Abstract
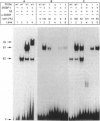
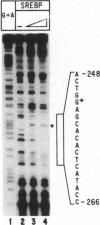
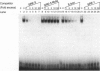
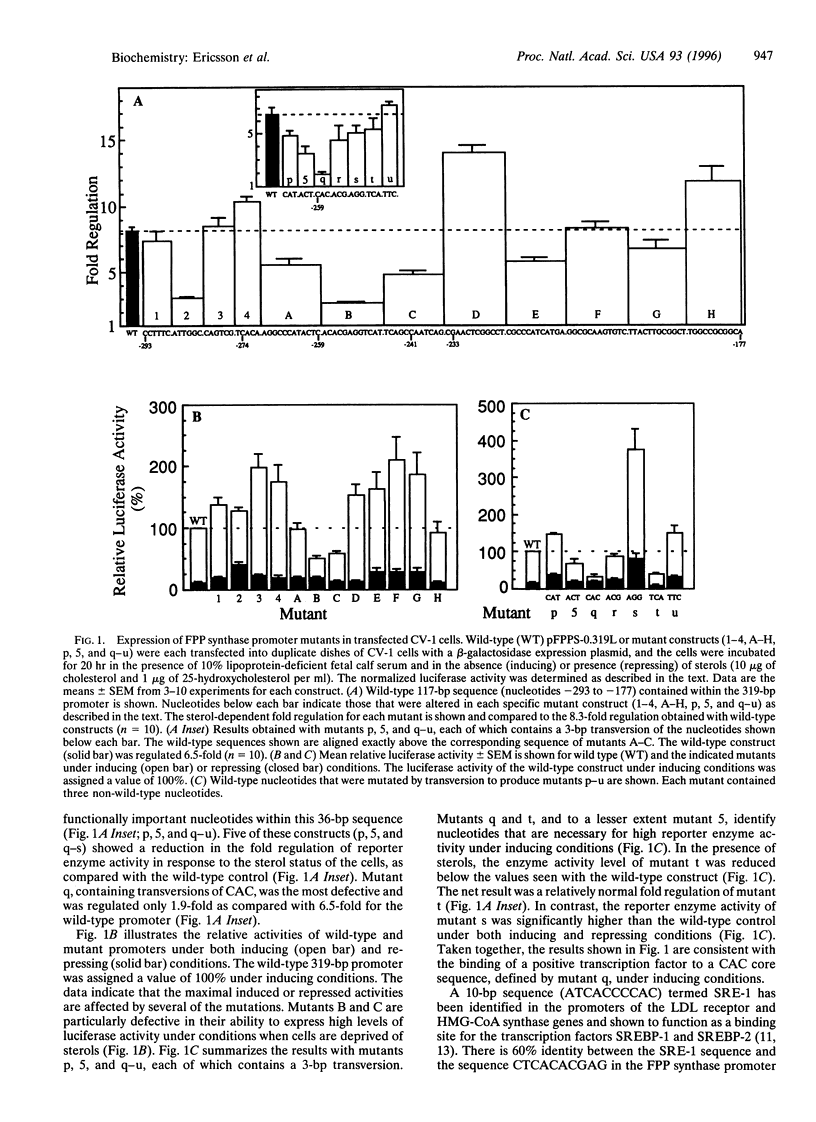
Sterol-regulated transcription of the gene for rat farnesyl diphosphate (FPP) synthase (geranyl-diphosphate:isopentenyl-diphosphate geranyltranstransferase, EC 2.5.1.10) is dependent in part on the binding of the ubiquitous transcription factor NF-Y to a 6-bp element within the proximal promoter. Current studies identify a second element in this promoter that is also required for sterol-regulated transcription in vivo. Mutation of three nucleotides (CAC) within this element blocks the 8-fold induction of FPP synthase promoter-reporter genes that normally occurs when the transfected cells are incubated in medium deprived of sterols. Gel mobility-shift assays demonstrate that the transcriptionally active 68-kDa fragment of the sterol regulatory element (SRE-1)-binding protein assays (SREBP-1) binds to an oligonucleotide containing the wild-type sequence but not to an oligonucleotide in which the CAC has been mutated. DNase 1 protection pattern (footprint) analysis indicates that SREBP-1 binds to nucleotides that include the CAC. Both the in vivo and in vitro assays are affected by mutagenesis of nucleotides adjacent to the CAC. Coexpression of SREBP with a wild-type FPP synthase promoter-reporter gene in CV-1 cells results in very high levels of reporter activity that is sterol-independent. In contrast, the reporter activity remained low when the promoter contained a mutation in the CAC trinucleotide. We conclude that sterol-regulated transcription of FPP synthase is controlled in part by the interaction of SREBP with a binding site that we have termed SRE-3. Identification of this element may prove useful in the identification of other genes that are both regulated by SREBP and involved in lipid biosynthesis.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambroziak J., Henry S. A. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J Biol Chem. 1994 May 27;269(21):15344–15349. [PubMed] [Google Scholar]
- Briggs M. R., Yokoyama C., Wang X., Brown M. S., Goldstein J. L. Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. I. Identification of the protein and delineation of its target nucleotide sequence. J Biol Chem. 1993 Jul 5;268(19):14490–14496. [PubMed] [Google Scholar]
- Clarke C. F., Tanaka R. D., Svenson K., Wamsley M., Fogelman A. M., Edwards P. A. Molecular cloning and sequence of a cholesterol-repressible enzyme related to prenyltransferase in the isoprene biosynthetic pathway. Mol Cell Biol. 1987 Sep;7(9):3138–3146. doi: 10.1128/mcb.7.9.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein J. L., Brown M. S. Regulation of the mevalonate pathway. Nature. 1990 Feb 1;343(6257):425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- Hua X., Yokoyama C., Wu J., Briggs M. R., Brown M. S., Goldstein J. L., Wang X. SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S. M., Ericsson J., Osborne T. F., Edwards P. A. NF-Y has a novel role in sterol-dependent transcription of two cholesterogenic genes. J Biol Chem. 1995 Sep 15;270(37):21445–21448. doi: 10.1074/jbc.270.37.21445. [DOI] [PubMed] [Google Scholar]
- Kim J. B., Spotts G. D., Halvorsen Y. D., Shih H. M., Ellenberger T., Towle H. C., Spiegelman B. M. Dual DNA binding specificity of ADD1/SREBP1 controlled by a single amino acid in the basic helix-loop-helix domain. Mol Cell Biol. 1995 May;15(5):2582–2588. doi: 10.1128/mcb.15.5.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocarek T. A., Schuetz E. G., Guzelian P. S. Regulation of phenobarbital-inducible cytochrome P450 2B1/2 mRNA by lovastatin and oxysterols in primary cultures of adult rat hepatocytes. Toxicol Appl Pharmacol. 1993 Jun;120(2):298–307. doi: 10.1006/taap.1993.1115. [DOI] [PubMed] [Google Scholar]
