Abstract
Purpose:
Infertility affects 10–15 % of the population, of which, approximately 40 % is due to male etiology consisting primarily of low sperm count (oligozoospermia) and/or abnormal sperm motility (asthenozoospermia). It has been demonstrated that mtDNA base substitutions can greatly influence semen quality.
Methods:
In the present study we performed a systematic sequence analysis of the mitochondrial cytochrome oxidase III (COIII) gene in 31 asthenozoospermic infertile men in comparaison to normozoospermic infertile men (n=33) and fertile men (n=150) from Tunisian population.
Results:
A novel m.9588G>A mutation was found in the mtDNA sperm’s in all asthenozoospermic patients and was absent in the normozoospermic and in fertile men. The m.9588G>A mutation substitutes a highly conserved Glutamate at position 128 to Lysine. In addition, PolyPhen-2 analysis predicted that this variant is “probably damaging”.
Keywords: Male infertility, Mitochondrial DNA, COIII gene, m.9588G > A mutation
Introduction
Mutations in the mitochondria have been implicated in a variety of human diseases ranging from degenerative diseases to infertility [1, 2]. This may be caused by the increase of the number of mutations in mitochondria, which is the result of several properties that make mtDNA distinct from cellular DNA. First, replication of mtDNA is rapid and lacks proofreading [3, 4] resulting in a 10–100 times higher of mitochondrial mutation rate than that of nuclear DNA [5, 6]. Second, because the lack of in adequate DNA-repair mechanisms in the mitochondria the mutation rate of mtDNA is 10_20 times higher than that of nuclear DNA [3]. Finally, the mode of inheritance of mitochondria is unique because the genetic information is inherited maternally. This creates an asymmetry in the natural selection process.
Current understanding of the role of mitochondrial mutations in human disease is expanding. Tissues that require high levels of respiratory energy, such as the brain, heart, skeletal muscle, kidney, liver, endocrine system, and other somatic tissues, can malfunction if they possess defects in their mitochondria [7]. Sperm are also included in this category. It has been proposed that the ATP produced by mitochondria is essential for sperm motility [8]. Other evidence supports an essential role for mitochondrial formation and function in spermatogenesis [1, 9–13].
The maturation of spermatogonia to a spermatozoon capable of fertilization involves rearrangement of mitochondria and development of a functional tail. Asthenozoospermia could potentially be caused by defects in tail formation in spermatozoa or by defects in the energy-producing machinery required to drive motility. In sperm, mitochondria are located around the midpiece and are arranged in a helix of 11–13 gyri (or individual spirals around the core), with two mitochondria per gyrus [14]. In the absence of glycolytic support, ATP generated from the mitochondria is delivered to the axoneme and is used for flagellar propulsion. Reductions in motility could potentially arise from defects in any of the 200–300 separate genes that are necessary for proper assembly and function of the sperm axoneme and tail [15]. In brief, mutations in mtDNA could contribute to a large percentage of asthenozoospermia. In fact mtDNA mutations affecting flagellar movement are a cause of sperm dysmotility. DNA rearrangements including point mutations and deletions of mtDNA have been reported in patients with low sperm motility who have asthenozoospermia and oligoasthenozoospermia [16–18].
In this study the human sperm mitochondrial COIII gene was analysed with the aim of identifying point mutations which may be associated with asthenozoospermia and male infertility.
Patients and methods
Semen collection and analysis
We included 64 male partners from consecutively enrolled couples who had their first infertility consultation in the Reproductive Biology laboratory at the faculty of medicine of Sfax (Tunisia). Because their female’s partners did not conceive after 2 year of marriage, both of them had undergone clinical examination. The results had shown that their female’s partners were normal.
Semen was collect by masturbation after a period of sexual abstinence of 3–5 days. The samples were analyzed following standard protocols according to WHO [19]. They were allowed to liquefy for 30 min at 37 °C. Then the number and percentage of motile spermatozoa was evaluated [19]. In particular, sperm-motility analysis was performed at room temperature, a minimum of four microscopy fields were systematically scanned and the motility of each spermatozoon graded as “a,” “b,” “c,” or “d,” according to whether it showed rapid progressive motility, slow or sluggish progressive motility, non progressive motility, or no motility at all.
Of the 64 patients, sperm count revealed normozoospermia (concentration of spermatozoa >20 × 106/mL and total progressive motility > 40 %) in 33 patients and asthenozoospermia (total progressive motility < 40 %) in 31 patients. As control group, we included 150 fertile and healthy males who fathered at least one child. All subjects gave an informed consent for molecular analysis of their blood samples.
