Abstract
Purpose
To determine the frequencies and the characteristics of Y chromosome microdeletions (pl) in infertile men from central China to perform appropriate therapeutic choices by updated multiplex-PCR.
Methods
In this study, we established a novel universal primer-multiplex-PCR (U-M-PCR) method to overcome the disadvantages of traditional multiplex PCR (M-PCR). We chose 15 sequence-tagged sites (STS) for detection of Y chromosome microdeletions. 540 infertile male patients and 100 healthy male controls were selected in the study.
Results
Of the 540 male infertility patients, 48 Y-chromosome microdeletions were detected, with a total deletion rate of 8.9 %. Of these deletions, the rate of AZFa deletions (sY84) was 0.5 % (3/540), the rate of AZFb deletions (sY143) was 0.7 % (4/540) and the rate of AZFc deletions (sY242, sY254 and sY255) was 7.6 % (41/540). Compared with AZF deletion rates by M-PCR, we found U-M-PCR could detect AZFc deletion more specifically (1.0 % & 7.6 %). No Y-chromosome microdeletions were detected in the 100 males with normal semen (the control group).
Conclusions
U-M-PCR method was more specific to detect AZFc microdeletions. It is necessary to use the U-M-PCR method to offer genetic screening and counseling to infertile men prior to intracytoplasmic sperm injection (ICSI) or in-vitro fertilization (IVF).
Keywords: Male infertility, Y-chromosome, Microdeletions, Multiplex PCR, AZFc
Introduction
Approximately 10 % of couples that are of childbearing age suffer from infertility, of which male spermatogenic disorders are one of the major causes. Y chromosome infertility is characterized by azoospermia (absence of sperm), severe oligozoospermia (<5 × 106 sperm/ml semen) and moderate oligozoospermia (5–20 × 106 sperm/ml semen) [1, 2], which account for 10 % of male fertility cases. In addition to vas deferens obstruction, infections of the reproductive system and gonadal endocrine disorders, genetics are also a crucial factor contributing to male infertility [3]. It is widely believed that at least three nonoverlapping regions of the human Y chromosome-AZFa, AZFb, and AZFc (“azoospermia factors” a, b, and c)—are essential for normal spermatogenesis [4]. Today the sequence of the MSY [5] and the molecular mechanism resulting in microdeletions have been clarified. This resulted in a new model of deletions in which the AZFb and AZFc regions are overlapping (called AZFd) [6, 7]. And the STS of AZFd were mainly including sY133, sY145, sY153, and sY152 [7]. Microdeletions at any subregion can result in a spermatogenic disorder [8].
In recent years, the technique of intracytoplasmic sperm injection (ICSI) offers new hope for men with severe oligozoospermia and azoospermia to achieve successful fertilizations and pregnancies [9]. However, the Yq microdeletions, especially deletion of AZFc region, might have a potential risk to be transmitted from infertile fathers to their male offspring, who could also experience infertility, with the use of the ICSI procedure [10]. Thus, it is important to evaluate Yq microdeletions in infertility in men before assisted reproduction to provide appropriate information to the patients.
In the past, conventional multiplex PCR (M-PCR) can amplify two or more DNA templates in one reaction, which is flexible and can effectively reduce costs. However, as new methodologies have been developed, traditional M-PCR has begun to show its inadequacies, which mainly include a complicated procedure, low detection sensitivity, amplification inhibition and inconsistent amplification efficiency [11]. To overcome the aforementioned shortcomings of traditional M-PCR, universal primers (UPs) were used in this study to establish a novel, stable and reliable UP-M-PCR system for the detection of Y-chromosome microdeletions. The present study aimed to introduce a novel UP-M-PCR method to detect Y choromosome microdeletion, which could provide a more accurate evidence for the diagnosis of these patients with severe oligozoospermia or azoospermia.
Materials and methods
Subjects
Molecular screening of Y-chromosome microdeletions were carried out in the 200 male infertility patients plus 35 healthy men from 2007 to 2009 by M-PCR, and then carried out in the 540 male infertility patients plus 100 healthy men from 2009 to 2013 by UP-M-PCR. They were between 24 and 39 years of age, with an average age of 34 years. All the patients were treated in the Reproductive Center at Renmin Hospital of Wuhan University. In total 200 cases, 18 patients had nonobstructive azoospermia (absence of sperm), and 182 patients had severe oligozoospermia (<5 × 106 sperm/ml). In total 540 cases, 54 patients had nonobstructive azoospermia (absence of sperm), and 486 patients had severe oligozoospermia (<5 × 106 sperm/ml), as is the standard set by Silber SJ’s book [12, 2].
Methods
DNA extraction from peripheral blood
Two milliliters of peripheral blood was drawn from each subject and treated with ethylenediaminetetraacetic acid (EDTA) to prevent coagulation. A whole-blood DNA extraction kit (Shanghai SBS Genetech Co., Ltd.) was used to extract DNA.
