Abstract
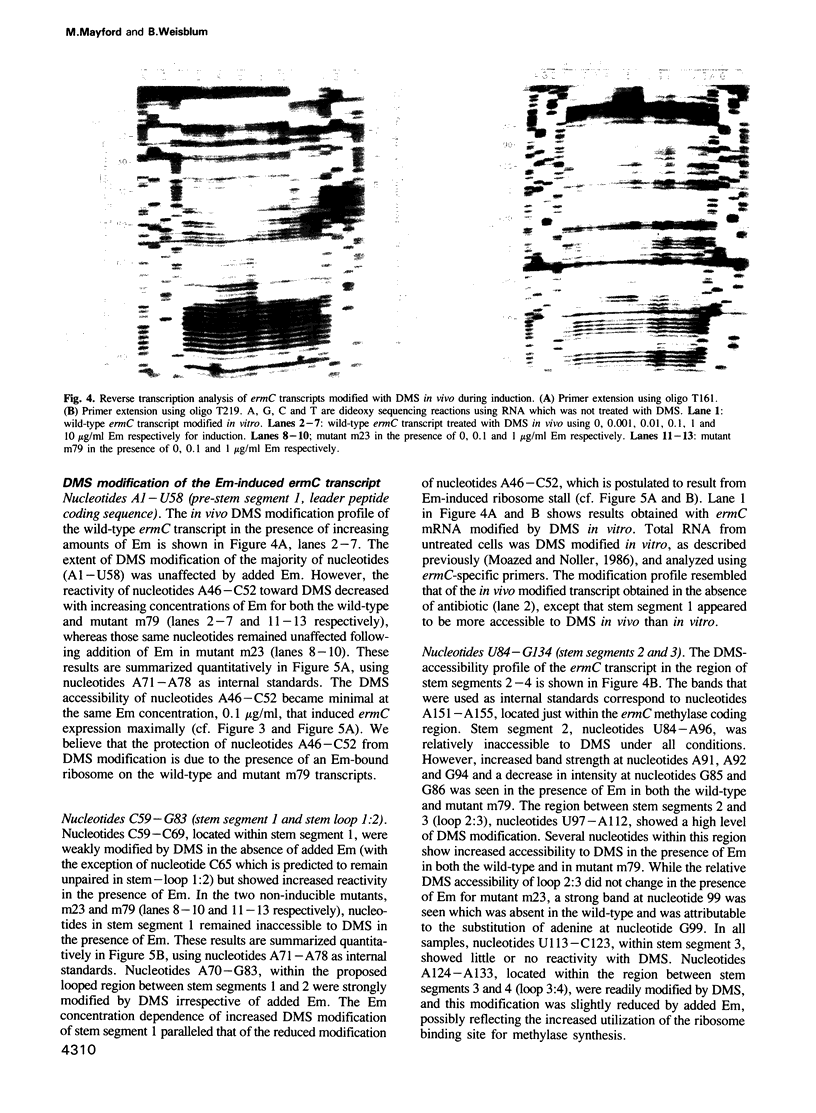
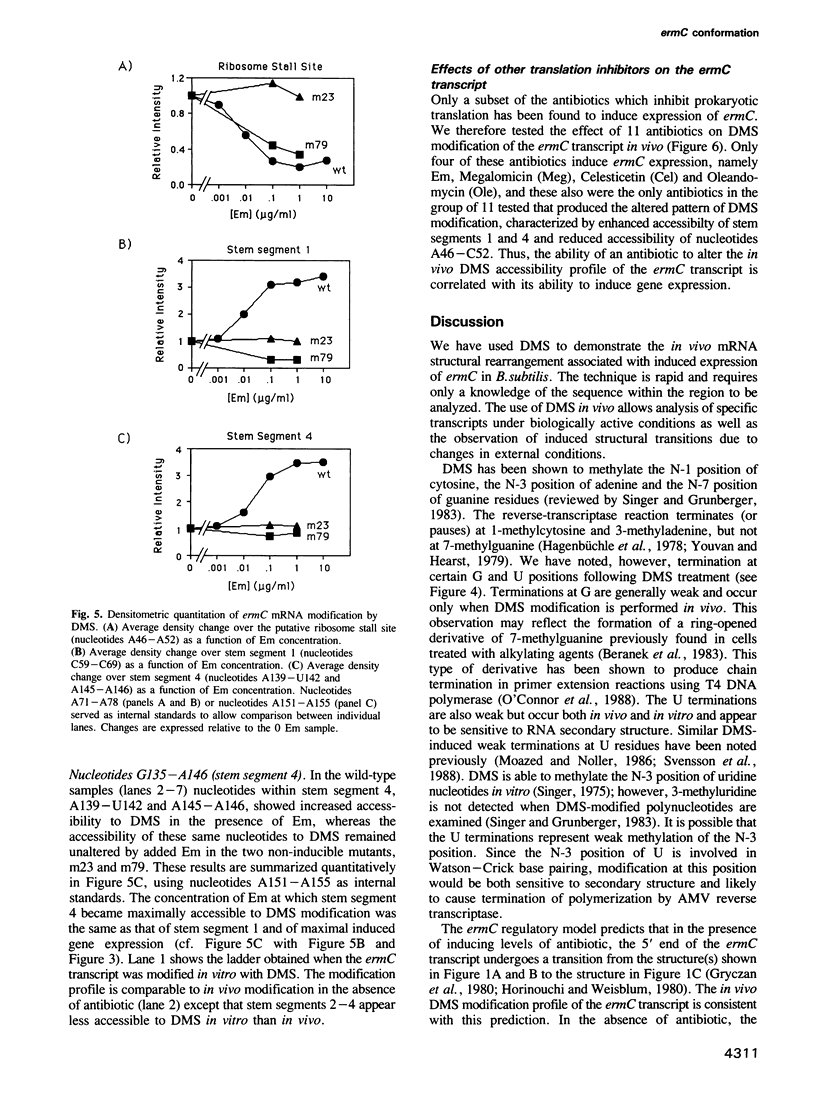
ermC is an inducible antibiotic resistance gene from Staphylococcus aureus, one of several whose expression is regulated at the level of mRNA secondary structure. During induction of ermC, the inhibition of a ribosome active in translation of a short leader peptide by low levels of antibiotic belonging to the macrolide-lincosamide-streptogramin b family is believed to cause a rearrangement in mRNA secondary structure. The resultant conformational isomerization unmasks the methylase ribosome binding site and initiator Met codon, causing increased translation of the ermC transcript. Expression of ermC can also be demonstrated in Bacillus subtilis carrying plasmid pE194. To probe the ermC transcript in vivo during induction, ermC was transferred to B. subtilis by transformation and the resultant transformants were treated with dimethyl sulfate which reacts with N-1 of adenine and N-3 of cytosine residues in a manner that is sensitive to secondary structure. The bases modified in vivo were detected by primer extension with reverse transcriptase using total cellular RNA as template and a complementary ermC-specific oligonucleotide as primer. Physical evidence was obtained for the secondary structural rearrangements predicted by the ermC regulatory model. Additionally, physical evidence was obtained demonstrating that during induction, the stalled ribosome protects codons 9 and 10 of the leader peptide from modification by dimethyl sulfate, in agreement with genetic data obtained previously that identified the integrity of codons 5-9 as critical for induction of ermC by erythromycin.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altuvia S., Locker-Giladi H., Koby S., Ben-Nun O., Oppenheim A. B. RNase III stimulates the translation of the cIII gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6511–6515. doi: 10.1073/pnas.84.18.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer D. H., Dubnau D. Induced mRNA stability in Bacillus subtilis. Proc Natl Acad Sci U S A. 1987 Jan;84(2):498–502. doi: 10.1073/pnas.84.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker P. B., Ruppert S., Schütz G. Genomic footprinting reveals cell type-specific DNA binding of ubiquitous factors. Cell. 1987 Nov 6;51(3):435–443. doi: 10.1016/0092-8674(87)90639-8. [DOI] [PubMed] [Google Scholar]
- Beranek D. T., Weis C. C., Evans F. E., Chetsanga C. J., Kadlubar F. F. Identification of N5-methyl-N5-formyl-2,5,6-triamino-4-hydroxypyrimidine as a major adduct in rat liver DNA after treatment with the carcinogens, N,N-dimethylnitrosamine or 1,2-dimethylhydrazine. Biochem Biophys Res Commun. 1983 Jan 27;110(2):625–631. doi: 10.1016/0006-291x(83)91195-6. [DOI] [PubMed] [Google Scholar]