- Maltese W. A. Posttranslational modification of proteins by isoprenoids in mammalian cells. FASEB J. 1990 Dec;4(15):3319–3328. doi: 10.1096/fasebj.4.15.2123808. [DOI] [PubMed] [Google Scholar]
- Osborne T. F., Bennett M., Rhee K. Red 25, a protein that binds specifically to the sterol regulatory region in the promoter for 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J Biol Chem. 1992 Sep 15;267(26):18973–18982. [PubMed] [Google Scholar]
- Osborne T. F. Single nucleotide resolution of sterol regulatory region in promoter for 3-hydroxy-3-methylglutaryl coenzyme A reductase. J Biol Chem. 1991 Jul 25;266(21):13947–13951. [PubMed] [Google Scholar]
- Puga A., Raychaudhuri B., Nebert D. W. Transcriptional derepression of the murine Cyp1a-1 gene by mevinolin. FASEB J. 1992 Jan 6;6(2):777–785. doi: 10.1096/fasebj.6.2.1311272. [DOI] [PubMed] [Google Scholar]
- Rosser D. S., Ashby M. N., Ellis J. L., Edwards P. A. Coordinate regulation of 3-hydroxy-3-methylglutaryl-coenzyme A synthase, 3-hydroxy-3-methylglutaryl-coenzyme A reductase, and prenyltransferase synthesis but not degradation in HepG2 cells. J Biol Chem. 1989 Jul 25;264(21):12653–12656. [PubMed] [Google Scholar]
- Sagami H., Morita Y., Ogura K. Purification and properties of geranylgeranyl-diphosphate synthase from bovine brain. J Biol Chem. 1994 Aug 12;269(32):20561–20566. [PubMed] [Google Scholar]
- Sanchez H. B., Yieh L., Osborne T. F. Cooperation by sterol regulatory element-binding protein and Sp1 in sterol regulation of low density lipoprotein receptor gene. J Biol Chem. 1995 Jan 20;270(3):1161–1169. doi: 10.1074/jbc.270.3.1161. [DOI] [PubMed] [Google Scholar]
- Schüller H. J., Hahn A., Tröster F., Schütz A., Schweizer E. Coordinate genetic control of yeast fatty acid synthase genes FAS1 and FAS2 by an upstream activation site common to genes involved in membrane lipid biosynthesis. EMBO J. 1992 Jan;11(1):107–114. doi: 10.1002/j.1460-2075.1992.tb05033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih H. M., Towle H. C. Definition of the carbohydrate response element of the rat S14 gene. Evidence for a common factor required for carbohydrate regulation of hepatic genes. J Biol Chem. 1992 Jul 5;267(19):13222–13228. [PubMed] [Google Scholar]
- Spear D. H., Ericsson J., Jackson S. M., Edwards P. A. Identification of a 6-base pair element involved in the sterol-mediated transcriptional regulation of farnesyl diphosphate synthase. J Biol Chem. 1994 Oct 7;269(40):25212–25218. [PubMed] [Google Scholar]
- Spear D. H., Kutsunai S. Y., Correll C. C., Edwards P. A. Molecular cloning and promoter analysis of the rat liver farnesyl diphosphate synthase gene. J Biol Chem. 1992 Jul 15;267(20):14462–14469. [PubMed] [Google Scholar]
- Tontonoz P., Kim J. B., Graves R. A., Spiegelman B. M. ADD1: a novel helix-loop-helix transcription factor associated with adipocyte determination and differentiation. Mol Cell Biol. 1993 Aug;13(8):4753–4759. doi: 10.1128/mcb.13.8.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Briggs M. R., Hua X., Yokoyama C., Goldstein J. L., Brown M. S. Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. II. Purification and characterization. J Biol Chem. 1993 Jul 5;268(19):14497–14504. [PubMed] [Google Scholar]
- Yokoyama C., Wang X., Briggs M. R., Admon A., Wu J., Hua X., Goldstein J. L., Brown M. S. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993 Oct 8;75(1):187–197. [PubMed] [Google Scholar]