Design of universal primers (UP)
ABI PRISM Primer Express software was used to design UPs. Factors such as the minimal number of genomic binding sites, a high GC content and a melting temperature (Tm) of approximately 60 °C were taken into consideration. According to these principles, the following UPs were designed: UP-F: TACAGTCGGACGCGTCCCTC; UP-R:CTGGTCCGTACTACCGTGCG. The universal primer was designed in the 5′-end of each specific primer pairs.
Design of primers
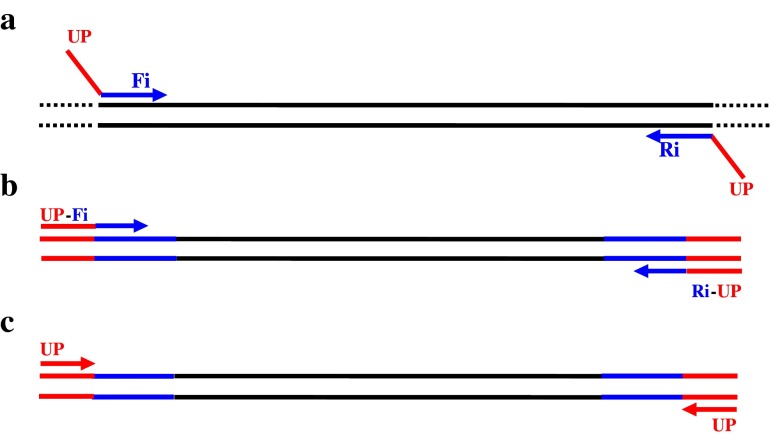
Six sequence-tagged sites (STS) recommended by the international authoritative guide (European Academy of Andrology, Institute of Molecular Genetics) [13] were used for the analysis of Y-chromosome microdeletions. 15 STS-specific primers of Y-chromosome regions AZFa, AZFb, AZFc and AZFd were recommended and could detect more Y microdeletions [7]. All primers were synthesized by Shanghai Invitrogen Biotechnology Co., Ltd. UPs were added to 5′-end of the specific primers to produce primers that included UPs, a specific target representing one of the 15 STSs and an internal control, the sex-determining region of the Y chromosome (SRY). The sequence of M-PCR primers was composed of the sequences of the UPs and one of the 15 STS-specific primers. The entire M-PCR reaction process is shown in Fig. 1. During the early cycles, the chimeric STS-specific primers (with the same UPs at both ends) primarily mediated the amplification of the DNA template. The denaturing temperature of this process was high, and therefore, UPs played almost no role in amplification (approximately the first ten cycles, Fig. 1a). With the depletion of the STS primers and the increasing number of DNA templates with the UPs sequences at 5′-ends (Fig. 1b), UPs became predominant through rapid PCR amplification to complete the entire PCR reaction (Fig. 1c).
Fig. 1.
The process of the M-PCR reaction using specific primers in combination with UPs. UP (red) represent universal primers, and Fi/Ri (blue) represent STS-specific primers. a During the early stages, chimeric-specific primers mediated the amplification of target DNA templates; b A certain number of target DNA templates with the UP sequences were obtained, and c UPs specifically amplified the target DNA templates
Sequence of specific primers and groupings of M-PCR
The sequence of the 15 STS-specific primers for the Y-chromosome regions AZFa, AZFb, AZFc and AZFd were located in the PubMed uniSTS database. Then UPs were added to the 5′-ends of the specific primers. Based on the size of amplified fragments, 4 stable and reliable UP-M-PCR reaction systems were established. The primer sequences and the groupings of the M-PCR systems are shown in detail in Table 1.
Table 1.
