Abstract
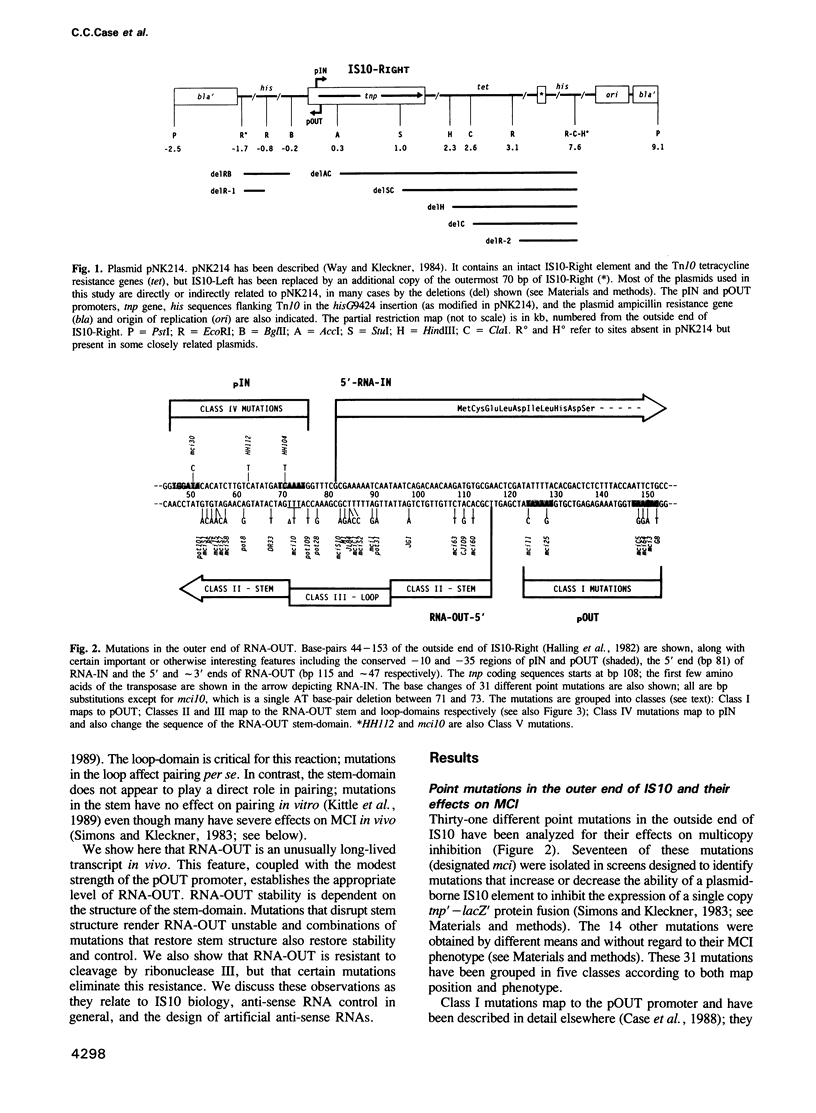
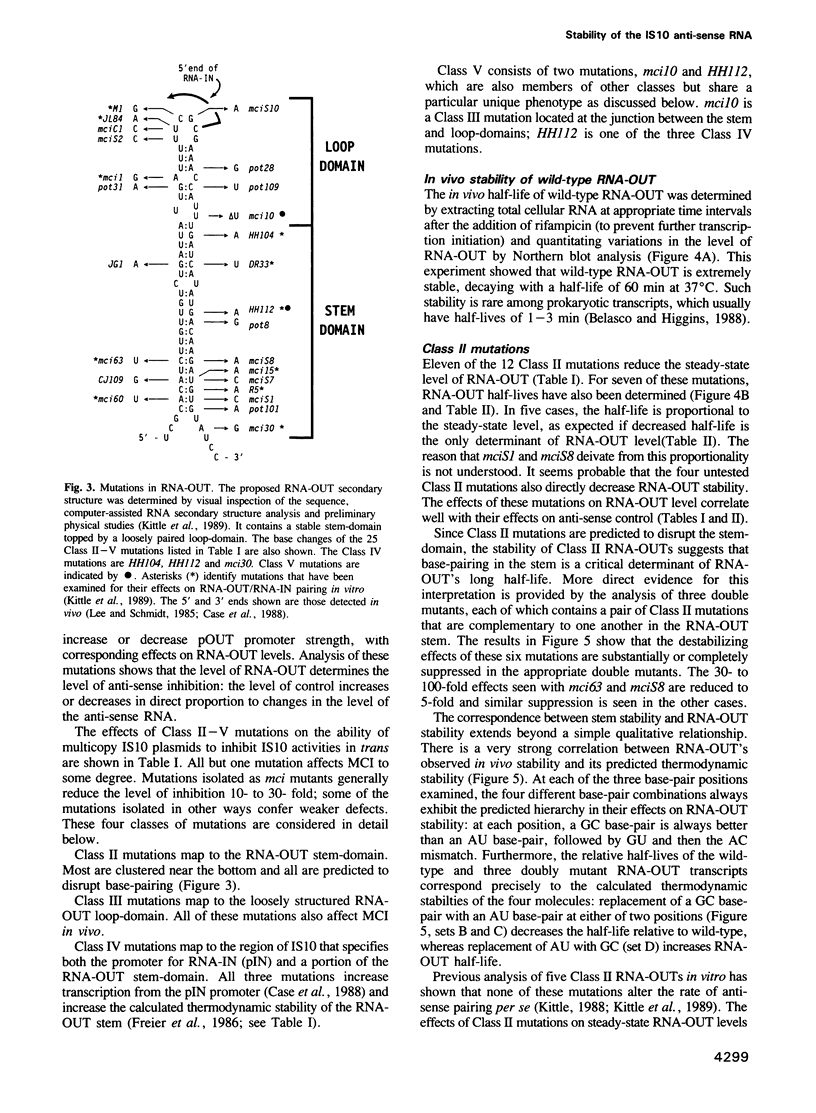
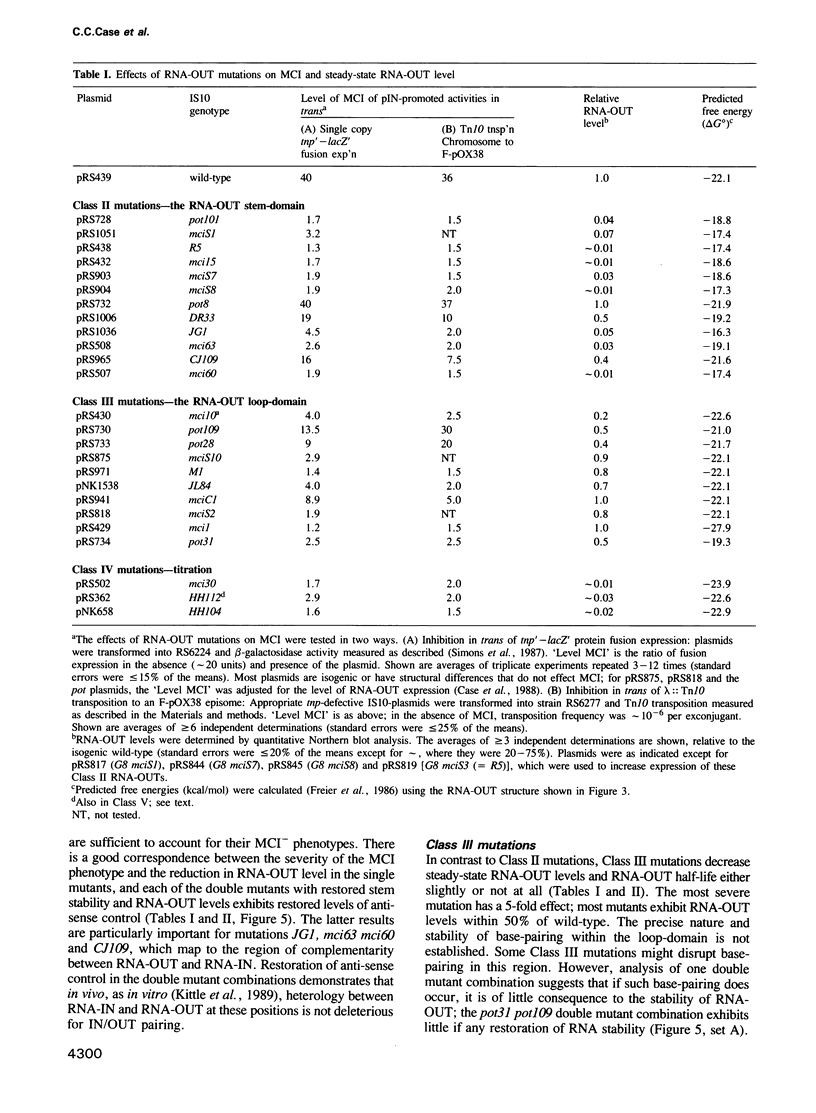
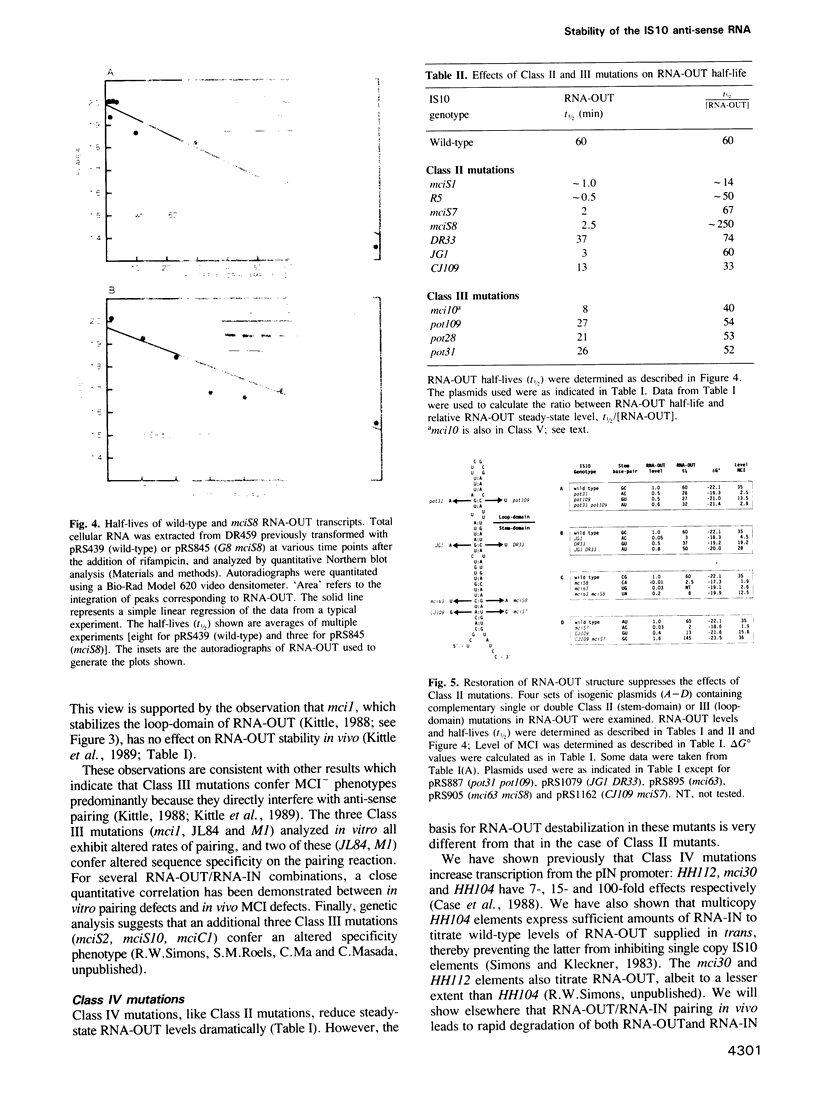
IS10 transposition is regulated by an approximately 70 nt anti-sense RNA, RNA-OUT. RNA-OUT folds into a duplex 'stem-domain' topped by a loosely paired 'loop-domain'. The loop-domain is critical for RNA-RNA pairing per se; pairing initiates by interaction of the RNA-OUT loop with the 5' end of the target mRNA. We show here that RNA-OUT is unusually stable in vivo (half-life 60 min) and that this stability is conferred by specific features of the RNA-OUT stem-domain. One critical feature is stable base-pairing: mutations that disrupt stem pairing destabilize RNA-OUT in vivo and abolish anti-sense control; combinations of mutations that restore pairing also restore both stability and control. We propose that the stem renders RNA-OUT resistant to 3' exoribonucleases. Other features of the stem-domain prevent this essential duplex from being an effective substrate for double-strand nucleases: two single base mutations disrupt antisense control by making RNA-OUT susceptible to RNase III. Mutations in the loop region have little effect on RNA-OUT stability. Implications for IS10 biology and the design of efficient anti-sense RNAs are discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Belasco J. G., Beatty J. T., Adams C. W., von Gabain A., Cohen S. N. Differential expression of photosynthesis genes in R. capsulata results from segmental differences in stability within the polycistronic rxcA transcript. Cell. 1985 Jan;40(1):171–181. doi: 10.1016/0092-8674(85)90320-4. [DOI] [PubMed] [Google Scholar]
