase containing LRR repeats, PSLS (Phospho sulfolactate synthase), Hsp70, PP2Ca and PP2Cb in one or more of the tissue/treatment combinations (Table S3). The upregulation of these genes was ascertained to help plants acclimate to stress conditions, such as drought in AtCBF3- or AtABF3-overexpressing transgenic rice [64]. These results suggest that Cd exposure more or less perturbs the expression of genes in drought, high salinity- and low-temperature stress signaling pathways, which results in similar morphological changes in response to both Cd and drought (Figure 1, Figure S1). Even though comparative analysis using microarray data revealed that 28.0–54.3% of the responsive transcripts (≥2-fold or ≤ 0.5-fold) responded to other abiotic stresses (drought, high salinity or low-temperature) in roots and shoots (Figure S4), Cd-responsive transcripts responded to drought/high-salinity stresses more than twice as much as to low temperature stress (Figure S4), suggesting there are higher levels of cross-talk between Cd and drought/high-salinity stresses than in the cross-talk between Cd and low temperature stress as expected from observation of wilted seedlings under Cd exposure (Figure 1). We also identified that 9% of Cd-upregulated transcripts were commonly upregulated among the four stresses, including SNAC1, ZEP252 and Myb4 (Table S5), which may confer general adaptation under abiotic stress.
Upregulation of genes in the defense system to Cd exposure and RNA-Seq revealed the regulated genes of multigenic families
Next, we investigated the expression of antioxidant and detoxification enzymes to confirm that our expression profiles reflected the response to Cd exposure.
i) Antioxidant enzymes for ROS-scavenging
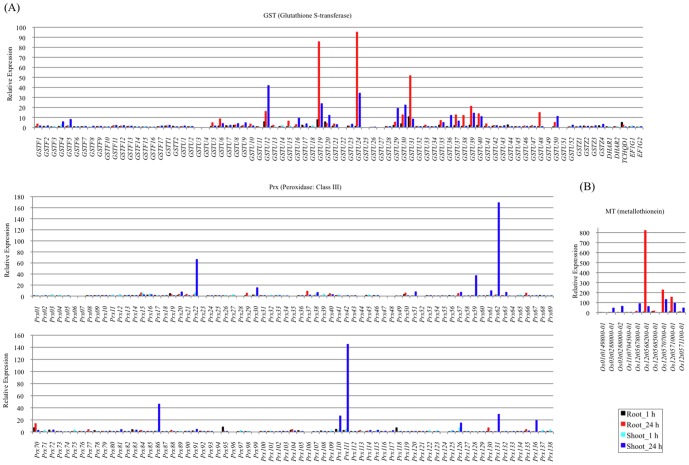
The production of ROS is unavoidable under various biotic and abiotic stresses including Cd exposure. The accumulation of ROS is largely counteracted by an intricate antioxidant defense system [6]. The enzymatic scavengers GST [65] (Pfam accession: PF02798.15, PF13417.1, PF13410.1, PF00043.20, http://pfam.sanger.ac.uk/) and class III Peroxidase (Prx) (E.C. 1.11.1.7) [66] have large and complex gene families that control key metabolic steps in many eukaryotic systems. Among 80 GST genes, the responsive genes tended to be more upregulated in roots at 24 h compared with the other tissue/treatment combinations. In particular, GSTU12 (shoot), GSTU19 (root and shoot), GSTU24 (root and shoot), GSTU30 (shoot), GSTU31 (root) and GSTU39 (root) were strongly upregulated [fold change (FC) > 20] (Figure 4A). GSTU19, GSTU24 and GSTU39 were found to be upregulated in the roots of rice seedlings under arsenate exposure using a microarray platform [67], suggesting that part of the Cd defense might be similar to the As defense in the roots of rice. Enhanced GST with glutathione peroxidase activity in transgenic tobacco increased glutathione-dependent peroxide scavenging and alterations in glutathione and ascorbate metabolism that led to reduced oxidative damage [68]. Class III Prx genes catalyze the reduction of H2O2 by transferring electrons to various donor molecules such as lignin precursors and secondary metabolites [69], [70]. Prx22, Prx59, Prx62, Prx86, Prx110, Prx111 and Prx131 among 138 Prxs were upregulated strongly (FC > 20) in shoots after 24 h of Cd exposure (Figure 4A). Prx86 and Prx111 were strongly upregulated in the plant defense against a gall midge [71] and against Xanthomonas oryzae and Magnaporthe grisea [72]. As many studies have described the diverse functions of Prx in Arabidopsis [8], [73], these genes might be expressed in a time and tissue specific manner in rice.
Figure 4. Relative expression of transcripts that may function in defense and detoxification under Cd treatment.
RPKM fold changes at 1 h and 24 h were calculated for Cd treated samples compared with non-treated samples (0 h). (A) Relative expression of the GST gene family, (B) the Prx gene family, and (C) the MT gene family are shown. The x-axis shows transcripts and y-axis shows relative expression. The black bar shows the relative expression in roots at 1 h, the red bar roots at 24 h, the light blue bar shoots at 1 h and the blue bar shoots at 24 h.