The sequences of the U-M-PCR primers
| Groups | Locus | AZF | Primer sequences | MW (bp) |
|---|---|---|---|---|
| UP | UP-F: TACAGTCGGACGCGT CCCTC | |||
| UP-R: CTGGTCCGTACTACCGTGCG | ||||
| TC | SRY | sY14 | F: TACAGTCGGACGCGT CCCTCGAATATTCCCGCTCTCCGGA | 510 |
| R: CTGGTCCGTACTACCGTGCGGCTGGTGCTCCATTCTTGAG | ||||
| I | sY254 | AZFc | F: TACAGTCGGACGCGT CCCTCGGGTGTTACCAGAAGGCAAA | 420 |
| R: CTGGTCCGTACTACCGTGCGGAACCGTATCTACCAAAGCAGC | ||||
| I | sY143 | AZFb | F: TACAGTCGGACGCGT CCCTC GCAGGATGAGAAGCAGGTAG | 351 |
| R: CTGGTCCGTACTACCGTGCGCCGTGTGCTGGAGACTAATC | ||||
| I | sY242 | AZFc | F: TACAGTCGGACGCGT CCCTCACACAGTAGCAGCGGGAGTT | 273 |
| R: CTGGTCCGTACTACCGTGCGTCTGCCACTAAACTGTAAGCTCC | ||||
| I | sY255 | AZFc | F: TACAGTCGGACGCGT CCCTCGTTACAGGATTCGGCGTGAT | 163 |
| R: CTGGTCCGTACTACCGTGCGCTCGTCATGTGCAGCCAC | ||||
| II | sY84 | AZFa | F: TACAGTCGGACGCGT CCCTCAGAAGGGTCTGAAAGCAGGT | 366 |
| R: CTGGTCCGTACTACCGTGCGGCCTACTACCTGGAGGCTTC | ||||
| II | sY239 | AZFc | F: TACAGTCGGACGCGT CCCTCCATTCATCTTCCCTTTTGAAGG | 241 |
| R: CTGGTCCGTACTACCGTGCGATGCAAGTCGCAGGAAATCT | ||||
| II | sY152 | AZFd | F: TACAGTCGGACGCGT CCCTCAAGACAGTCTGCCATGTTTCA | 165 |
| R: CTGGTCCGTACTACCGTGCGACAGGAGGGTACTTAGCAGT | ||||
| III | sY86 | AZFa | F: TACAGTCGGACGCGT CCCTCGTGACACACAGACTATGCTTC | 360 |
| R: CTGGTCCGTACTACCGTGCGACACACAGAGGGACAACCCT | ||||
| III | sY127 | AZFb | F: TACAGTCGGACGCGT CCCTCGGCTCACAAACGAAAAGAAA | 314 |
| R: CTGGTCCGTACTACCGTGCGCTGCAGGCAGTAATAAGGGA | ||||
| III | sY145 | AZFd | F:TACAGTCGGACGCGTCCCTCCAACACAAAAACACTCATATACTCG | 183 |
| R: CTGGTCCGTACTACCGTGCGTTGAGAATAATTGTATGTTACGGG | ||||
| III | sY124 | AZFb | F: TACAGTCGGACGCGT CCCTCCAGGCAGGACAGCTTAAAAG | 149 |
| R: CTGGTCCGTACTACCGTGCGACTGTGGCAAAGTTGCTTTC | ||||
| IV | sY134 | AZFb | F: TACAGTCGGACGCGT CCCTCGTCTGCCTCACCATAAAACG | 341 |
| R: CTGGTCCGTACTACCGTGCGACCACTGCCAAAACTTTCAA | ||||
| IV | sY82 | AZFa | F: TACAGTCGGACGCGT CCCTCATCCTGCCCTTCTGAATCTC | 304 |
| R: CTGGTCCGTACTACCGTGCGCAGTGTCCACTGATGGATGA | ||||
| IV | sY128 | AZFb | F: TACAGTCGGACGCGT CCCTCGGATGAGACATTTTTGTGGG | 268 |
| R: CTGGTCCGTACTACCGTGCGAGCCCAATGTAAACTGGACA | ||||
| IV | sY133 | AZFb | F: TACAGTCGGACGCGT CCCTCATTTCTCTGCCCTTCACCAG | 217 |
| R: CTGGTCCGTACTACCGTGCGTGATGATTGCCTAAAGGGAA |
The table lists detailed information on the sequences of the primers and the size of the amplified target DNA fragments. The same UPs were added at the 5′ end of each STS-specific primer. According to the size of the amplified fragments, M-PCRs were divided into four groups: I (SRY, sY254, sY143, sY242, sY255), II (SRY, sY84, sY239, sY152), III (SRY, sY86, sY127, sY145, sY124) and IV (SRY, sY134, sY82, sY128, sY133). TC represented for Internal control, and MW for Molecular Weight
Electrophoresis of the PCR amplification products
10 μl of the PCR amplification products was separated by electrophoresis on a 4 % agarose gel (0.5 μl/ml ethidium bromide) at 100 V for 45 min. The results were analyzed under UV light. The PCR bands were missing in patients with Y-chromosome microdeletions. To exclude false- negative results, the missing band was further verified with simple PCR to ensure the accuracy of the UP-M-PCR detection.
Results
Specificity of compound specific primers
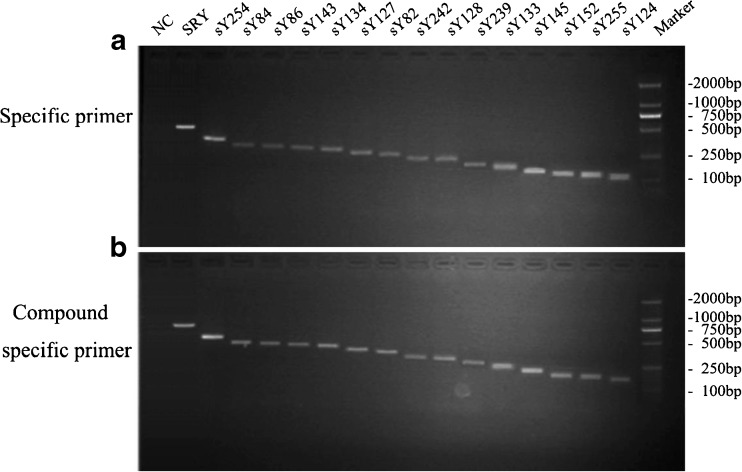
Keeping the concentration of templates at 50 ng/ul, and the 15 STS (SRY, sY254, sY84, sY86, sY143, sY134, sY127, sY82, sY242, sY128, sY239, sY133, sY145, sY152, sY255, sY124) were amplified with singlet PCR by specific primers (Fig. 2a). Meanwhile, the same samples were also amplified with the new designed compound universal primer pairs (Table 1), which were originated from specific primers. The result showed that compound universal primer tests displayed to get equivalent intensities of bands on gels with primers alone tests (Fig. 2b). Thus showed that the set of compound specific primers worked efficiently and had the same specificity as the specific primers alone. Because the compound specific primers contained a common sequence (20 bp) at the 5′-end, so the bands on gel were a little higher too (Fig. 2b).
