Abstract
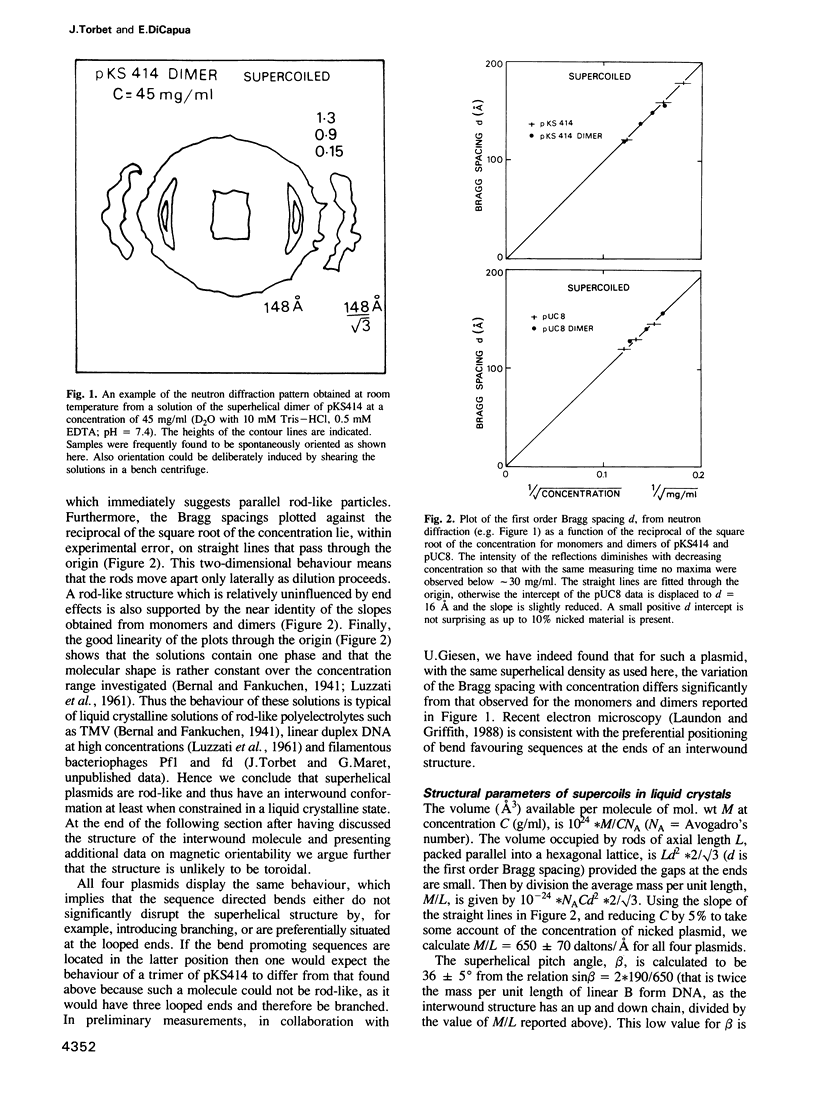
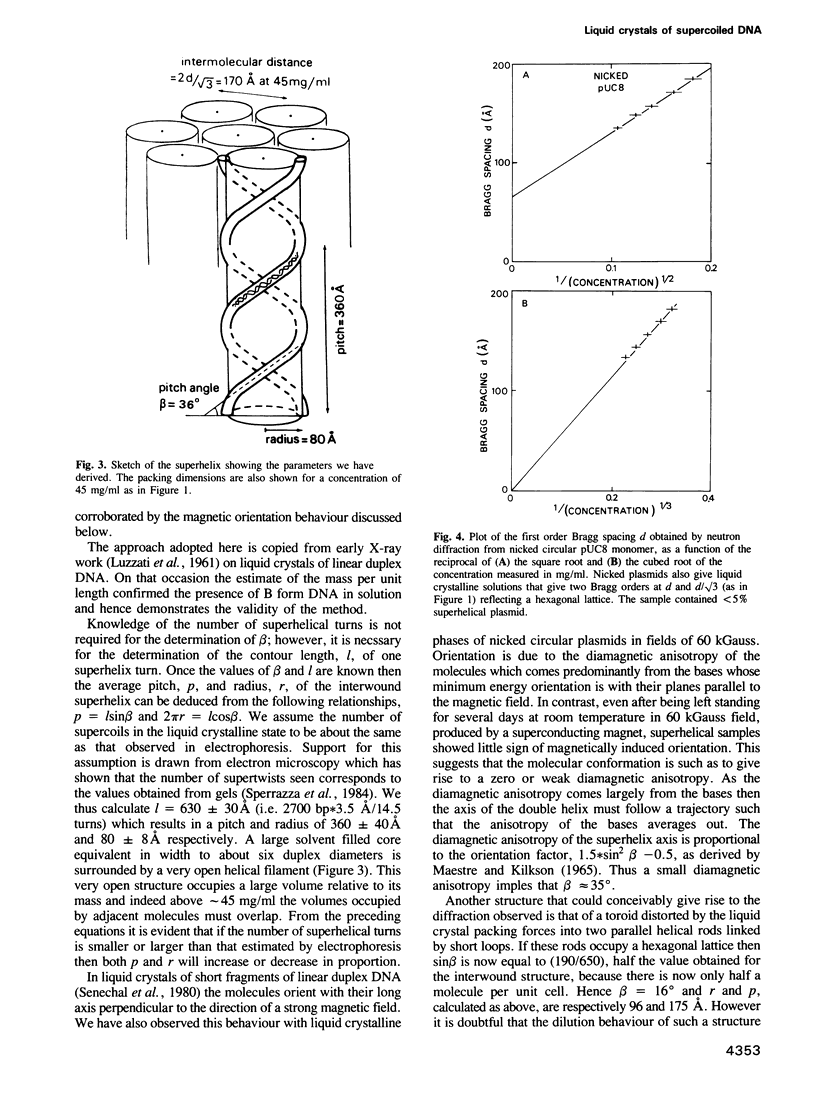
Two structures have been proposed for supercoiled DNA: it is idealized either as a toroidal ring or as a rod of two interwound duplex chains. The latter model is the most widely depicted but the evidence remains controversial. We have worked with monomers and dimers of two plasmids, pUC8 and pKS414, of similar size and natural superhelical density. pKS414 contains a bend promoting sequence whereas pUC8 does not. In concentrated solutions these plasmids form a partially ordered liquid crystalline phase which is found, using neutron diffraction, to consist of a hexagonally packed assembly of parallel rod-like particles. This shape strongly suggests an interwound conformation for which some structural parameters are deduced. The mass/unit length obtained by combining the area of the hexagonal lattice and the concentration is approximately 3.6 times that of linear DNA. This implies a shallow superhelical pitch angle approximately 36 degrees which, when combined with the known number of supercoil turns, yields the pitch approximately 360 A and radius approximately 80 A for the supercoil. Oriented X-ray fibre diffraction patterns at 92% relative humidity indicate a B type duplex structure. Nicked circular plasmids also form liquid crystals but their behaviour, as a function of concentration, differs from that of the superhelical plasmids.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson P., Bauer W. Supercoiling in closed circular DNA: dependence upon ion type and concentration. Biochemistry. 1978 Feb 21;17(4):594–601. doi: 10.1021/bi00597a006. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Banerjee A. K., He R., Walker J. K. New wrinkles on polynucleotide duplexes. J Biomol Struct Dyn. 1983 Oct;1(2):437–452. doi: 10.1080/07391102.1983.10507453. [DOI] [PubMed] [Google Scholar]
- Benham C. J., Brady G. W., Fein D. B. X-ray scattering from randomly oriented superhelices. Circular superhelical DNA. Biophys J. 1980 Mar;29(3):351–366. doi: 10.1016/S0006-3495(80)85139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliska J. B., Cozzarelli N. R. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol. 1987 Mar 20;194(2):205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
- Bourguignon M. F., Bourgaux P. Electron microscopic study of the tertiary structure of covalently closed polyoma virus DNA. Biochim Biophys Acta. 1968 Dec 17;169(2):476–487. doi: 10.1016/0005-2787(68)90056-7. [DOI] [PubMed] [Google Scholar]