Extraction of total DNA
DNA from all experimental samples was isolated using DNA isolation kit from Qiagen (QIAamp DNA Mini Kit). Briefly, the spermatozoa pellet was resuspended in disterile water and admixed with lysis solution containing 100 mg/mL proteinase K and 40 mmol/L dithiothreitol. Lysis was performed at 55 °C for 2–3 h with gentle agitation. Magnetic glass particles (MGPs) were added to the lysates to bind the DNA. DNA, which was immobilized on MGPs, was washed and eluted from the particles.
PCR amplification of the mitochondrial COIII gene
The mitochondrial COIII gene and the flanking regions were amplified using a thermal cycler (GeneAmp PCR System 9700 (Applied Biosystems) in a final volume of 50 μL using 200 ng DNA, 8 pmol of each primer (mt-14 F: 5′ CCCACCAATCACATGCCTAT 3′, mt-14R: 5′ TGTAGCCGTTGAGTTGTGGT 3′), 2 mM MgCl2, 500 μM dNTP, 1 × PCR buffer and 2 U Taq DNA polymerase). The conditions for PCR amplification were as follows: initial denaturation at 95 °C for 5 min followed by 35 cycles of denaturation (94 °C, 1 min), annealing (56.5 °C, 1 min), extension (72 °C, 1 min) and a final extension at 72 °C for 5 min.
Sequencing
After PCR amplification, PCR products were purified using NucleoSpin (MACHEREY-NAGEL) and sequenced with the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (ABI PRISM/Biosystems).
The blast homology searches were performed using the program available at the National Center for Biotechnology Information Web site in comparison with the updated consensus ambridge sequence (GenBank Accession NC_012920). Regions containing putative novel variations were amplified and sequenced again on both strands to exclude that they were PCR artifacts.
Sequence alignment
The sequence alignment of the mitochondrial COIII gene was performed using the Clustal W program (http://www.ebi.ac.uk/Tools/msa/clustalw2). Sequences from the species were obtained from NCBI.
The pathogenicity prediction of the m.9588G > A mutation and prediction software of hydrophobicity and secondary structure MT-COIII protein