Fig. 2.
Detection of the specificity of UP-M-PCR. a Comparison of specificity between specific primers and compound specific primers. A amplicon fragments of specific primer;B amplicon fragments of UP and compound specific primer; lane 1, negative control (NC) without template; the last lane, 100 bp DNA Marker
Optimization of the UP-M-PCR
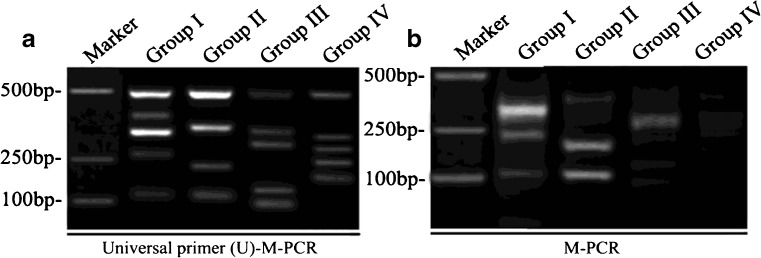
The concentrations of primers strongly influence the efficiency and disparity of PCR reaction, which is very important for the PCR reaction, especially in multiplex PCR. The final optimized concentration of universal primer (UP) was 500 nmol/L in UP-M-PCR, while the compound specific primers were 25 nmol/L(about 1/20 of UP) in singlet PCR that can ensure an efficient amplification for all these primers, but in 16-plex UP-M-PCR, because of the interaction and the difference of work efficiency among primers, all compound specific primers at 25 nmol/L could not get an equivalent amount of amplified products, thus there need an adjustment. The final optimized concentration of the compound specific primers were 10 nmol/L for sY143-F/R and sY84-F/R (about 1/50 of UP), 15 nmol/L for sY127-F/R and sY133-F/R(about 1/30 of UP), 50 nmol/L for sY86-F/R, sY255-F/R and sY124-F/R(about 1/10 of UP), and 25 nmol/L for all other primers (including SRY-F/R, sY254-F/R, sY242-F/R, sY239-F/R, sY152-F/R, sY145-F/R, sY134-F/R, sY82-F/R and sY128-F/R). To find the best annealing temperature, a gradient temperature PCR from 56 to 64 °C has been performed. At last the optimum annealing temperature was chosen at 60°C. Similarly the best extension temperature was chosen at 70°C. The time for annealing and extension was chosen at 50s. We obtained the DNA from the healthy men, and performed the M-PCR amplification by compound universal primers (Fig. 3a) or no compound primers (Fig. 3b). The data showed that the U-M-PCR by compound universal primers was more specific compared to M-PCR, which indicated that using U-M-PCR could detect the Y chromosome microdeletions in the inferitility male patients.
Fig. 3.
Optimization of the UP-M-PCR. A Lane 1, 100 bp DNA Marker; Lane 2 is group I (SRY, sY254, sY143, sY242, sY255); Lane 3 is group II (SRY, sY84, sY239, sY152); Lane 4 is group III (SRY, sY86, sY127, sY145, sY124) and Lane 5 is group IV (SRY, sY134, sY82, sY128, sY133). B Normal multiplex PCR
The results of Y-chromosome microdeletions detected by UP-M-PCR in the 540 male infertility patients
UP-M-PCR was used for the rapid detection of microdeletions of the 15 STSs in the four regions of AZF gene family on the Y chromosome: AZFa, AZFb, AZFc and AZFd. UP-M-PCR reactions for groups I, II, III and IV were carried out for each sample (Fig. 4). Of the 540 male infertility patients, 48 patients were found to have Y-chromosome microdeletions, with a deletion rate of 8.9 % (48/540). Of these 48, 41 patients had AZFc deletions alone with a deletion rate of 7.6 % (41/540), 4 patients had AZFb deletions with a deletion rate of 0.7 % (4/540), and 3 patients had AZFa deletions alone with a deletion rate of 0.5 % (3/540) (Table 2, Fig. 4). No joint deletions or large deletions were found. To exclude false-negative results, a simple PCR assay was carried out for each of the deletion sites of the seven samples. No amplifications were found (data not show), confirming the accuracy of UP-M-PCR detection. In addition, no AZF gene deletions were found in the control group of 100 normal, healthy males (those with normal sperm counts). In order to substantiate the more specificity of U-M-PCR, we compared the AZF deletion rates by two different methods-M-PCR and U-M-PCR. The data showed that there were no difference between AZFa and AZFb deletions (P > 0.05), but significant different between AZFc deletions (P < 0.001) (Table 3.)