- Brady G. W., Fein D. B., Lambertson H., Grassian V., Foos D., Benham C. J. X-ray scattering from the superhelix in circular DNA. Proc Natl Acad Sci U S A. 1983 Feb;80(3):741–744. doi: 10.1073/pnas.80.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady G. W., Fein D. B. X-ray diffraction studies of circular superhelical DNA at 300-10,000-A resolution. Nature. 1976 Nov 18;264(5583):231–234. doi: 10.1038/264231a0. [DOI] [PubMed] [Google Scholar]
- Brady G. W., Foos D., Benham C. J. Evidence for an interwound form for the superhelix in circular DNA. Biopolymers. 1984 Dec;23(12):2963–2966. doi: 10.1002/bip.360231219. [DOI] [PubMed] [Google Scholar]
- Brady G. W., Satkowski M., Foos D., Benham C. J. Environmental influences on DNA superhelicity. The effect of ionic strength on superhelix conformation in solution. J Mol Biol. 1987 May 5;195(1):185–191. doi: 10.1016/0022-2836(87)90335-4. [DOI] [PubMed] [Google Scholar]
- Brandes R., Kearns D. R. Magnetic ordering of DNA liquid crystals. Biochemistry. 1986 Oct 7;25(20):5890–5895. doi: 10.1021/bi00368a008. [DOI] [PubMed] [Google Scholar]
- Böttger M., Kuhn W. Sedimentation analysis of conformation changes of circular PM 2 DNA in relation to the ionic strength. Biochim Biophys Acta. 1971 Dec 30;254(3):407–411. doi: 10.1016/0005-2787(71)90872-0. [DOI] [PubMed] [Google Scholar]
- Campbell A. M. Conformational variation in superhelical deoxyribonucleic acid. Biochem J. 1978 Apr 1;171(1):281–283. doi: 10.1042/bj1710281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diekmann S., Wang J. C. On the sequence determinants and flexibility of the kinetoplast DNA fragment with abnormal gel electrophoretic mobilities. J Mol Biol. 1985 Nov 5;186(1):1–11. doi: 10.1016/0022-2836(85)90251-7. [DOI] [PubMed] [Google Scholar]
- Gray H. B., Jr Sedimentation coefficient of polyoma virus DNA. Biopolymers. 1967;5(10):1009–1019. doi: 10.1002/bip.1967.360051012. [DOI] [PubMed] [Google Scholar]
- Hsieh C. H., Griffith J. D. The terminus of SV40 DNA replication and transcription contains a sharp sequence-directed curve. Cell. 1988 Feb 26;52(4):535–544. doi: 10.1016/0092-8674(88)90466-7. [DOI] [PubMed] [Google Scholar]
- Hull R., Shepherd R. J. The structure of cauliflower mosaic virus genome. Virology. 1977 Jun 1;79(1):216–230. doi: 10.1016/0042-6822(77)90346-4. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUZZATI V., NICOLAIEFF A., MASSON F. [Structure of desoxyribonucleic acid in solution. Study by the diffusion of x-rays at small angles]. J Mol Biol. 1961 Apr;3:185–201. doi: 10.1016/s0022-2836(61)80045-4. [DOI] [PubMed] [Google Scholar]
- Laundon C. H., Griffith J. D. Curved helix segments can uniquely orient the topology of supertwisted DNA. Cell. 1988 Feb 26;52(4):545–549. doi: 10.1016/0092-8674(88)90467-9. [DOI] [PubMed] [Google Scholar]