- Belasco J. G., Higgins C. F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988 Dec 10;72(1-2):15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Belasco J. G., Nilsson G., von Gabain A., Cohen S. N. The stability of E. coli gene transcripts is dependent on determinants localized to specific mRNA segments. Cell. 1986 Jul 18;46(2):245–251. doi: 10.1016/0092-8674(86)90741-5. [DOI] [PubMed] [Google Scholar]
- Brawerman G. Determinants of messenger RNA stability. Cell. 1987 Jan 16;48(1):5–6. doi: 10.1016/0092-8674(87)90346-1. [DOI] [PubMed] [Google Scholar]
- Case C. C., Roels S. M., González J. E., Simons E. L., Simons R. W. Analysis of the promoters and transcripts involved in IS10 anti-sense RNA control. Gene. 1988 Dec 10;72(1-2):219–236. doi: 10.1016/0378-1119(88)90147-3. [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Beatty J. T., Cohen S. N., Belasco J. G. An intercistronic stem-loop structure functions as an mRNA decay terminator necessary but insufficient for puf mRNA stability. Cell. 1988 Feb 26;52(4):609–619. doi: 10.1016/0092-8674(88)90473-4. [DOI] [PubMed] [Google Scholar]
- Davis M. A., Simons R. W., Kleckner N. Tn10 protects itself at two levels from fortuitous activation by external promoters. Cell. 1985 Nov;43(1):379–387. doi: 10.1016/0092-8674(85)90043-1. [DOI] [PubMed] [Google Scholar]
- Donovan W. P., Kushner S. R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1986 Jan;83(1):120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Lundblad V., Hanley-Way S., Halling S. M., Kleckner N. Three Tn10-associated excision events: relationship to transposition and role of direct and inverted repeats. Cell. 1981 Jan;23(1):215–227. doi: 10.1016/0092-8674(81)90286-5. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarneros G. Retroregulation of bacteriophage lambda int gene expression. Curr Top Microbiol Immunol. 1988;136:1–19. doi: 10.1007/978-3-642-73115-0_1. [DOI] [PubMed] [Google Scholar]