Fig. 4.
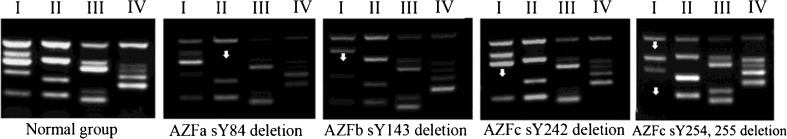
The gel electrophoresis results for the Y chromosome UP-M-PCR of the samples from the control group and infertility patients. Compared with the control group, AZFa sY84 microdeletions were detected in three patients, AZFb sY143 microdeletions were detected in four patients and AZFc sY242, sY254, sY255 microdeletions were detected in 41 patients
Table 2.
Y chromosome microdeletions in patients
| Groups | Number of subjects | AZFa deletion | AZFb deletion | AZFc deletion |
|---|---|---|---|---|
| Male infertility group | 540 | 3 (0.5 %) | 4 (0.7 %) | 41 (7.6 %) |
| Normal control group | 100 | 0 | 0 | 0 |
Y chromosome microdeletions rates of male infertility and normal control. Statistical analyses show that the Y-chromosome microdeletion rates in the 540 male infertility patients were 0.5 % for AZFa, 0.7 % for AZFb, and 7.6 % for AZFc
Table 3.
Compared with AZF deletion rates by two different methods-M-PCR and U-M-PCR
| Methods | Number of subjects | AZFa deletion | AZFb deletion | AZFc deletion |
|---|---|---|---|---|
| M-PCR group | 200 | 1 (0.5 %) | 1 (0.5 %) | 2 (1.0 %) |
| U-M-PCR group | 540 | 3 (0.5 %) | 4 (0.7 %) | 41 (7.6 %) |
| P value | 0.42 | 0.38 | <0.001 |
Fisher’s exact test statistical analyses show that the Y-chromosome microdeletion rates were similar between AZFa and AZFb (P > 0.05). There were statistical difference between AZFc (P < 0.001)
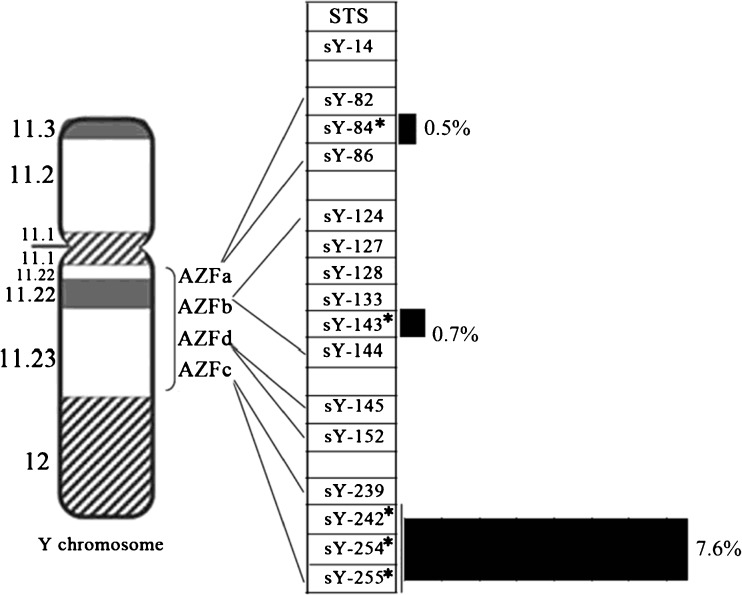
The characteristic of Y chromosome microdeletions in infertile men from central of China
The above data showed that Y chromosome microdeletions from central of China mainly put together in the sY84, sY143, sY242, sY254 and sY255 sites, which distributed in different AZF regions. In addition, the deletion rate of them were also distinct, particularly high in AZFc region (Fig. 5).
Fig. 5.