Genes encoding various kinds of ROS-scavenging enzymes, such as glutaredoxin (Grx) [74], Trx [75], PrxR [76], monodehydroascorbate reductase (MDAR) [77], alternative oxidase (AOX) [78], Cat (PF00199), and alpha-dioxygenase (α-DOX1) [79] were also upregulated (FC > 3) (Figure S5), suggesting activation of several antioxidative systems in the plant cells under Cd exposure. Most of these genes tended to be upregulated more at 24 h compared with 1 h after Cd exposure. α-DOX1, which plays a role in protecting tissues from oxidative damage [80], was the most upregulated (∼94-fold) in shoots at 24 h under Cd exposure. α-DOX1 is well-known to be upregulated and show increased activity under oxidative- and heavy metal-stresses [80]. Tissue-specific upregulation was observed for some genes, such as Os01g0194600 (Grx) in roots and α-DOX1 in shoots. Even though the genes were from the same family, we observed tissue-specific expression for different PrxR and AOX genes. One of the two genes in each family was upregulated more in roots compared with shoots and the other gene showed an opposite upregulation pattern (Figure S5). Respiratory burst oxidase homolog (Rboh: NADPH oxidase) genes that produce ROS [6], [81] were also upregulated (Figure S5). These results indicate that the treatment evokes ROS in rice cells. However, while the gene expression profiles of the whole multigenic family have not been well investigated under Cd exposure, the regulated genes in the multigenic family were elucidated clearly by RNA-Seq analysis. Through the use of gene transfer technology to manipulate the activities of these antioxidant enzymes, ROS-scavenging systems in plants might be used to increase plant stress tolerance.
ii) Detoxification enzyme through chelation
The chelation of heavy metals by particular ligands is a well-characterized mechanism of Cd detoxification that has evolved in plants. The two best-characterized heavy metal-binding ligands in plant cells are PC and MT, which are involved in detoxification [9]. PC is synthesized from reduced glutathione (GSH) in a transpeptidation reaction [82]. PC synthase (PF05023.9) and GSH synthase (PF04107, PF03917, PF03199) genes did not show much response to Cd exposure in this study, but cad1, a PC deficiency mutant in Arabidopsis, is Cd sensitive [83]. MTs have been found in diverse organisms including mammals, plants, and fungi as well as some prokaryotes and function in maintaining homeostasis of essential metals and metal detoxification [1], [9]. MTs are rich in cysteine residues that coordinate multiple metal atoms such as cadmium, zinc and copper. Eleven MT genes were found to be differentially expressed during growth and development, in various tissues and during biotic and abiotic stresses [84], [85]. Here, 9 of these 11 MT genes were strongly or weakly upregulated in at least one set of conditions (Figure 4B). Interestingly, two sets of three MT genes (Os12g0567800, Os12g0568200, Os12g0568500 and Os12g0570700, Os12g0571000, Os12g0571100) reside on chromosome 12 within 40-kb and 15-kb regions, respectively. As the clustering of MTs is well-known in mouse and human [86], these might have evolved from gene duplications in rice. These clustered genes were upregulated at different levels in at least two of the tissue/treatment combinations (Figure 4B). Os12g0568200 showed the strongest upregulation (821-fold) among them, in roots at 24 h under Cd exposure (Figure 4B). Simultaneously, cis-regulatory changes and minor changes in the regulatory regions of genes may occur during evolution [87], [88], resulting in diversity of gene expression levels. Members of the rice MT gene clusters differed in their tissue expression patterns, suggesting that each gene may perform different functions in specific tissues. The responsive transcripts of antioxidative and detoxification enzymes differed in their expression patterns among metal stresses (Figure S6, Table S4). Their upregulation indicated that our expression profiles under Cd exposure obtained by RNA-Seq analysis were reliable.
Responses of various metal transporter genes under Cd and other metal stresses
To confirm whether other ions affect the expression of Cd-responsive genes, we investigated the expression of metal transporter genes from the HMA, MatE (multi antimicrobial extrusion protein) [89], Zip, CDF, NRAMP, and PDR families, as well as LCT1 [12] and LCT1 homologues (Os06g0576200, Os06g0674000), because at least one gene from each of these families and LCT1 have been reported to function in Cd transport. As LCT1 did not have any Pfam domain for heavy metal binding, we searched for homologues in RAP DB using a BLASTP search (threshold of 1e-03). These gene families contain specific metal ion binding domains [PF01554 (MatE), PF08370 (PDR_assoc), PF01545 (Cation_efflux), PF02535 (Zip), PF00403 (HMA), PF01566 (Nramp)], which may function in Cd transport. In total, 88.9% of the transporters (168 transcripts) were responsive to Cd exposure, with 35.7% (60 transcripts) in the HMA family, 29.8% (50 transcripts) in MatE, 10.7% (18 transcripts) in Zip, 7.7% (13 transcripts) in Cation_efflux and Nramp, 6.0% (10 transcripts) in PDR_assoc and 2.4% (4 transcripts) being LCT1 and LCT1 homologue. PDR9 [11], LCT1 [12] and HMA3 [13], which are important of Cd translocation in rice, were identified as the responsive transcripts. Transcripts upregulated more than 5-fold in one or more of the tissue/treatment combinations are shown in Table 2. Os02t0585200 containing an HMA domain was the most upregulated in roots (87-fold), and Os01t0609900 containing a PDR_assoc domain was the most upregulated in shoots (53-fold) at 24 h during Cd exposure. These results emphasized the potential of the RNA-Seq strategy to reveal novel Cd-responsive transporter transcripts in rice. It should be mentioned that the expression levels were diverse among the gene families, suggesting they may have differentiated functions in transporting various metal ions.
