Abstract
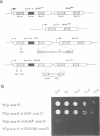
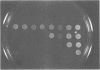
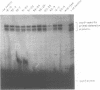
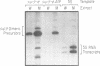
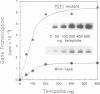
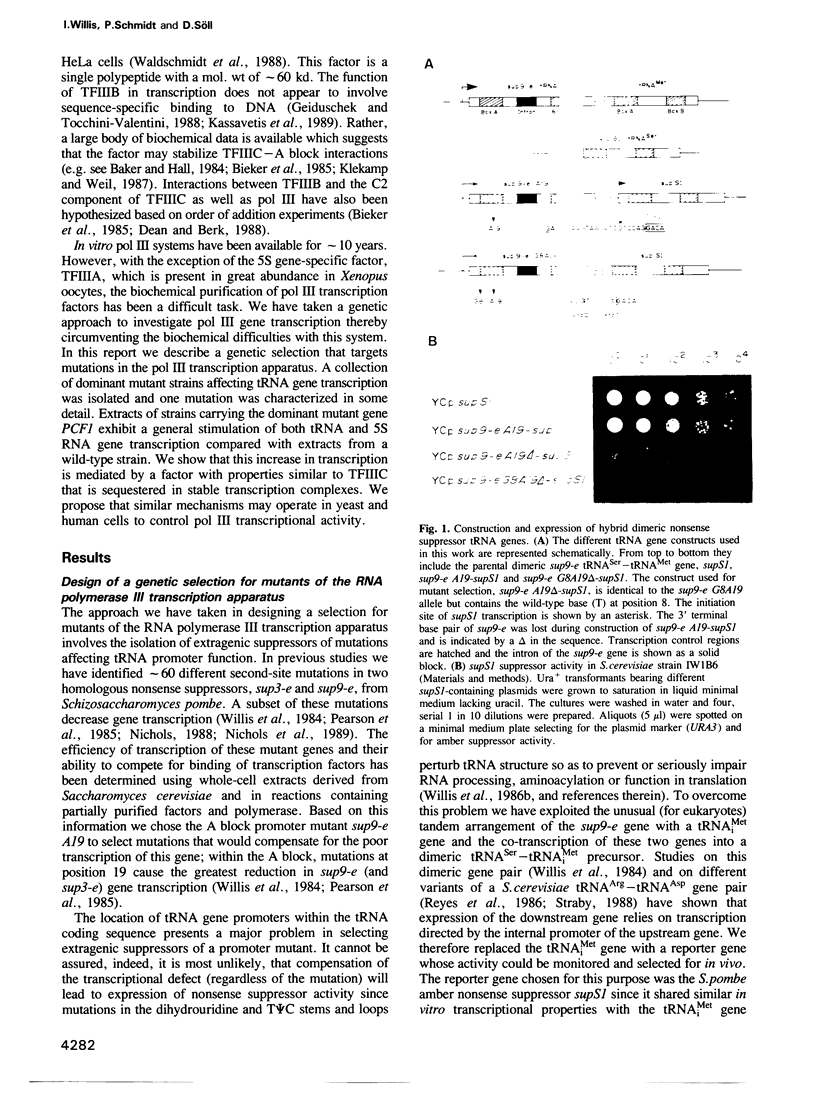
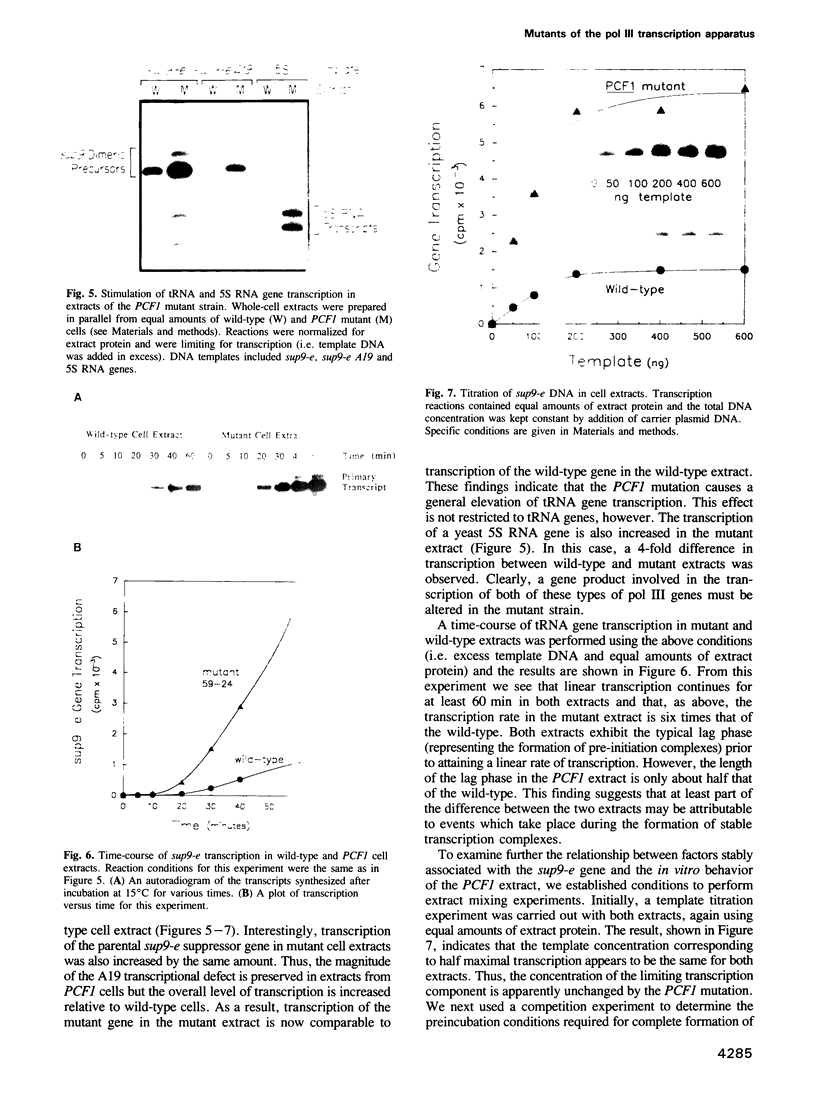
A genetic approach has been developed to study transcription by RNA polymerase III. A pair of Schizosaccharomyces pombe nonsense suppressor tRNA genes were arranged in tandem such that expression of the downstream (supS1) tRNA suppressor was dependent upon transcription initiated by the internal promoter of the upstream (sup9-e) gene. Dominant mutant strains of Saccharomyces cerevisiae were isolated that suppress in trans the effect of an A block promoter mutation (A19) in the sup9-e gene and restore supS1 suppressor activity. Fifteen mutant strains, eight of which were independently isolated, all have elevated steady-state levels of sup9-e A19 RNA consistent with an increase in gene transcription. Extracts of a strain carrying the dominant mutant gene, PCF1, show a general 6-fold stimulation in transcription of mutant (A19) and wild-type tRNA genes and increase 5S gene transcription 4-fold compared with extracts from a wild-type strain. A transcription factor exclusion assay was used to show that the PCF1 mutation affects two distinct stages in transcription: one prior to and one after stable complex formation; and that these effects are mediated by a component of the stable complex. Further evidence of an effect during complex assembly was obtained in a time-course experiment that showed a shortened lag phase in the PCF1 extract. The results indicate that PCF1 is either a component of the stable complex or a positive regulator of its activity.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison D. S., Goh S. H., Hall B. D. The promoter sequence of a yeast tRNAtyr gene. Cell. 1983 Sep;34(2):655–664. doi: 10.1016/0092-8674(83)90398-7. [DOI] [PubMed] [Google Scholar]
- Baker R. E., Camier S., Sentenac A., Hall B. D. Gene size differentially affects the binding of yeast transcription factor tau to two intragenic regions. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8768–8772. doi: 10.1073/pnas.84.24.8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker R. E., Gabrielsen O., Hall B. D. Effects of tRNATyr point mutations on the binding of yeast RNA polymerase III transcription factor C. J Biol Chem. 1986 Apr 25;261(12):5275–5282. [PubMed] [Google Scholar]