The distribution of Y-chromosome microdeletions in the 48 patients. The distribution of the Y-chromosome microdeletions in the 48 patients showed that deletions were found in all regions, 0.5 % for AZFa, 0.7 % for AZFb and 7.6 % for AZFc, of which AZFc had the highest deletion rate. Note: Asterisks (*) and black rectangles indicate deletion rate of AZFa, AZFb and AZFc, respectively
Discussion
Nucleic acid analysis is becoming increasingly important for the detection of Y-chromosome microdeletions, and PCR provides a powerful tool for the analysis. Conventional PCR method can specifically amplify a single DNA template whereas M-PCR can specifically amplify two or more DNA templates in one reaction. Thus, M-PCR is more widely used in scientific and clinical research for genetic testing of microbial pathogens [14], gene expression [15], whole genome sequencing [16], forensic analysis and paternity testing [17]. Currently, M-PCR is mainly utilized in the analysis and detection of Y-chromosome microdeletions and uses specific primers to amplify the STS fragments on different regions of the Y chromosome. However, with advances in research technology, the inadequacies of the M-PCR system cannot be ignored. First, the primer-primer interaction results in the self-inhibition of amplification. Second, the overall efficiency of the PCR amplification is low. Third, the different parameters of different primers lead to inconsistencies in the amplification efficiencies of the various DNA templates. The above shortcomings and inadequacies have limited the further development and broad application of M-PCR.
In this study, a novel UP-M-PCR technique was adopted to solve the issues with M-PCR amplification. In this study, UP was introduced to the field of Y-chromosome M-PCR detection. This new UP-M-PCR system can further enhance the sensitivity and specificity of the detection of Y-chromosome microdeletions. Improving the detection rate of Y-chromosome microdeletions in different countries and regions has great significance for identifying deletions before clinical intracytoplasmic sperm injection (ICSI) technology assisted fertility treatment, genetic testing for male patients with oligozoospermia and azoospermia, screening for sperm before it is deposited into sperm banks and population genetics researches.
The human Y chromosome is the primary target for male infertility and population genetics research. Large differences in Y-chromosome microdeletions have been found in different countries. Y chromosome deletions encompassing the AZFc region have been reported in 13 % of azoospermic men and 7 % of severely oligozoospermic men. The most frequently deleted region is AZFc (65–70 %), followed by deletions of the AZFb and AZFb + c or AZFa + b + c regions (25–30 %), whereas deletions of the AZFa region are extremely rare (5 %) [6]. Puzuka et al. reported a study of 105 infertile Latvian males, and AZFc deletions were detected in three males while AZF (a + b + c) was completely deleted in two males [18]. A study in India showed that the rate of Y-chromosome microdeletions was 10.5 % in 200 patients [13]. A study of 187 Turkish patients with infertility showed that Y-chromosome microdeletions were detected in seven patients, with a deletion rate of 3.93 %. Moreover, AZFc microdeletions were the most common [19]. Mirfakhraieř [20] studied 100 infertile Iranian males suffering from azoospermia and found that 12 % of the patients had Y-chromosome microdeletions. A 10-year retrospective study [21] suggested that AZFc deletions are closely related to phenotypic variation in sperm, whereas complete deletions of AZFb and AZFc are associated with Sertoli-cell-only syndrome. Another study conducted at the Xiangya School of Medicine showed that 12 of 101 patients had AZFc microdeletions, for a total deletion rate of 12 % [22]. Of these patients, one had both an AZFc and AZFd co-deletion. Deletions of both AZFb and AZFc together occur by two major mechanisms involving homologous recombination between P5/distal P1 or between P4/distal P1 [4] Complete removal of the AZFa and AZFb regions is associated with severe testicular phenotype. Complete removal of the AZFc region causes a variable phenotype that may range from azoospermia to oligozoospermia [6]. Langer [2] reported that an idicYp or isoYp in eight (2.7 %) of 293 men with nonobstructive azoospermia, but in none of 288 men with severe or moderate oligozoospermia. In the large majority of cases, idicYp or isoYp formation precludes sperm production. In this respect, the consequences of idicYp or isoYp formation appear to be more severe than those of AZFc deletions, which are found in men with either severe oligozoospermia or nonobstructive azoospermia.
In the past, six STSs strongly recommended by European Molecular Genetics Quality Network and European Academy of Andrology, which could lead to undetection of some microdeletions. Our study selected 15 STSs in the detection of AZF microdeletion. Compared with STSs in the minimum detection system (sY84 and sY86 for AZFa, sY127 and sY134 for AZFb, sY254 and sY255 for AZFc) [23]. 15 STSs could cover all the sparse region of the Yq chromosome. Particularly, sY82, sY84 and sY86 were used for detecting AZFa; sY127, sY134, and sY143 for AZFb; sY134, sY242, sY254, sY255 and sY239 for AZFc region and sY152, sY145 for AZFd region. Of the above studies, AZFb deletions were the most common (66.67 %) followed by AZFc deletions (41.67 %), AZFd deletions (33.33 %) and AZFa deletions (8.33 %) [24]. Above studies suggest that in different countries and regions, not only are the rate of Y chromosome deletions different but also are the deletion sites. In Chinese men, the updated data showed that the total AZF deletions was 11.55 %, and AZFc deletions represented the most frequent finding (5.9 %), followed by the AZFb region (1.1 %), AZFa (0.9 %) and AZFab/bc/abc (2.3 %) [25]. In the present study, the microdeletion rate of AZF was 8.9 %, followed by 0.5 % of AZFa, 0.7 % of AZFb and 7.6 % of AZFc. Among all the regions, microdeletions in the AZFc region were the most common, and sY254 and sY255 for deleted in infertility men presented a higher prevalence of deletions than other STSs, followed by sY242. Other more popular sites were sY84 in AZFa region, and sY143 in AZFb region. There were no patients with joint deletions or large deletions. Males with normal sperm counts had no-Y chromosome microdeletions. Compared with previous reports from China [26], the rate of Y-chromosome microdeletions in this study was lower, which may be due to the regional limitation and an insufficient sample size.