Table 2. Cadmium-responsive metal ion transporters identified in rice.
| Root | Shoot | ||||
| Family | Transcript | 1 h | 24 h | 1 h | 24 h |
| HMA | Os02t0585200-01 | 10.78 | 87.00 | 0.27 | 2.11 |
| Os03t0152000-01 | 7.68 | 34.42 | 0.09 | 0.91 | |
| Os02t0584800-01 | 3.15 | 33.27 | 0.15 | 0.78 | |
| Os02t0585100-00 | 5.67 | 23.19 | 1.03 | 5.24 | |
| Os02t0584700-01 | 3.32 | 17.49 | 0.23 | 1.09 | |
| Os03t0372600-00 | 2.48 | 9.90 | 1.21 | 2.41 | |
| Os02t0530100-02 | 0.98 | 7.17 | 0.55 | 0.42 | |
| Os02t0530100-01 | 0.92 | 7.03 | 0.56 | 0.45 | |
| Os01t0976300-01 | 3.07 | 6.36 | 0.96 | 13.77 | |
| Os04t0244800-01 | 5.67 | 3.67 | 1.95 | 5.61 | |
| Os06t0542300-01 | 1.39 | 2.35 | 0.62 | 15.21 | |
| Os06t0665800-01 | 1.06 | 2.33 | 1.51 | 5.90 | |
| Os08t0403300-00 | 0.49 | 1.73 | 1.07 | 6.41 | |
| Os03t0178100-00 | 1.10 | 0.67 | 1.85 | 5.25 | |
| MatE | Os10t0344000-01 | 1.05 | 12.19 | 0.63 | 13.65 |
| Os03t0188100-01 | 1.75 | 9.77 | 1.22 | 14.48 | |
| Os10t0345100-01 | 2.63 | 4.53 | 2.04 | 6.69 | |
| Os04t0571600-01 | 10.32 | 2.55 | 1.13 | 0.25 | |
| Os03t0572900-01 | 1.18 | 0.97 | 1.45 | 7.38 | |
| Os07t0502200-01 | 2.45 | 1.30 | 2.65 | 6.24 | |
| Os01t0504500-02 | 0.97 | 0.90 | 1.10 | 5.39 | |
| Os02t0676400-00 | 7.77 | 0.73 | 1.12 | 1.13 | |
| Zip | Os03t0411800-01 | 1.21 | 10.56 | 1.20 | 14.60 |
| Os01t0972200-00 | 5.06 | 1.69 | 1.39 | 2.07 | |
| Cation_efflux | Os01t0130000-01 | 1.45 | 1.02 | 1.46 | 6.70 |
| Os01t0130000-02 | 1.28 | 0.92 | 1.30 | 5.82 | |
| Nramp | Os07t0258400-02 | 0.66 | 5.05 | 1.08 | 1.50 |
| PDR_assoc | Os01t0342750-01 | 0.75 | 5.82 | 0.91 | 2.47 |
| CUFF.28142.2 | 2.97 | 2.43 | 2.47 | 45.91 | |
| Os01t0609900-02 | 1.75 | 2.14 | 1.77 | 53.64 | |
| Os01t0609300-01 | 1.55 | 2.03 | 1.03 | 32.64 | |
| Os08t0384500-01 | 1.51 | 1.30 | 1.59 | 5.20 | |
| Os01t0609200-00 | 6.67 | 0.74 | 1.04 | 1.15 | |
| LCT1 | CUFF.25087.1 | 1.11 | 1.57 | 1.87 | 5.65 |
| CUFF.25087.2 | 1.08 | 1.48 | 1.96 | 6.04 | |
| CUFF.25087.3 | 1.12 | 1.49 | 1.95 | 5.97 | |
Metal ion transporters containing Pfam domains [PF01554 (MatE), PF08370 (PDR_assoc), PF01545 (Cation_efflux), PF02535 (Zip), PF00403 (HMA), PF01566 (Nramp)], LCT1 (unannotated transcripts by RAP, identified by the Cufflinks program) and LCT1 homologues upregulated more than 5-fold in one or more of the tissue/treatments combinations are shown. Bold characters show fold changes greater than 5 in upregulated transcripts. CUFF.28142.2 (chr07: 20207865..20213557) CUFF.25087.1 (chr06: 22566131..22572032), CUFF.25087.2 (chr06: 22566593..22572032) and CUFF.25087.3 (chr06: 22567729..22571545) were identified by the Cufflinks program.
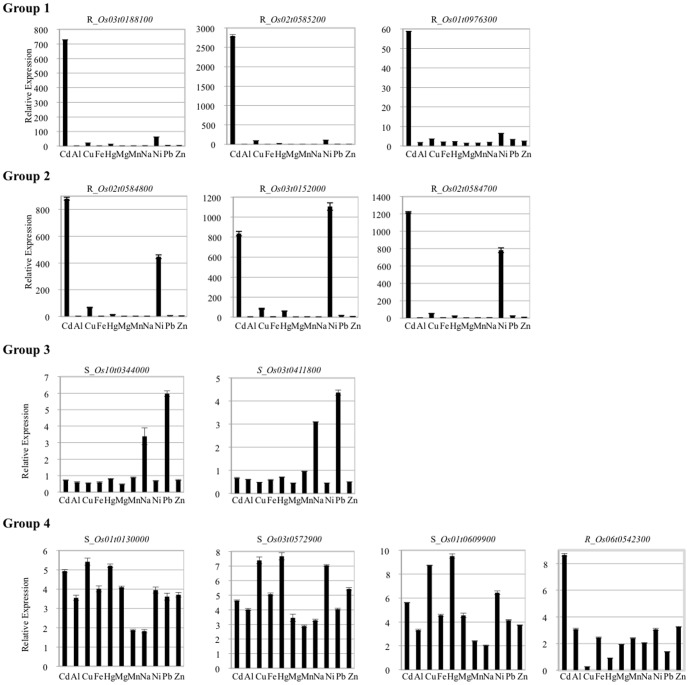
Next, the expression of 12 randomly selected upregulated transporter genes was analyzed by qRT-PCR in seedlings treated with various metals, such as Zn, Mg and Pb (Figure 5, Table S4). The seedlings were grown in water for 10 days to avoid priming effects before transferring to various media. The expression patterns of the Cd-upregulated transporter transcripts were classified into four groups: three transcripts (MatE, HMAs) upregulated significantly under Cd exposure (group 1), three transcripts (HMAs) upregulated significantly under both Cd and the other treatments (group 2), two transcripts (MatE, Zip) upregulated under the other treatments but not Cd (group 3), and four transcripts (Cation efflux, MatE, PDR_assoc, HMA) upregulated under general stress (group 4), though their expression might have been affected by ion concentration, treatment time, the balance of other ions, tissue and growth stage. The transcripts of group 1 might respond specifically to Cd. In our conditions, the transcripts of group 2 were not only upregulated by Cd, but also by the essential metal Ni, suggesting they might function in Ni transport. The nickel transporter TjZnt in the Ni hyperaccumulator Thlaspi japonicum has been reported to have Cd2+ transport ability [90]. The transcripts of group 3 did not respond to Cd in water substituted for nutrient-rich medium, suggesting they were easily affected by other metal ions. The transcripts of group 4 might respond to general stress. The expression of most of these was affected to different degrees by exposure to other metal ions, suggesting specific systems for transporting Cd may have not developed in rice because Cd is a non-essential metal for the plant.