- Baker R. E., Hall B. D. Structural features of yeast tRNA genes which affect transcription factor binding. EMBO J. 1984 Dec 1;3(12):2793–2800. doi: 10.1002/j.1460-2075.1984.tb02211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieker J. J., Martin P. L., Roeder R. G. Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell. 1985 Jan;40(1):119–127. doi: 10.1016/0092-8674(85)90315-0. [DOI] [PubMed] [Google Scholar]
- Camier S., Gabrielsen O., Baker R., Sentenac A. A split binding site for transcription factor tau on the tRNA3Glu gene. EMBO J. 1985 Feb;4(2):491–500. doi: 10.1002/j.1460-2075.1985.tb03655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean N., Berk A. J. Ordering promoter binding of class III transcription factors TFIIIC1 and TFIIIC2. Mol Cell Biol. 1988 Aug;8(8):3017–3025. doi: 10.1128/mcb.8.8.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P., Coppo A., Fruscoloni P., Benedetti P., Di Segni G., Tocchini-Valentini G. P. Comparative mutational analysis of wild-type and stretched tRNA3(Leu) gene promoters. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8763–8767. doi: 10.1073/pnas.84.24.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folk W. R., Hofstetter H. A detailed mutational analysis of the eucaryotic tRNAmet1 gene promoter. Cell. 1983 Jun;33(2):585–593. doi: 10.1016/0092-8674(83)90439-7. [DOI] [PubMed] [Google Scholar]
- Gaynor R. B., Feldman L. T., Berk A. J. Transcription of class III genes activated by viral immediate early proteins. Science. 1985 Oct 25;230(4724):447–450. doi: 10.1126/science.2996135. [DOI] [PubMed] [Google Scholar]
- Geiduschek E. P., Tocchini-Valentini G. P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–914. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- Hoeffler W. K., Kovelman R., Roeder R. G. Activation of transcription factor IIIC by the adenovirus E1A protein. Cell. 1988 Jun 17;53(6):907–920. doi: 10.1016/s0092-8674(88)90409-6. [DOI] [PubMed] [Google Scholar]
- Hoeffler W. K., Roeder R. G. Enhancement of RNA polymerase III transcription by the E1A gene product of adenovirus. Cell. 1985 Jul;41(3):955–963. doi: 10.1016/s0092-8674(85)80076-3. [DOI] [PubMed] [Google Scholar]
- Hottinger-Werlen A., Schaack J., Lapointe J., Mao J., Nichols M., Söll D. Dimeric tRNA gene arrangement in Schizosaccharomyces pombe allows increased expression of the downstream gene. Nucleic Acids Res. 1985 Dec 20;13(24):8739–8747. doi: 10.1093/nar/13.24.8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottinger H., Stadelmann B., Pearson D., Frendewey D., Kohli J., Söll D. The Schizosaccharomyces pombe sup3-i suppressor recognizes ochre, but not amber codons in vitro and in vivo. EMBO J. 1984 Feb;3(2):423–428. doi: 10.1002/j.1460-2075.1984.tb01823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Riggs D. L., Negri R., Nguyen L. H., Geiduschek E. P. Transcription factor IIIB generates extended DNA interactions in RNA polymerase III transcription complexes on tRNA genes. Mol Cell Biol. 1989 Jun;9(6):2551–2566. doi: 10.1128/mcb.9.