- Dubnau D., Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J Mol Biol. 1971 Mar 14;56(2):209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- Dubnau D. Induction of ermC requires translation of the leader peptide. EMBO J. 1985 Feb;4(2):533–537. doi: 10.1002/j.1460-2075.1985.tb03661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Grandi G., Hahn J., Grandi R., Dubnau D. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res. 1980 Dec 20;8(24):6081–6097. doi: 10.1093/nar/8.24.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbüchle O., Santer M., Steitz J. A., Mans R. J. Conservation of the primary structure at the 3' end of 18S rRNA from eucaryotic cells. Cell. 1978 Mar;13(3):551–563. doi: 10.1016/0092-8674(78)90328-8. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol. 1982 May;150(2):804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7079–7083. doi: 10.1073/pnas.77.12.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T., Cech T. R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci U S A. 1985 Feb;82(3):648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C. W., Cantor C. R. Structure of ribosome-bound messenger RNA as revealed by enzymatic accessibility studies. J Mol Biol. 1985 Jan 20;181(2):241–251. doi: 10.1016/0022-2836(85)90088-9. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mayford M., Weisblum B. Messenger RNA from Staphylococcus aureus that specifies macrolide-lincosamide-streptogramin resistance. Demonstration of its conformations and of the leader peptide it encodes. J Mol Biol. 1985 Oct 20;185(4):769–780. doi: 10.1016/0022-2836(85)90061-0. [DOI] [PubMed] [Google Scholar]
- Mayford M., Weisblum B. ermC leader peptide. Amino acid sequence critical for induction by translational attenuation. J Mol Biol. 1989 Mar 5;206(1):69–79. doi: 10.1016/0022-2836(89)90524-x. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986 Dec 26;47(6):985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H. F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986 Feb 5;187(3):399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Müllner E. W., Kühn L. C. A stem-loop in the 3' untranslated region mediates iron-dependent regulation of transferrin receptor mRNA stability in the cytoplasm. Cell. 1988 Jun 3;53(5):815–825. doi: 10.1016/0092-8674(88)90098-0. [DOI] [PubMed] [Google Scholar]
- Narayanan C. S., Dubnau D. An in vitro study of the translational attenuation model of ermC regulation. J Biol Chem. 1987 Feb 5;262(4):1756–1765. [PubMed] [Google Scholar]