Figure 5. Expression patterns of Cd-upregulated heavy metal ion transporters in various medium conditions by qRT-PCR analysis.
The expression of Cd-upregulated heavy metal ion transporters was investigated in various liquid media containing different kinds of metal ions by qRT-PCR analysis. The x-axis shows treatments and the y-axis shows relative expression. Transcript expression levels were normalized using an internal control (ubiquitin 1) and plotted relative to expression in water (control) at 24 h in roots (R) and shoots (S). The transcripts were classified into four groups based on their expression patterns.
Conclusions
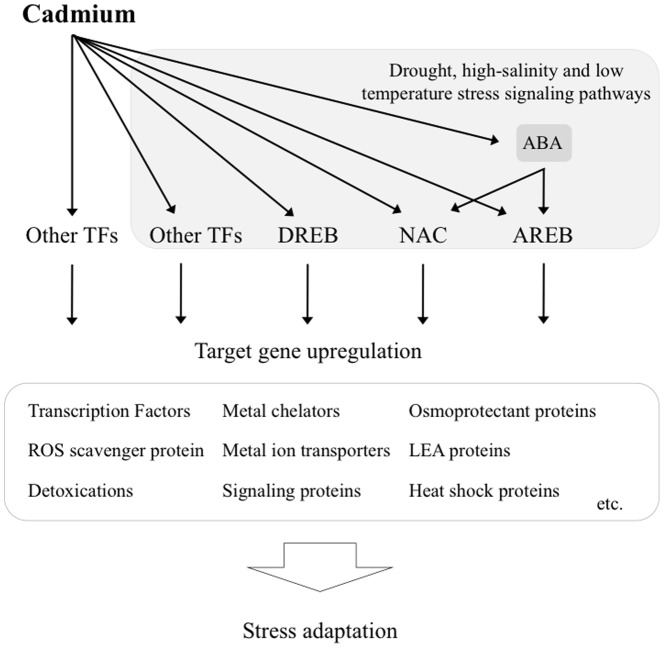
In summary, the sequencing of mRNAs uncovered new genes involved in signal transduction, antioxidation, detoxification and metal transport, and enabled us to develop new hypotheses for the transcriptional network underlying the response to Cd exposure. A highlight was the discovery of a relationship between Cd stress and drought stress. The data suggested that the Cd stress signaling pathway is involved in controlling drought stress signaling pathways that might confer to tolerance to Cd exposure. It is quite possible that the network underlying Cd stress responses and tolerance, which plants have developed to adapt to other stresses, could help to acclimate to Cd exposure because it is a non-essential metal. We have summarized the scheme of signal transduction for acclimation to Cd exposure in rice in Figure 6. As ROS are a toxic byproduct of aerobic metabolism, but also act as signaling molecules in the complex signaling network of cells [5], the generated ROS might act as messengers to activate gene expression in both Cd and abiotic stress pathways. Understanding the relationships between the transduction of different stress signals is useful to develop transgenic rice that show enhanced stress tolerance. Establishing the exact composition and organization of the transcriptional network underlying the response to Cd exposure will provide a robust tool for manipulating the stress tolerance of crops in the future.
Figure 6. Overview of Cd-dependent signaling cascade that affects drought, high-salinity and low temperature stress signaling pathways.
Many abiotic stress-related transcription factors including DREB, bZIP and NAC may function in the signal transduction pathway under Cd exposure. These TFs may regulate the expression of many downstream genes for stress tolerance and responses.
Supporting Information
Changes in rice morphology after 24 h drought stress. Rice seedlings grown by hydroponic culture in nutrient media were subjected to drought stress treatment by transferring them to a case without media. The rice seedlings began to show signs of wilting in shoots after 1 h and the changes gradually became more prominent, such that after 24 h of drought stress the shoots were completely wilted compared with control rice not removed from the nutrient media.
(TIFF)
Identification of GO terms enriched in Cd-responsive transcripts. Significant GO terms identified by GO enrichment analysis based on the most enriched biological processes associated with variations under Cd exposure are shown in a heatmap of responsive transcripts (left: upregulated transcripts, right: downregulated transcripts). The bar with red-black gradation indicates the level of significance of GO enrichment with the extremes representing statistically significant (red) and non-significant (black) GO terms.