The assessment of the damaging effect of missense mutation was performed using PolyPhen-2 software (http://genetics.bwh.harvard.edu/pph2/). This software calculates the probability that a given mutation is damaging and reports estimates of false positive (the chance that the mutation is classified as damaging when it is in fact non damaging) and true positive (the chance that the mutation is classified as damaging when it is indeed damaging) rates. For a false positive rate of 20 %, PolyPhen-2 achieved true positive prediction rates of 92 % and 73 % on HumDiv and HumVar datasets, respectively [20]. PolyPhen uses the predicted hydrophobic and transmembrane (PHAT) matrix score to evaluate the possible functional effect of a substitution in the transmembrane region.
For the prediction of possible changes in hydrophobicity and changes in configuration of transmembrane domains of the COIII protein due to sequence variation, we used TopPred online prediction software (http://bioweb.pasteur.fr/seqanal/interfaces/toppred.html).
Results
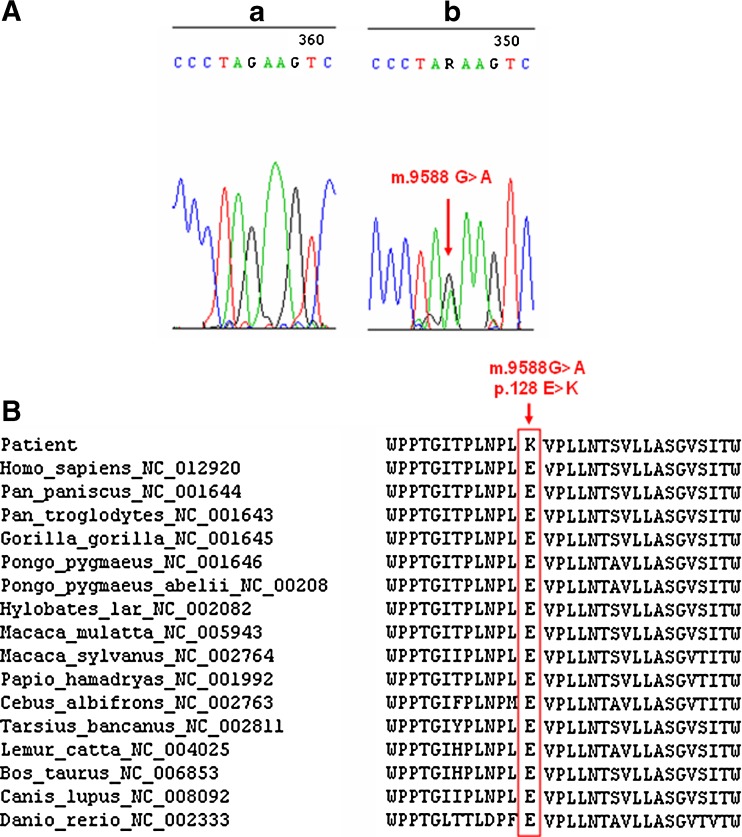
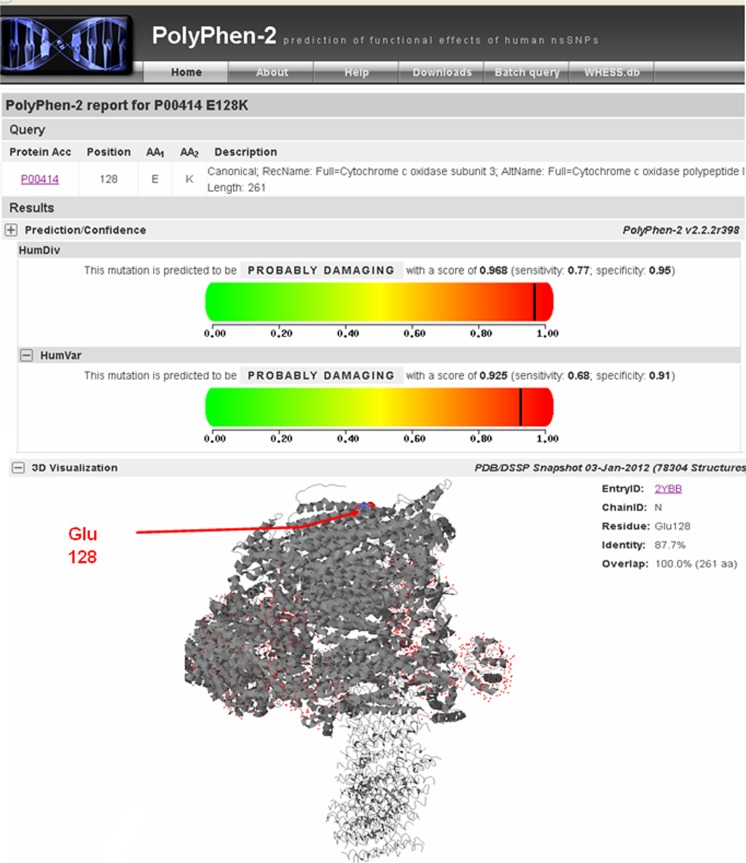
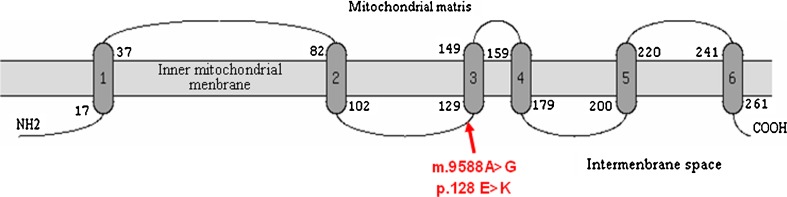
The sequencing analysis of the MT-COIII gene in asthenozoospermic and normozoospermic infertile man revealed the presence of a variant at nucleotide position 9588 (G > A) in all asthenozoospermic samples analyzed. This mutation (Fig. 1a) was absent in the normozoospermic and in 150 Tunisian fertile individuals. Interestingly, the A allele never before has been reported, either as a polymorphism or in association with a disease, anywhere in the world. The mutation substitutes the Glutamate residue at position 128 to lysine. The affected aminoacid is located in a highly conserved domain of the COIII polypeptide in many species (Fig. 1b). In addition, PolyPhen-2 analysis predicted that this variant is “probably damaging” with scores of 0.968 and 0.925 on HumDiv and HumVar models, respectively (Fig. 2) and showed that the Glu128 is located in the transmembrane functional domain of the mitochondrial COIII protein (Fig. 3).