- Narayanan C. S., Dubnau D. Demonstration of erythromycin-dependent stalling of ribosomes on the ermC leader transcript. J Biol Chem. 1987 Feb 5;262(4):1766–1771. [PubMed] [Google Scholar]
- Narayanan C. S., Dubnau D. Evidence for the translational attenuation model: ribosome-binding studies and structural analysis with an in vitro run-off transcript of ermC. Nucleic Acids Res. 1985 Oct 25;13(20):7307–7326. doi: 10.1093/nar/13.20.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick H., Gilbert W. Detection in vivo of protein-DNA interactions within the lac operon of Escherichia coli. 1985 Feb 28-Mar 6Nature. 313(6005):795–798. doi: 10.1038/313795a0. [DOI] [PubMed] [Google Scholar]
- O'Connor T. R., Boiteux S., Laval J. Ring-opened 7-methylguanine residues in DNA are a block to in vitro DNA synthesis. Nucleic Acids Res. 1988 Jul 11;16(13):5879–5894. doi: 10.1093/nar/16.13.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler P., Weisblum B. Erythromycin-induced ribosome stall in the ermA leader: a barricade to 5'-to-3' nucleolytic cleavage of the ermA transcript. J Bacteriol. 1989 Dec;171(12):6680–6688. doi: 10.1128/jb.171.12.6680-6688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler P., Weisblum B. Erythromycin-induced stabilization of ermA messenger RNA in Staphylococcus aureus and Bacillus subtilis. J Mol Biol. 1988 Oct 20;203(4):905–915. doi: 10.1016/0022-2836(88)90116-7. [DOI] [PubMed] [Google Scholar]
- Shivakumar A. G., Hahn J., Dubnau D. Studies on the synthesis of plasmid-coded proteins and their control in Bacillus subtilis minicells. Plasmid. 1979 Apr;2(2):279–289. doi: 10.1016/0147-619x(79)90046-5. [DOI] [PubMed] [Google Scholar]
- Singer B. Methylation and ethylation of uridylic acid and thymidylic acid. Reactivity of the ring and phosphate as a function of pH and alkyl group. Biochemistry. 1975 Oct 7;14(20):4353–4357. doi: 10.1021/bi00691a001. [DOI] [PubMed] [Google Scholar]
- Stern S., Moazed D., Noller H. F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Svensson P., Changchien L. M., Craven G. R., Noller H. F. Interaction of ribosomal proteins, S6, S8, S15 and S18 with the central domain of 16 S ribosomal RNA. J Mol Biol. 1988 Mar 20;200(2):301–308. doi: 10.1016/0022-2836(88)90242-2. [DOI] [PubMed] [Google Scholar]
- Youvan D. C., Hearst J. E. Reverse transcriptase pauses at N2-methylguanine during in vitro transcription of Escherichia coli 16S ribosomal RNA. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3751–3754. doi: 10.1073/pnas.76.8.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]