(TIFF)
qRT-PCR analysis of Cd-responsive genes that may function in drought. The expression of Cd-responsive drought-related genes was investigated under Cd exposure up to 24 h (black) and drought at 3 h (gray) in qRT-PCR analysis. The x-axis shows treatments and the y-axis shows relative expression. Transcript expression levels were normalized using an internal control (ubiquitin 1) and plotted relative to expression in non-treated samples (control) in roots (R) and shoots (S).
(TIFF)
Venn diagram analysis of Cd and other stress responsive transcripts. The resulting four-way Venn diagrams for roots and shoots show the number of transcripts responsive (≥2-fold or ≤ 0.5-fold) to Cd (24 h), drought, high-salinity and low temperature relative to the control (0 d).
(TIFF)
Expression analysis of gene families that may function in defense against Cd stress. The graph shows the expression of ROS-scavenging enzyme genes and respiratory burst oxidase homolog (Rboh) genes under Cd exposure in RNA-Seq analysis. The x-axis shows genes and y-axis shows relative expression. The black bar shows the relative expression in roots at 1 h, the red bar roots at 24 h, the light blue bar shoots at 1 h and the blue bar shoots at 24 h.
(TIFF)
Expression patterns of Cd-upregulated antioxidative and detoxification enzymes in various medium conditions by qRT-PCR analysis. The expression of Cd-upregulated antioxidative and detoxification enzymes was investigated in various liquid media containing different kinds of metal ions by qRT-PCR analysis. The x-axis shows treatments and the y-axis shows relative expression. Transcript expression levels were normalized using an internal control (ubiquitin 1) and plotted relative to expression in water (control) at hour 24 in roots (R) and shoots (S). The transcripts were classified into four groups based on their expression patterns.
(TIFF)
Mapping of RNA-Seq reads obtained from root and shoot samples into the reference IRGSP-1.0 genome sequence.
(XLS)
Expression (RPKM value) of Ca-responsive transcripts.
(XLS)
Expression of abiotic stress-related rice genes under Cd exposure.
(XLS)
PCR primers for qRT-PCR analysis.
(XLS)
Commonly upregulated transcripts among stresses in roots and shoots.
(XLS)
Acknowledgments
We thank Ms. F. Aota, Ms. K. Ohtsu, and Ms. K. Yamada for technical assistance.
Funding Statement
This work was supported by a grant from the Ministry of Agriculture, Forestry and Fisheries of Japan (Genomics for Agricultural Innovation, RTR-0001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Clemens S, Palmgren MG, Krämer U (2002) A long way ahead: understanding and engineering plant metal accumulation. Trends Plant Sci 7: 309–315. [DOI] [PubMed] [Google Scholar]
- 2. Roth U, von Roepenack-Lahaye E, Clemens S (2006) Proteome changes in Arabidopsis thaliana roots upon exposure to Cd2+. J Exp Bot 57: 4003–4013. [DOI] [PubMed] [Google Scholar]
- 3. Zhang M, Liu X, Yuan L, Wu K, Duan J, et al. (2012) Transcriptional profiling in cadmium-treated rice seedling roots using suppressive subtractive hybridization. Plant Physiol Biochem 50: 79–86. [DOI] [PubMed] [Google Scholar]
- 4. Halliwell B, Gutteridge JM (1984) Oxygen-toxicity, oxygen radicals, transition-metals and disease. Biochem J 219: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, et al. (2011) ROS signaling: the new wave? Trends Plant Sci 16: 300–309. [DOI] [PubMed] [Google Scholar]
- 6. Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9: 490–498. [DOI] [PubMed] [Google Scholar]
- 7. Frova C (2003) The plant glutathione transferase gene family: genomic structure, functions, expression and evolution. Physiol Plant 119: 469–479. [Google Scholar]
- 8. Cosio C, Dunand C (2009) Specific functions of individual class III peroxidase genes. J Exp Bot 60: 391–408. [DOI] [PubMed] [Google Scholar]
- 9. Cobbett C, Goldsbrough P (2002) Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu Rev Plant Biol 53: 159–182. [DOI] [PubMed] [Google Scholar]
- 10. Kramer U, Talke IN, Hanikenne M (2007) Transition metal transport. FEBS Lett 581: 2263–2272. [DOI] [PubMed] [Google Scholar]
- 11. Moons A (2003) Ospdr9, which encodes a PDR-type ABC transporter, is induced by heavy metals, hypoxic stress and redox perturbations in rice roots. FEBS Lett 553: 370–376. [DOI] [PubMed] [Google Scholar]
- 12. Uraguchi S, Kamiya T, Sakamoto T, Kasai K, Sato Y, et al. (2011) Low-affinity cation transporter (OsLCT1) regulates cadmium transport into rice grains. Proc Natl Acad Sci USA 108: 20959–20964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ueno D, Yamaji N, Kono I, Huang CF, Ando T, et al. (2010) Gene limiting cadmium accumulation in rice. Proc Natl Acad Sci USA 107: 16500–16505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Korshunova YO, Eide D, Clark WG, Guerinot ML, Pakrasi HB (1999) The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol Biol 40: 37–44. [DOI] [PubMed] [Google Scholar]
- 15. Grossoehme NE, Akilesh S, Guerinot ML, Wilcox DE (2006) Metal-binding thermodynamics of the histidine-rich sequence from the metal-transport protein IRT1 of Arabidopsis thaliana . Inorg Chem 45: 8500–8508. [DOI] [PubMed] [Google Scholar]
- 16. Pedas P, Ytting CK, Fuglsang AT, Jahn TP, Schjoerring JK, et al. (2008) Manganese efficiency in barley: identification and characterization of the metal ion transporter HvIRT1. Plant Physiol 148: 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee S, An G (2009) Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ 32: 408–416. [DOI] [PubMed] [Google Scholar]
- 18. Satoh-Nagasawa N, Mori M, Nakazawa N, Kawamoto T, Nagato Y, et al. (2012) Mutations in rice (Oryza sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol 53: 213–224. [DOI] [PubMed] [Google Scholar]
- 19. Sasaki A, Yamaji N, Yokosho K, Ma JF (2012) Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 24: 2155–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ogawa I, Nakanishi H, Mori S, Nishizawa N (2009) Time course analysis of gene regulation under cadmium stress in rice. Plant and Soil 325: 97–108. [Google Scholar]
- 21. Oono Y, Kawahara Y, Kanamori H, Mizuno H, Yamagata H, et al. (2011) mRNA-seq reveals a comprehensive transcriptome profile of rice under phosphate stress. Rice 4: 50–65. [Google Scholar]
- 22. Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshida S, Forno AD, Cock HJ, Gomez AK (1976) Laboratory Manual for Physiological Studies of Rice, 3rd edn. Manila, The Philippines.