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klekamp M. S., Weil P. A. Partial purification and characterization of the Saccharomyces cerevisiae transcription factor TFIIIB. J Biol Chem. 1986 Feb 25;261(6):2819–2827. [PubMed] [Google Scholar]
- Klekamp M. S., Weil P. A. Properties of yeast class III gene transcription factor TFIIIB. Implications regarding mechanism of action. J Biol Chem. 1987 Jun 5;262(16):7878–7883. [PubMed] [Google Scholar]
- Klekamp M. S., Weil P. A. Specific transcription of homologous class III genes in yeast-soluble cell-free extracts. J Biol Chem. 1982 Jul 25;257(14):8432–8441. [PubMed] [Google Scholar]
- Krupp G., Thurianx P., Willis I., Gamulin V., Söll D. First identification of an amber nonsense mutation in Schizosaccharomyces pombe: major differences in the efficiency of homologous versus heterologous yeast suppressor tRNA genes. Mol Gen Genet. 1985;201(1):82–87. doi: 10.1007/BF00397990. [DOI] [PubMed] [Google Scholar]
- Ottonello S., Rivier D. H., Doolittle G. M., Young L. S., Sprague K. U. The properties of a new polymerase III transcription factor reveal that transcription complexes can assemble by more than one pathway. EMBO J. 1987 Jul;6(7):1921–1927. doi: 10.1002/j.1460-2075.1987.tb02452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson D., Willis I., Hottinger H., Bell J., Kumar A., Leupold U., Söll D. Mutations preventing expression of sup3 tRNASer nonsense suppressors of Schizosaccharomyces pombe. Mol Cell Biol. 1985 Apr;5(4):808–815. doi: 10.1128/mcb.5.4.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieler T., Hamm J., Roeder R. G. The 5S gene internal control region is composed of three distinct sequence elements, organized as two functional domains with variable spacing. Cell. 1987 Jan 16;48(1):91–100. doi: 10.1016/0092-8674(87)90359-x. [DOI] [PubMed] [Google Scholar]
- Reyes V. M., Newman A., Abelson J. Mutational analysis of the coordinate expression of the yeast tRNAArg-tRNAAsp gene tandem. Mol Cell Biol. 1986 Jul;6(7):2436–2442. doi: 10.1128/mcb.6.7.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruet A., Camier S., Smagowicz W., Sentenac A., Fromageot P. Isolation of a class C transcription factor which forms a stable complex with tRNA genes. EMBO J. 1984 Feb;3(2):343–350. doi: 10.1002/j.1460-2075.1984.tb01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S. J., Garcia A. D. Transcription of the Drosophila melanogaster 5S RNA gene requires an upstream promoter and four intragenic sequence elements. Mol Cell Biol. 1988 Mar;8(3):1266–1274. doi: 10.1128/mcb.8.