Fig. 1.
a Sequencing electropherogram showing the presence of the novel m.9588G > A mutation in the mitochondrial COIII gene in a representative patient and its absence in a control subject. b Sequence alignment of the mitochondrial COIII gene in different species. The Glutamate at position 128 is highly conserved throughout evolution. Amino acid change of interest is framed and indicated with arrow
Fig. 2.
a Results of the PolyPhen-2 analysis predicting the pathogenicity the p.128 N > S substitution on the mitochondrial COIII protein. b Location of the Glu128 residue in the 3D structure of the mitochondrial COIII protein
Fig. 3.
Predicted transmembrane structures of human MT-COIII protien by the TopPred program
The m.9588G > A mutation in the mitochondrial COIII gene was found in Heteroplasmic state in all asthenozoospermic samples analyzed; this is confirm the patogenicty of this mutation because the most pathogenic mtDNA mutations coexist with the wild-type sequence: in heteroplasmy state.
Moreover, we detected 14 known substitutions in the COIII gene (Table 1) which were previously reported in the Human Mitochondrial Database (http://www.mitomap.org). Among these variations, four substitutions were responsible for an amino acids change in several mitochondrial subunits: m.9477 G > A (p.91 V > I), m. 9301C > T (p.32A > V), m. 9336A > G (p.44 M > V) and m. 9390A > G (p.62 T > A). These substitutions were reported as polymorphisms without negative effects on mitochondrial function.
Table 1.
Punctual mitochondrial variants detected in the COIII gene in asthenospermic patients, and each line in the table is from a different patient
| Locus | Nucleotide change | Position | Aminoacid change |
|---|---|---|---|
| MT-COIII | A > G | 9347 | syn |
| MT-COIII | T > C | 9540 | syn |
| MT-COIII | G > A | 9755 | syn |
| MT-COIII | C > T | 9818 | syn |
| MT-COIII | G > A | 9932 | syn |
| MT-COIII | G > A | 9477 | V > I |
| MT-COIII | A > G | 9494 | syn |
| MT-COIII | C > T | 9301 | A > V |
| MT-COIII | C > T | 9302 | syn |
| MT-COIII | A > G | 9336 | M > V |
| MT-COIII | A > G | 9390 | T > A |
| MT-COIII | A > G | 9425 | syn |
| MT-COIII | T > C | 9656 | syn |
| MT-COIII | T > C | 9770 | syn |
Discussion
As recently summarised by Moore and Reijo-Pera, 10–15 % of human couples are affected by infertility, and approximately 50 % of these cases of infertility should be attributed to men [24]. Sperm motility is one of the major determinants of male fertility. Furthermore, it has been demonstrated that mtDNA base substitutions can greatly influence semen quality and motility [21–23]. Therefore, we investigated in the present study the relationship between the sperm mtDNA COIII gene polymorphisms and male infertility.
We investigated COIII gene in sperm mtDNA of Tunisian infertile and fertile men and showed the presence of an undescribed missense mutation (m.9588G > A) in all asthenozoospermic infertile men. This mutation (m.9588G > A) was absent in normozoospermic patients and in fertile men, suggesting that it could be a cause of low sperm motility in asthenospermic patients
The m.9588G > A mutation found in the present study substitutes the Glutamate residue at position 128 to Lysine. The affected amino acid is located in a highly conserved domain of the COIII protein. In addition, PolyPhen-2 analysis predicted that this variant is “probably damaging” and showed that the Glu 128 is located in the transmembrane functional domain of COIII protein.
A similar study from India showed a novel missense mutation (C11994T) in the ND4 gene of oligoasthenozoospermic, but no in 150 proven-fertile men (normozoospermic) [24]. This mutation (m.11994C > T) is located in the second base of a codon, resulting in an amino-acid change from threonine to isoleucine at amino-acid position 412 of the ND4 gene. The authors conclude that this novel missense mutation C11994T in the mitochondrial ND4 gene as a cause of low sperm motility in the Indian subcontinent. This novel mutation (C11994T) was absent in oligoasthenozoospermic infertile patient from Portugal suggesting that such mutation may be population specific [25]. In another reports, Spiropoulos J et al. correlated low sperm motility with high levels of mutant mtDNA in males who had inherited the A3243G mtDNA mutation from their mother [22]; similarly, Holyoake et al., found the two most common substitutions at 9055 (MT-ATP6) and 11719 (ND4) in men with a significantly higher frequency of reduced sperm motility [23]. Also, Kumar et al. showed that the T9098C transition was present only in infertile cases with a significant difference in comparaison to controls [23].