- Halling S. M., Simons R. W., Way J. C., Walsh R. B., Kleckner N. DNA sequence organization of IS10-right of Tn10 and comparison with IS10-left. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2608–2612. doi: 10.1073/pnas.79.8.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M. N., Hayashi M. Cloned DNA sequences that determine mRNA stability of bacteriophage phi X174 in vivo are functional. Nucleic Acids Res. 1985 Aug 26;13(16):5937–5948. doi: 10.1093/nar/13.16.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M. N., Hayashi M., Müller U. R. Role for the J-F intercistronic region of bacteriophages phi X174 and G4 in stability of mRNA. J Virol. 1983 Oct;48(1):186–196. doi: 10.1128/jvi.48.1.186-196.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King T. C., Sirdeskmukh R., Schlessinger D. Nucleolytic processing of ribonucleic acid transcripts in procaryotes. Microbiol Rev. 1986 Dec;50(4):428–451. doi: 10.1128/mr.50.4.428-451.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Schmidt F. J. Characterization of the in vivo RNA product of the pOUT promoter of IS10R. J Bacteriol. 1985 Nov;164(2):556–562. doi: 10.1128/jb.164.2.556-562.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S. M., Wu T. H., Chiang C. H., Susskind M. M., McClure W. R. Control of gene expression in bacteriophage P22 by a small antisense RNA. I. Characterization in vitro of the Psar promoter and the sar RNA transcript. Genes Dev. 1987 Apr;1(2):197–203. doi: 10.1101/gad.1.2.197. [DOI] [PubMed] [Google Scholar]
- Lin-Chao S., Bremer H. Activities of the RNAI and RNAII promoters of plasmid pBR322. J Bacteriol. 1987 Mar;169(3):1217–1222. doi: 10.1128/jb.169.3.1217-1222.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Chao S., Bremer H. Effect of the bacterial growth rate on replication control of plasmid pBR322 in Escherichia coli. Mol Gen Genet. 1986 Apr;203(1):143–149. doi: 10.1007/BF00330395. [DOI] [PubMed] [Google Scholar]