Acknowledgments
This research supported by the forty-ninth issue of the China Postdoctoral Science Foundation, first-class funding (Grant No. 20110490119), National Natural Science Foundation of China (Grant No. 81100959) and National Clinical Key Specialty Construction Projects of the Health Ministry (2010).
Author contributions
ZHY carried out the multiplex PCR, performed the statistical analysis and drafted the manuscript. SFJ and TYQ collected the samples and extracted genomic DNA from peripheral blood. LY designed the study and helped to draft the manuscript. All authors read and approved the final manuscript.
Competing financial interests
The authors have nothing to disclose.
Footnotes
Capsule Traditional multiplex PCR (M-PCR) has been widely used to detect Y-chromosome microdeletions, however, its specificity is not very high. In the present study, we established a novel universal primer-multiplex-PCR (UP-M-PCR) method to overcome the disadvantage. On the other hand, we detected 15 STS to find all the possible deletion sites, which is more accurate for diagnosis of male infertility compared to the past used 6 STS.
In the current manuscript, we eatablished 4 stable and reliable UP-M-PCR reaction systems to detected the 15 sequence-tagged sites (STS) for the AZFa, AZFb, AZFc and AZFd regions of the Y-chromosome. Of the 540 male infertility patients, 48 Y-chromosome microdeletions were detected, with a total deletion rate of 8.9 %. Of these deletions, the rate of AZFa deletions (sY84) was 0.5 % (3/540), the rate of AZFb deletions (sY143) was 0.7 % (4/540) and the rate of AZFc deletions (sY242, sY254 and sY255) was 7.6 % (41/540). However, there were only 0.5 % and 1 % of AZFb and AZFc ’s respectively by traditional M-PCR. Thus data showed the great advantages of UP-M-PCR, which could provide a more accurate evidence for the diagnosis of these patients with severe oligozoospermia or azoospermia.
References
- 1.Reijo R, Alagappan RK, Patrizio P, Page DC. Severe oligozoospermia resulting from deletions of azoospermia factor gene on Y chromosome. Lancet. 1996;347(9011):1290–1293. doi: 10.1016/S0140-6736(96)90938-1. [DOI] [PubMed] [Google Scholar]
- 2.Lange J, Skaletsky H, van Daalen SK, Embry SL, Korver CM, Brown LG, et al. Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell. 2009;138(5):855–869. doi: 10.1016/j.cell.2009.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Efstratiou A, George RC. Laboratory guidelines for the diagnosis of infections caused by Corynebacterium diphtheriae and C. ulcerans. World Health Organization. Commun Dis Public Health/PHLS. 1999;2(4):250–257. [PubMed] [Google Scholar]
- 4.Repping S, Skaletsky H, Lange J, Silber S, Van Der Veen F, Oates RD, et al. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am J Hum Genet. 2002;71(4):906–922. doi: 10.1086/342928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423(6942):825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- 6.Jungwirth A, Giwercman A, Tournaye H, Diemer T, Kopa Z, Dohle G, et al. European Association of Urology guidelines on male infertility: the 2012 update. Eur Urol. 2012;62(2):324–332. doi: 10.1016/j.eururo.2012.04.048. [DOI] [PubMed] [Google Scholar]
- 7.Muslumanoglu MH, Turgut M, Cilingir O, Can C, Ozyurek Y, Artan S. Role of the AZFd locus in spermatogenesis. Fertil Steril. 2005;84(2):519–522. doi: 10.1016/j.fertnstert.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 8.Cai ZM. [Y chromosome microdeletion and male infertility: past, present and future]. Zhonghua nan ke xue. Natl J Androl. 2010;16(5):387–394. [PubMed] [Google Scholar]
- 9.Demir B, Arikan II, Bozdag G, Esinler I, Karakoc Sokmensuer L, Gunalp S. ICSI outcome of patients with severe oligospermia vs. non-obstructive azoospermia. Clin Exp Obstet Gynecol. 2012;39(2):141–143. [PubMed] [Google Scholar]
- 10.Patrat C, Bienvenu T, Janny L, Faure AK, Fauque P, Aknin-Seifer I, et al. Clinical data and parenthood of 63 infertile and Y-microdeleted men. Fertil Steril. 2010;93(3):822–832. doi: 10.1016/j.fertnstert.2008.10.033. [DOI] [PubMed] [Google Scholar]