Our findings showed also the presence of 14 known substitutions in the COIII gene reported in the Human Mitochondrial Database (http://www.mitomap.org). Among these variations, 4 substitutions were responsible for an amino acids change in several mitochondrial subunits. In a recent Indian study, the authors revealed 36 substitutions in an oligoasthenoteratozoospermic (OAT) man who also had varicocele of the left testis (8 in COXI, 13 in COXII, 5 in ATPase8, and 10 in ATPase6) [24]. The fourth of these substitutions (n = 9) were missense mutations (3 in COXI, 3 in COXII, 1 in ATPase8, and 2 in ATPase6). The authors suggested that the diminished sperm motility would be due to a 2-nucleotide deletion in COXII mitochondrial gene. In another report it was shown significant nucleotide changes in mitochondrial genes (ATPase6, ATPase8, ND2, ND3, ND4 and ND5) in sperm mtDNA of infertile men [23]. Otherwise, Güney et al. identified 38 different nucleotide substitutions, of which 15 caused an amino acid change. In addition, 12 were considered novel mutations. C8927G, A9041G, C9105G novel polymorphisms were found in the ATPase6 gene, and T14969C, C15143T, T15282G, A15296C, T15804A, and G15806C novel polymorphisms were found in the Cytb gene [25]. Three other polymorphisms (T4114G, G4153A and C4159A novel polymorphism) were found in the ND1 gene.
To elucidate the effect and functional importance of m.9588G > A mutation, functional studies using the cybrid cells like rho-0 (a cell culture model used for the study of mitochondrial disorders), should be undertaken to evaluate the eventual change of Cytochrome c oxidase subunitIII (COIII) activity and of protein synthesis, and the impact of novel mutation on respiratory chain function.
In conclusion, we reported in this study an undescribed heteroplasmic missense mutation (m.9588G > A) in sperm mtDNA COIII gene in all asthenozoospermic infertile patients who is absent in normozoospermic and in 150 feriles men and we suggest that this mutation may affect the electrons transfer from reduced cytochrome c to molecular oxygen in mitochondria. We also suggest that this novel m.9588G > A mutation should routinely be screened in asthenozoospermic infertile individuals to give some insight into the aetiology of motility disorders in infertile men. Assessment of identified mtDNA polymorphisms should be also undertaken in clinical male infertility cases.
Acknowledgments
We thank the patient and their family for their cooperation in the present study. This work was supported by The Ministry of the Higher Education and the Scientific Research in Tunisia.
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Capsule
A systematic sequence analysis of the mitochondrial (COIII) gene have been studied in 64 Tunisian infertile men in order to assess the incidence of mtDNA base substitutions; that can greatly influence semen quality.
References
- 1.Følgero T, Bertheussen K, Lindal S, Torbergsen T, Oian P. Mitochondrial disease and reduced sperm motility. Hum Reprod. 1993;8:1863–1868. doi: 10.1093/oxfordjournals.humrep.a137950. [DOI] [PubMed] [Google Scholar]
- 2.Wallace DC. Mitochondrial DNA, variation in human evolution, degenerative disease, and aging. Am J Hum Genet. 1995;57:201–223. [PMC free article] [PubMed] [Google Scholar]
- 3.Clayton DA, Doda JN, Friedberg EC. The absence of a pyrimidine dimer repair mechanism in mammalian mitochondria. Proc Natl Acad Sci U S A. 1974;71:2777–2781. doi: 10.1073/pnas.71.7.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukunaga M, Yielding K. Fate during cell growth of yeast mitochondrial and nuclear DNA after photolytic attachment of the monoazide analog of ethidium bromide. Biochem Biophys Res Commun. 1979;90:582–586. doi: 10.1016/0006-291X(79)91275-0. [DOI] [PubMed] [Google Scholar]
- 5.Richter C, Park JW, Ames BN. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc Natl Acad Sci U S A. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagley P, Zhang C, Martinus RD, Vaillant F, Linnane AW. Mitochondrial DNA mutation and human aging: molecular biology, bioenergetics, and redox therapy. In: DiMauro 1993