- Maurer R., Meyer B., Ptashne M. Gene regulation at the right operator (OR) bacteriophage lambda. I. OR3 and autogenous negative control by repressor. J Mol Biol. 1980 May 15;139(2):147–161. doi: 10.1016/0022-2836(80)90302-2. [DOI] [PubMed] [Google Scholar]
- Morisato D., Way J. C., Kim H. J., Kleckner N. Tn10 transposase acts preferentially on nearby transposon ends in vivo. Cell. 1983 Mar;32(3):799–807. doi: 10.1016/0092-8674(83)90066-1. [DOI] [PubMed] [Google Scholar]
- Mott J. E., Galloway J. L., Platt T. Maturation of Escherichia coli tryptophan operon mRNA: evidence for 3' exonucleolytic processing after rho-dependent termination. EMBO J. 1985 Jul;4(7):1887–1891. doi: 10.1002/j.1460-2075.1985.tb03865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Higgins C. F. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell. 1987 Dec 24;51(6):1131–1143. doi: 10.1016/0092-8674(87)90599-x. [DOI] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Robinson E. C., Hiles I. D., Higgins C. F. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell. 1987 Jan 30;48(2):297–310. doi: 10.1016/0092-8674(87)90433-8. [DOI] [PubMed] [Google Scholar]
- Nordström K., Wagner E. G., Persson C., Blomberg P., Ohman M. Translational control by antisense RNA in control of plasmid replication. Gene. 1988 Dec 10;72(1-2):237–240. doi: 10.1016/0378-1119(88)90148-5. [DOI] [PubMed] [Google Scholar]
- Roberts T. M., Kacich R., Ptashne M. A general method for maximizing the expression of a cloned gene. Proc Natl Acad Sci U S A. 1979 Feb;76(2):760–764. doi: 10.1073/pnas.76.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R. W., Hoopes B. C., McClure W. R., Kleckner N. Three promoters near the termini of IS10: pIN, pOUT, and pIII. Cell. 1983 Sep;34(2):673–682. doi: 10.1016/0092-8674(83)90400-2. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Kleckner N. Biological regulation by antisense RNA in prokaryotes. Annu Rev Genet. 1988;22:567–600. doi: 10.1146/annurev.ge.22.120188.003031. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Kleckner N. Translational control of IS10 transposition. Cell. 1983 Sep;34(2):683–691. doi: 10.1016/0092-8674(83)90401-4. [DOI] [PubMed] [Google Scholar]