- Livolant F. Cholesteric organization of DNA in vivo and in vitro. Eur J Cell Biol. 1984 Mar;33(2):300–311. [PubMed] [Google Scholar]
- Maestre M. F., Kilkson R. Intrinsic Birefringence of Multiple-Coiled DNA, Theory and Applications. Biophys J. 1965 May;5(3):275–287. doi: 10.1016/s0006-3495(65)86716-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M., Thomas C. A., Jr The P22 bacteriophage DNA molecule. II. Circular intracellular forms. J Mol Biol. 1968 Oct 14;37(1):41–61. doi: 10.1016/0022-2836(68)90072-7. [DOI] [PubMed] [Google Scholar]
- Ryder K., Silver S., DeLucia A. L., Fanning E., Tegtmeyer P. An altered DNA conformation in origin region I is a determinant for the binding of SV40 large T antigen. Cell. 1986 Mar 14;44(5):719–725. doi: 10.1016/0092-8674(86)90838-x. [DOI] [PubMed] [Google Scholar]
- Shure M., Vinograd J. The number of superhelical turns in native virion SV40 DNA and minicol DNA determined by the band counting method. Cell. 1976 Jun;8(2):215–226. doi: 10.1016/0092-8674(76)90005-2. [DOI] [PubMed] [Google Scholar]
- Spengler S. J., Stasiak A., Cozzarelli N. R. The stereostructure of knots and catenanes produced by phage lambda integrative recombination: implications for mechanism and DNA structure. Cell. 1985 Aug;42(1):325–334. doi: 10.1016/s0092-8674(85)80128-8. [DOI] [PubMed] [Google Scholar]
- Sperrazza J. M., Register J. C., 3rd, Griffith J. Electron microscopy can be used to measure DNA supertwisting. Gene. 1984 Nov;31(1-3):17–22. doi: 10.1016/0378-1119(84)90190-2. [DOI] [PubMed] [Google Scholar]
- Stenzel T. T., Patel P., Bastia D. The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell. 1987 Jun 5;49(5):709–717. doi: 10.1016/0092-8674(87)90547-2. [DOI] [PubMed] [Google Scholar]
- Strzelecka T. E., Davidson M. W., Rill R. L. Multiple liquid crystal phases of DNA at high concentrations. Nature. 1988 Feb 4;331(6155):457–460. doi: 10.1038/331457a0. [DOI] [PubMed] [Google Scholar]
- Wu H. Y., Shyy S. H., Wang J. C., Liu L. F. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988 May 6;53(3):433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Direct evidence for DNA bending at the lambda replication origin. Science. 1987 Apr 24;236(4800):416–422. doi: 10.1126/science.2951850. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Pheiffer B. H. Helical parameters of DNA do not change when DNA fibers are wetted: X-ray diffraction study. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2703–2707. doi: 10.1073/pnas.76.6.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]