- 24. Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. [Google Scholar]
- 25. Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, et al. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mortazavi A, Williams BA, Mccue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- 30. Lee K, Bae DW, Kim SH, Han HJ, Liu X, et al. (2010) Comparative proteomic analysis of the short-term responses of rice roots and leaves to cadmium. J Plant Physiol 167: 161–168. [DOI] [PubMed] [Google Scholar]
- 31. Shah K, Kumar RG, Verma S, Dubey RS (2001) Effect of cadmium on lipid peroxidation, superoxide anion generation and activities of antioxidant enzymes in growing rice seedlings. Plant Sci 161: 1135–1144. [Google Scholar]
- 32. Perfus-Barbeoch L, Leonhardt N, Vavasseur A, Forestier C (2002) Heavy metal toxicity: cadmium permeates through calcium channels and disturbs the plant water status. Plant J 32: 539–548. [DOI] [PubMed] [Google Scholar]
- 33. Lu TT, Lu GJ, Fan DL, Zhu CR, Li W, et al. (2010) Function annotation of the rice transcriptome at single-nucleotide resolution by RNA-seq. Genome Res 20: 1238–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jang IC, Oh SJ, Seo JS, Choi WB, Song SI, et al. (2003) Expression of a bifunctional fusion of the Escherichia coli genes for trehalose-6-phosphate synthase and trehalose-6-phosphate phosphatase in transgenic rice plants increases trehalose accumulation and abiotic stress tolerance without stunting growth. Plant Physiol 131: 516–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mundy J, Chua NH (1988) Abscisic-acid and water-stress induce the expression of a novel rice gene. Embo J 7: 2279–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yamaguchi-Shinozaki K, Mundy J, Chua NH (1990) 4 tightly linked Rab genes are differentially expressed in rice. Plant Mol Biol 14: 29–39. [DOI] [PubMed] [Google Scholar]
- 37. Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803. [DOI] [PubMed] [Google Scholar]
- 38. Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, et al. (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33: 751–763. [DOI] [PubMed] [Google Scholar]
- 39. Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, et al. (2006) Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol 47: 141–153. [DOI] [PubMed] [Google Scholar]
- 40. Chen JQ, Meng XP, Zhang Y, Xia M, Wang XP (2008) Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol Lett 30: 2191–2198. [DOI] [PubMed] [Google Scholar]
- 41. Xiang Y, Tang N, Du H, Ye HY, Xiong LZ (2008) Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148: 1938–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zou MJ, Guan YC, Ren HB, Zhang F, Chen F (2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66: 675–683. [DOI] [PubMed] [Google Scholar]
- 43. Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, et al. (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27: 325–333. [DOI] [PubMed] [Google Scholar]
- 44. Lefebvre V, North H, Frey A, Sotta B, Seo M, et al. (2006) Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J 45: 309–319. [DOI] [PubMed] [Google Scholar]
- 45. Qin XQ, Zeevaart JAD (2002) Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol 128: 544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tan BC, Joseph LM, Deng WT, Liu LJ, Li QB, et al. (2003) Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J 35: 44–56. [DOI] [PubMed] [Google Scholar]
- 47. Parcy F, Valon C, Raynal M, Gaubier-Comella P, Delseny M, et al. (1994) Regulation of gene expression programs during Arabidopsis seed development: roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6: 1567–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ingram J, Bartels D (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 377–403. [DOI] [PubMed] [Google Scholar]
- 49. Xiao BZ, Huang YM, Tang N, Xiong LZ (2007) Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor Appl Genet 115: 35–46. [DOI] [PubMed] [Google Scholar]
- 50. Takasaki H, Maruyama K, Kidokoro S, Ito Y, Fujita Y, et al. (2010) The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol Genet Genomics 284: 173–183. [DOI] [PubMed] [Google Scholar]
- 51. Hu HH, Dai MQ, Yao JL, Xiao BZ, Li XH, et al. (2006) Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA 103: 12987–12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sperotto RA, Ricachenevsky FK, Duarte GL, Boff T, Lopes KL, et al. (2009) Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor. Planta 230: 985–1002. [DOI] [PubMed] [Google Scholar]
- 53. Song SY, Chen Y, Chen J, Dai XY, Zhang WH (2011) Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234: 331–345. [DOI] [PubMed] [Google Scholar]
- 54. Tran LSP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, et al. (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Oh SJ, Kim YS, Kwon CW, Park HK, Jeong JS, et al. (2009) Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol 150: 1368–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Xu DQ, Huang J, Guo SQ, Yang X, Bao YM, et al. (2008) Overexpression of a TFIIIA-type zinc finger protein gene ZFP252 enhances drought and salt tolerance in rice (Oryza sativa L.). FEBS Lett 582: 1037–1043. [DOI] [PubMed] [Google Scholar]