- Simons R. W. Naturally occurring antisense RNA control--a brief review. Gene. 1988 Dec 10;72(1-2):35–44. doi: 10.1016/0378-1119(88)90125-4. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Gruissem W. Control of plastid gene expression: 3' inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987 Dec 24;51(6):1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gauss P., Thermes C., Groebe D. R., Gayle M., Guild N., Stormo G., d'Aubenton-Carafa Y., Uhlenbeck O. C., Tinoco I., Jr CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1364–1368. doi: 10.1073/pnas.85.5.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- Way J. C., Kleckner N. Essential sites at transposon Tn 10 termini. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3452–3456. doi: 10.1073/pnas.81.11.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womble D. D., Sampathkumar P., Easton A. M., Luckow V. A., Rownd R. H. Transcription of the replication control region of the IncFII R-plasmid NR1 in vitro and in vivo. J Mol Biol. 1985 Feb 5;181(3):395–410. doi: 10.1016/0022-2836(85)90228-1. [DOI] [PubMed] [Google Scholar]
- Wong H. C., Chang S. Identification of a positive retroregulator that stabilizes mRNAs in bacteria. Proc Natl Acad Sci U S A. 1986 May;83(10):3233–3237. doi: 10.1073/pnas.83.10.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. H., Liao S. M., McClure W. R., Susskind M. M. Control of gene expression in bacteriophage P22 by a small antisense RNA. II. Characterization of mutants defective in repression. Genes Dev. 1987 Apr;1(2):204–212. doi: 10.1101/gad.1.2.204. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]