- 11.Xu W, Zhai Z, Huang K, Zhang N, Yuan Y, Shang Y, et al. A novel universal primer-multiplex-PCR method with sequencing gel electrophoresis analysis. PLoS ONE. 2012;7(1):e22900. doi: 10.1371/journal.pone.0022900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silber SJ, Disteche CM. In: Y chromosome infertility. Pagon RA, Adam MP, Bird TD, Dolan CR, Fong CT, Stephens K, editors. Seattle: GeneReviews; 1993. [Google Scholar]
- 13.Sachdeva K, Saxena R, Majumdar A, Chadda S, Verma IC. Use of ethnicity-specific sequence tag site markers for Y chromosome microdeletion studies. Genet Test Mol Biomark. 2011;15(6):451–459. doi: 10.1089/gtmb.2010.0159. [DOI] [PubMed] [Google Scholar]
- 14.Pinar A, Bozdemir N, Kocagoz T, Alacam R. Rapid detection of bacterial atypical pneumonia agents by multiplex PCR. Cent Eur J Public Health. 2004;12(1):3–5. doi: 10.1007/s10389-003-0003-4. [DOI] [PubMed] [Google Scholar]
- 15.Ding C, Cantor CR. A high-throughput gene expression analysis technique using competitive PCR and matrix-assisted laser desorption ionization time-of-flight MS. Proc Natl Acad Sci U S A. 2003;100(6):3059–3064. doi: 10.1073/pnas.0630494100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tettelin H, Radune D, Kasif S, Khouri H, Salzberg SL. Optimized multiplex PCR: efficiently closing a whole-genome shotgun sequencing project. Genomics. 1999;62(3):500–507. doi: 10.1006/geno.1999.6048. [DOI] [PubMed] [Google Scholar]
- 17.Inagaki S, Yamamoto Y, Doi Y, Takata T, Ishikawa T, Imabayashi K, et al. A new 39-plex analysis method for SNPs including 15 blood group loci. Forensic Sci Int. 2004;144(1):45–57. doi: 10.1016/j.forsciint.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Puzuka A, Pronina N, Grinfelde I, Erenpreiss J, Lejins V, Bars J, et al. Y chromosome–a tool in infertility studies of Latvian population. Genetika. 2011;47(3):394–400. [PubMed] [Google Scholar]
- 19.Akin H, Onay H, Turker E, Ozkinay F. Primary male infertility in Izmir/Turkey: a cytogenetic and molecular study of 187 infertile Turkish patients. J Assist Reprod Genet. 2011;28(5):419–423. doi: 10.1007/s10815-011-9542-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirfakhraie R, Mirzajani F, Kalantar SM, Montazeri M, Salsabili N, Pourmand GR, et al. High prevalence of AZFb microdeletion in Iranian patients with idiopathic non-obstructive azoospermia. Indian J Med Res. 2010;132:265–270. [PubMed] [Google Scholar]
- 21.Ferlin A, Arredi B, Speltra E, Cazzadore C, Selice R, Garolla A, et al. Molecular and clinical characterization of Y chromosome microdeletions in infertile men: a 10-year experience in Italy. J Clin Endocrinol Metab. 2007;92(3):762–770. doi: 10.1210/jc.2006-1981. [DOI] [PubMed] [Google Scholar]
- 22.Fu J, Li L, Lu G. Relationship between microdeletion on Y chromosome and patients with idiopathic azoospermia and severe oligozoospermia in the Chinese. Chin Med J. 2002;115(1):72–75. [PubMed] [Google Scholar]
- 23.Aknin-Seifer IE, Touraine RL, Lejeune H, Laurent JL, Lauras B, Levy R. A simple, low cost and non-invasive method for screening Y-chromosome microdeletions in infertile men. Hum Reprod. 2003;18(2):257–261. doi: 10.1093/humrep/deg067. [DOI] [PubMed] [Google Scholar]
- 24.Mirfakhraie R, Mirzajani F, Kalantar SM, Montazeri M, Salsabili N, Pourmand GR, et al. High prevalence of AZFb microdeletion in Iranian patients with idiopathic non-obstructive azoospermia. Indian J Med Res. 2011;132:265–270. [PubMed] [Google Scholar]
- 25.Fu L, Xiong DK, Ding XP, Li C, Zhang LY, Ding M, et al. Genetic screening for chromosomal abnormalities and Y chromosome microdeletions in Chinese infertile men. J Assist Reprod Genet. 2012;29(6):521–527. doi: 10.1007/s10815-012-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bu Y, Huang H, Wu HP, Zhang XD, Zhou GH, Cui YX, et al. Direct multiplex-PCR from whole blood for rapid detection of Y chromosome microdeletions. Zhonghua yi xue yi chuan xue za zhi = Zhonghua yixue yichuanxue zazhi. Chin J Med Genet. 2008;25(4):406–409. [PubMed] [Google Scholar]