- 57. Taji T, Ohsumi C, Seki M, Iuchi S, Yamaguchi-Shinozaki K, et al. (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana . Plant J 29: 417–426. [DOI] [PubMed] [Google Scholar]
- 58. Sun XM, Zhang JX, Zhang HJ, Ni YW, Zhang Q, et al. (2010) The responses of Arabidopsis thaliana to cadmium exposure explored via metabolite profiling. Chemosphere 78: 840–845. [DOI] [PubMed] [Google Scholar]
- 59. Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14: 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chen SL, Kao CH (1995) Cd induced changes in proline level and peroxidase-activity in roots of rice seedlings. Plant Growth Regul 17: 67–71. [Google Scholar]
- 61. Shah K, Dubey RS (1997) Effect of cadmium on proline accumulation and ribonuclease activity in rice seedlings: role of proline as a possible enzyme protectant. Biol Plant 40: 121–130. [Google Scholar]
- 62. Ye HY, Du H, Tang N, Li XH, Xiong LZ (2009) Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol Biol 71: 291–305. [DOI] [PubMed] [Google Scholar]
- 63. Park MR, Yun KY, Mohanty B, Herath V, Xu FY, et al. (2010) Supra-optimal expression of the cold-regulated OsMyb4 transcription factor in transgenic rice changes the complexity of transcriptional network with major effects on stress tolerance and panicle development. Plant Cell Environ 33: 2209–2230. [DOI] [PubMed] [Google Scholar]
- 64. Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY, et al. (2005) Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol 138: 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Soranzo N, Gorla MS, Mizzi L, De Toma G, Frova C (2004) Organisation and structural evolution of the rice glutathione S-transferase gene family. Mol Genet Genomics 271: 511–521. [DOI] [PubMed] [Google Scholar]
- 66. Passardi F, Longet D, Penel C, Dunand C (2004) The class III peroxidase multigenic in land plants family in rice and its evolution. Phytochemistry 65: 1879–1893. [DOI] [PubMed] [Google Scholar]
- 67. Huang TL, Quynh TTN, Fu SF, Lin CY, Chen YC, et al. (2012) Transcriptomic changes and signalling pathways induced by arsenic stress in rice roots. Plant Mol Biol 80: 587–608. [DOI] [PubMed] [Google Scholar]
- 68. Roxas VP, Lodhi SA, Garrett DK, Mahan JR, Allen RD (2000) Stress tolerance in transgenic tobacco seedlings that overexpress glutathione S-transferase/glutathione peroxidase. Plant Cell Physiol 41: 1229–1234. [DOI] [PubMed] [Google Scholar]
- 69. Hiraga S, Sasaki K, Ito H, Ohashi Y, Matsui H (2001) A large family of class III plant peroxidases. Plant Cell Physiol 42: 462–468. [DOI] [PubMed] [Google Scholar]
- 70. Passardi F, Penel C, Dunand C (2004) Performing the paradoxical: how plant peroxidases modify the cell wall. Trends Plant Sci 9: 534–540. [DOI] [PubMed] [Google Scholar]
- 71. Liu X, Williams CE, Nemacheck JA, Wang H, Subramanyam S, et al. (2010) Reactive oxygen species are involved in plant defense against a gall midge. Plant Physiol 152: 985–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ahn IP, Kim S, Kang S, Suh SC, Lee YH (2005) Rice defense mechanisms against Cochliobolus miyabeanus and Magnaporthe grisea are distinct. Phytopathology 95: 1248–1255. [DOI] [PubMed] [Google Scholar]
- 73. Valerio L, De Meyer M, Penel C, Dunand C (2004) Expression analysis of the Arabidopsis peroxidase multigenic family. Phytochemistry 65: 1331–1342. [DOI] [PubMed] [Google Scholar]
- 74. Rouhier N, Couturier J, Jacquot JP (2006) Genome-wide analysis of plant glutaredoxin systems. J Exp Bot 57: 1685–1696. [DOI] [PubMed] [Google Scholar]
- 75. Nuruzzaman M, Gupta M, Zhang CJ, Wang L, Xie WB, et al. (2008) Sequence and expression analysis of the thioredoxin protein gene family in rice. Mol Genet Genomics 280: 139–151. [DOI] [PubMed] [Google Scholar]
- 76. Lee KO, Jang HH, Jung BG, Chi YH, Lee JY, et al. (2000) Rice 1Cys-peroxiredoxin over-expressed in transgenic tobacco does not maintain dormancy but enhances antioxidant activity. FEBS Lett 486: 103–106. [DOI] [PubMed] [Google Scholar]
- 77. Obara K, Sumi K, Fukuda H (2002) The use of multiple transcription starts causes the dual targeting of Arabidopsis putative monodehydroascorbate reductase to both mitochondria and chloroplasts. Plant Cell Physiol 43: 697–705. [DOI] [PubMed] [Google Scholar]
- 78. Considine MJ, Holtzapffel RC, Day DA, Whelan J, Millar AH (2002) Molecular distinction between alternative oxidase from monocots and dicots. Plant Physiol 129: 949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Koeduka T, Matsui K, Akakabe Y, Kajiwara T (2002) Catalytic properties of rice alpha-oxygenase - A comparison with mammalian prostaglandin H synthases. J Biol Chem 277: 22648–22655. [DOI] [PubMed] [Google Scholar]
- 80. Koeduka T, Matsui K, Hasegawa M, Akakabe Y, Kajiwara T (2005) Rice fatty acid alpha-dioxygenase is induced by pathogen attack and heavy metal stress: activation through jasmonate signaling. J Plant Physiol 162: 912–920. [DOI] [PubMed] [Google Scholar]
- 81. Tirajoh A, Aung TST, McKay AB, Plant AL (2005) Stress-responsive alpha-dioxygenase expression in tomato roots. J Exp Bot 56: 713–723. [DOI] [PubMed] [Google Scholar]