- 7.Johns DR. Mitochondrial DNA, and disease. N Engl J Med. 1995;333:638–644. doi: 10.1056/NEJM199509073331007. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell JA, Nelson L, Hafez E. Motility of spermatozoa. In: Hafez ESE, editor. Human semen and fertility regulation in men. St. Louis: C. V. Mosby; 1976. pp. 83–106. [Google Scholar]
- 9.Kao SH, Chao HT, Wei YH. Mitochondrial deoxyribonucleic acid 4977-bp deletion is associated with diminished fertility and motility of human sperm. Biol Reprod. 1995;52:729–736. doi: 10.1095/biolreprod52.4.729. [DOI] [PubMed] [Google Scholar]
- 10.Frank SA, Hurst LD. Mitochondria and male disease. Nature. Enriquez: Guan MX; 1996. p. 224–383. [DOI] [PubMed]
- 11.St. John JC, Cooke ID, Barratt CL. Mitochondrial mutations and male infertility. Nat Med. 1997;3:124–125. doi: 10.1038/nm0297-124c. [DOI] [PubMed] [Google Scholar]
- 12.Zeviani M, Antozzi C. Mitochondrial disorders. Mol Hum Reprod. 1997;3:133–148. doi: 10.1093/molehr/3.2.133. [DOI] [PubMed] [Google Scholar]
- 13.Ruiz-Pesini E, Lapena A, Diez-Sanchez C, Perez-Martos A, Enriquez J, Diaz M, Urries A, Montoro L, Lopez-Perez M, Enriquez J. Human mitochondrial DNA haplogroups associated with high or reduced spermatozoa motility. Am J Hum Genet. 2000;67:682–696. doi: 10.1086/303040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zamboni L. Physiology and pathophysiology of the human spermatozoon: the role of electron microscopy. J Electron Microscopy Technique. 1991;17:412–436. doi: 10.1002/jemt.1060170405. [DOI] [PubMed] [Google Scholar]
- 15.Oates R. Immotile cilia syndromes. Paper presented at a conference of the American Society of Reproductive Medicine: Male Reproduction—Basic, Genetic, and Clinical Information for the Next Millennium, San Francisco, 1998; October 3–4.
- 16.Folgero T, Bertheussen K, Lindal S, Tobergsen T, Oian P. Mitochondrial disease and reduced sperm motility. Hum Reprod. 1993;8:1863–1868. doi: 10.1093/oxfordjournals.humrep.a137950. [DOI] [PubMed] [Google Scholar]
- 17.Lestienne P, Reynier P, Chretien MF, Penisson-Besnier I, Malthièry Y, Rohmer V. Oligoasthenospermia associated with multiple mitochondrial DNA rearrangements. Mol Hum Reprod. 1997;3:811–814. doi: 10.1093/molehr/3.9.811. [DOI] [PubMed] [Google Scholar]
- 18.Kao SH, Chao HT, Wei YH. Multiple deletions of mitochondrial DNA are associated with the decline of motility and fertility of human spermatozoa. Mol Hum Reprod. 1998;4:657–666. doi: 10.1093/molehr/4.7.657. [DOI] [PubMed] [Google Scholar]
- 19.WHO laboratory manual for the examination of human semen and sperm-cervical interaction. 3. Cambridge, United Kingdom: Cambridge University Press; 1992. [Google Scholar]
- 20.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pereira L, Goncalves J, Bandelt HJ. Mutation C11994T in the mitochondrial ND4 gene is not a cause of low sperm motility in Portugal. Fertil Steril. 2008;89:738–741. doi: 10.1016/j.fertnstert.2007.03.048. [DOI] [PubMed] [Google Scholar]
- 22.Spiropoulos J, Turnbull DM, Chinnery PF. Can mitochondrial DNA mutations cause sperm dysfunction? Mol Hum Reprod. 2002;8:719–721. doi: 10.1093/molehr/8.8.719. [DOI] [PubMed] [Google Scholar]
- 23.Kumar R, Venkatesh S, Kumar M, Tanwar M, Shamsi MB, Kumar R. et al. Oxidative stress and sperm mitochondrial DNA mutation in idiopathic oligoasthenozoospermic (OA) infertile men. Indian J Biochem Biophys. (2009) (in press). [PubMed]
- 24.Thangaraj K, Joshi MB, Reddy AG, Rasalkar AA, Singh L. Sperm mitochondrial mutations as a cause of low sperm motility. J Androl. 2003;24:388–392. doi: 10.1002/j.1939-4640.2003.tb02687.x. [DOI] [PubMed] [Google Scholar]
- 25.Güney AI, Javadova D, Kırac D, Ulucan K, Koc G, Ergec D, Tavukcu H, Tarcan T. Detection of Y chromosome microdeletions and mitochondrial DNA mutations in male infertility patients. GRM. 2012;1525:1676–5680. doi: 10.4238/2012.April.27.2. [DOI] [PubMed] [Google Scholar]