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp S. J., Schaack J., Cooley L., Burke D. J., Söll D. Structure and transcription of eukaryotic tRNA genes. CRC Crit Rev Biochem. 1985;19(2):107–144. doi: 10.3109/10409238509082541. [DOI] [PubMed] [Google Scholar]
- Stillman D. J., Better M., Geiduschek E. P. Electron-microscopic examination of the binding of a large RNA polymerase III transcription factor to a tRNA gene. J Mol Biol. 1985 Sep 20;185(2):451–455. doi: 10.1016/0022-2836(85)90417-6. [DOI] [PubMed] [Google Scholar]
- Stråby K. B. A yeast tRNA(Arg) gene can act as promoter for a 5' flank deficient, non-transcribable tRNA(SUP)6 gene to produce biologically active suppressor tRNA. Nucleic Acids Res. 1988 Apr 11;16(7):2841–2857. doi: 10.1093/nar/16.7.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler B. M. Transcription of Neurospora crassa 5 S rRNA genes requires a TATA box and three internal elements. J Mol Biol. 1987 Aug 20;196(4):801–811. doi: 10.1016/0022-2836(87)90406-2. [DOI] [PubMed] [Google Scholar]
- Waldschmidt R., Jahn D., Seifart K. H. Purification of transcription factor IIIB from HeLa cells. J Biol Chem. 1988 Sep 15;263(26):13350–13356. [PubMed] [Google Scholar]
- Willis I., Frendewey D., Nichols M., Hottinger-Werlen A., Schaack J., Söll D. A single base change in the intron of a serine tRNA affects the rate of RNase P cleavage in vitro and suppressor activity in vivo in Saccharomyces cerevisiae. J Biol Chem. 1986 May 5;261(13):5878–5885. [PubMed] [Google Scholar]
- Willis I., Hottinger H., Pearson D., Chisholm V., Leupold U., Söll D. Mutations affecting excision of the intron from a eukaryotic dimeric tRNA precursor. EMBO J. 1984 Jul;3(7):1573–1580. doi: 10.1002/j.1460-2075.1984.tb02013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis I., Nichols M., Chisholm V., Söll D., Heyer W. D., Szankasi P., Amstutz H., Munz P., Kohli J. Functional complementation between mutations in a yeast suppressor tRNA gene reveals potential for evolution of tRNA sequences. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7860–7864. doi: 10.1073/pnas.83.20.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga S. K., Boulanger P. A., Berk A. J. Resolution of human transcription factor TFIIIC into two functional components. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3585–3589. doi: 10.1073/pnas.84.11.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]