- 82. Grill E, Loffler S, Winnacker EL, Zenk MH (1989) Phytochelatins, the heavy-metal-binding peptides of plants, are synthesized from glutathione by a specific gamma-glutamylcysteine dipeptidyl transpeptidase (Phytochelatin Synthase). Proc Natl Acad Sci USA 86: 6838–6842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ha SB, Smith AP, Howden R, Dietrich WM, Bugg S, et al. (1999) Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe. Plant Cell 11: 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gautam N, Verma PK, Verma S, Tripathi RD, Trivedi PK, et al. (2012) Genome-wide identification of rice class I metallothionein gene: tissue expression patterns and induction in response to heavy metal stress. Funct Integr Genomics 12: 635–647. [DOI] [PubMed] [Google Scholar]
- 85. Zhou GK, Xu YF, Li J, Yang LY, Liu JY (2006) Molecular analyses of the metallothionein gene family in rice (Oryza sativa L.). J Biochem Mol Biol 39: 595–606. [DOI] [PubMed] [Google Scholar]
- 86. Palmiter RD (1998) The elusive function of metallothioneins. Proc Natl Acad Sci USA 95: 8428–8430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Huang XH, Kurata N, Wei XH, Wang ZX, Wang A, et al. (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wittkopp PJ, Kalay G (2012) Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat Rev Genet 13: 59–69. [DOI] [PubMed] [Google Scholar]
- 89. Li LG, He ZY, Pandey GK, Tsuchiya T, Luan S (2002) Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. J Biol Chem 277: 5360–5368. [DOI] [PubMed] [Google Scholar]
- 90. Mizuno T, Usui K, Horie K, Nosaka S, Mizuno N, et al. (2005) Cloning of three ZIP/NRAMP transporter genes from a Ni hyperaccumulator plant Thlaspi japonicum and their Ni2+-transport abilities. Plant Physiol Biochem 43: 793–801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Changes in rice morphology after 24 h drought stress. Rice seedlings grown by hydroponic culture in nutrient media were subjected to drought stress treatment by transferring them to a case without media. The rice seedlings began to show signs of wilting in shoots after 1 h and the changes gradually became more prominent, such that after 24 h of drought stress the shoots were completely wilted compared with control rice not removed from the nutrient media.
(TIFF)
Identification of GO terms enriched in Cd-responsive transcripts. Significant GO terms identified by GO enrichment analysis based on the most enriched biological processes associated with variations under Cd exposure are shown in a heatmap of responsive transcripts (left: upregulated transcripts, right: downregulated transcripts). The bar with red-black gradation indicates the level of significance of GO enrichment with the extremes representing statistically significant (red) and non-significant (black) GO terms.
(TIFF)
qRT-PCR analysis of Cd-responsive genes that may function in drought. The expression of Cd-responsive drought-related genes was investigated under Cd exposure up to 24 h (black) and drought at 3 h (gray) in qRT-PCR analysis. The x-axis shows treatments and the y-axis shows relative expression. Transcript expression levels were normalized using an internal control (ubiquitin 1) and plotted relative to expression in non-treated samples (control) in roots (R) and shoots (S).
(TIFF)
Venn diagram analysis of Cd and other stress responsive transcripts. The resulting four-way Venn diagrams for roots and shoots show the number of transcripts responsive (≥2-fold or ≤ 0.5-fold) to Cd (24 h), drought, high-salinity and low temperature relative to the control (0 d).
(TIFF)
Expression analysis of gene families that may function in defense against Cd stress. The graph shows the expression of ROS-scavenging enzyme genes and respiratory burst oxidase homolog (Rboh) genes under Cd exposure in RNA-Seq analysis. The x-axis shows genes and y-axis shows relative expression. The black bar shows the relative expression in roots at 1 h, the red bar roots at 24 h, the light blue bar shoots at 1 h and the blue bar shoots at 24 h.
(TIFF)
Expression patterns of Cd-upregulated antioxidative and detoxification enzymes in various medium conditions by qRT-PCR analysis. The expression of Cd-upregulated antioxidative and detoxification enzymes was investigated in various liquid media containing different kinds of metal ions by qRT-PCR analysis. The x-axis shows treatments and the y-axis shows relative expression. Transcript expression levels were normalized using an internal control (ubiquitin 1) and plotted relative to expression in water (control) at hour 24 in roots (R) and shoots (S). The transcripts were classified into four groups based on their expression patterns.
(TIFF)
Mapping of RNA-Seq reads obtained from root and shoot samples into the reference IRGSP-1.0 genome sequence.
(XLS)
Expression (RPKM value) of Ca-responsive transcripts.
(XLS)
Expression of abiotic stress-related rice genes under Cd exposure.
(XLS)
PCR primers for qRT-PCR analysis.
(XLS)
Commonly upregulated transcripts among stresses in roots and shoots.
(XLS)