Abstract
One major challenge in nanomedicine is how to selectively deliver nanoparticles to diseased tissues. Nanoparticle delivery system requires targeting for specific delivery to pathogenic sites when enhanced permeability and retention (EPR) is not suitable or inefficient. Functionalizing nanoparticles is a widely-used technique that allows for conjugation with targeting ligands, which possess inherent ability to direct selective binding to cell types or states and, therefore, confer “smartness” to nanoparticles. This review illustrates methods of ligand-nanoparticle functionalization, provides a cross-section of various ligand classes, including small molecules, peptides, antibodies, engineered proteins, or nucleic acid aptamers, and discusses some unconventional approaches currently under investigation.
Introduction
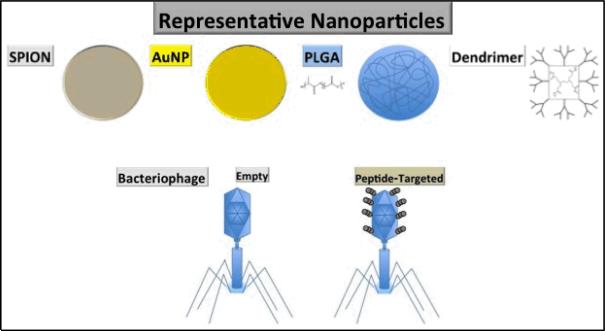
Research aimed at the better translation of benchside innovations to novel and more effective treatments in the clinic have increasingly turned to targeted nanoparticle platforms. Nanoparticles are an attractive choice because they can carry chemotherapeutic warheads, serve as imaging agents, and act as the active therapeutic agent themselves (e.g. the magnetic-induced hyperthermia using superparamagnetic iron oxide nanoparticles), as well as many more applications whose mention is beyond the scope of this article (Figure 1).
Figure 1.
Frequently utilized nanoparticles.
The ideal nanoparticle-based therapeutics should have specific targeting to pathologic tissues, which minimizes or avoids off-target effects of the active therapeutic agents on healthy tissues. Much research has conjugated targeting ligands specific to cell surface components that are unique to, or upregulated in, dysplastic and pathologic tissues to nanoparticle surfaces. These targeting ligands fall into several general classes: small molecules, polypeptide-based peptides, protein domains, antibodies, and nucleic acid-based aptamers [1]. At times, ligands from multiple classes (chimeras), or multiple ligands within the same class but with different targets (multi-valency and multi-specificity) have been implemented to enhance nanoparticle targeting. Each ligand class has particular advantages, disadvantages, unique attributes, and conjugation strategies that will be discussed further in the following sections.
The scope of this review covers advances made in the nanoparticle targeting field over the last four years. Tables bearing publication information regarding various targeting ligands, nanoparticles, and conjugation chemistries are provided to guide discussion of current approaches. The purpose of this review is to provide the reader with an overview of current nanoparticle targeting research and distill this information into an accessible form conducive to the design of desired targeting approaches.
Chemistry of Conjugation
When utilizing nanoparticles for targeted delivery with any of the aforementioned ligands, it is often necessary to chemically modify the surface of the nanoparticles with an appropriate chemistry to introduce reactive moieties, thereby providing functional groups that can be conjugated to a targeting ligand of choice. It is important that the selective ligand has a functional group that can be used for conjugation as well. The conjugation of a targeting ligand to chemically modified nanoparticles will allow for selective delivery of the desired nanoparticle therapeutics.
Most of the conjugation chemistries that are used to modify nanoparticles are covalent. Some of the most prevalent covalent reactions that are utilized in conjugating nanoparticles to targeting ligands include chemical reactions that use carbonyl reactive groups (i.e., carbonyl reacts with hydrazide or alkyoxyamine to form hydrazone or oxime bond), amine reactive groups (i.e., amine reacts with activated carboxylate or imidoester to form amide or amidine bond), sulfhydryl reactive groups (thiol reacts with maleimide, haloacetyl, pyridyl disulfide or gold surface, to form thioester, disulfide, or gold-thiol bond), and a type of orthogonal reaction known as Click Chemistry (i.e., azide reacts with phosphine or alkyne to form amide bond or triazole ring) (Tables 1, 2).
Table 1.
Conjugation reactions, linkages formed, and their stability*.
| Type of Conjugation | Linkage | Stability under physiological conditions |
|---|---|---|
| Covalent | ||
| NH2/COOH | Amide bond | Stable |
| Thiol/Maleimide | Thio-ether bond | Stable |
| Thiol/Thiol | Disulfide bond | Cleaved under reducing conditions |
| Hydrazide/Aldehyde | Hydrazone | Acid labile |
| Gold/Thiol | Gold-thiol bond | Stable |
| Click Chemistry i.e. Azide/Alkyne | Triazole ring | Stable |
| Noncovalent | ||
| Biotin/(Strept)avidin | Non-covalent Almost irreversible | Stable |
Hermanson, Greg. Bioconjugate Techniques. San Diego: Academic Press Inc., 1996.
Table 2.
Covalent conjugation reactions represented by a schematic.
| Type of Covalent Conjugation | Diagram |
|---|---|
| Hydrazide-Aldehyde |
|
| Amine-Carboxyl |
|
| Thiol-Maleimide |
|
| Thiol-Thiol |
|
| Gold-Thiol |
|
| Click Chemistry |
|
In addition to the many covalent reactions that are used to conjugate nanoparticles to targeting ligands, there is one non-covalent interaction that is commonly used as well. This is the interaction between (strept)avidin and biotin, the strongest known noncovalent interaction with a Kd of (10−14−10−15 M). With its almost irreversible binding, this noncovalent interaction can be readily used to conjugate nanoparticles to targeting ligands.
A basic schematic of these covalent conjugation chemistries and their reactions with each other are listed in Table 2, below. The applications of these conjugation reactions can be seen in subsequent sections, which further describe the use of specific targeting ligand classes on nanoparticles. While the majority of nanoparticle modifications involve the chemistries described in Table 2, other chemistries allow specific release of ligand or drug from the nanoparticle upon internalization via intracellular physiological properties, such as acidic pH, redox sensitivity, protease digestion, for example. While the pH is most commonly taken into account, redox and protease sensitivity should be considered as well.
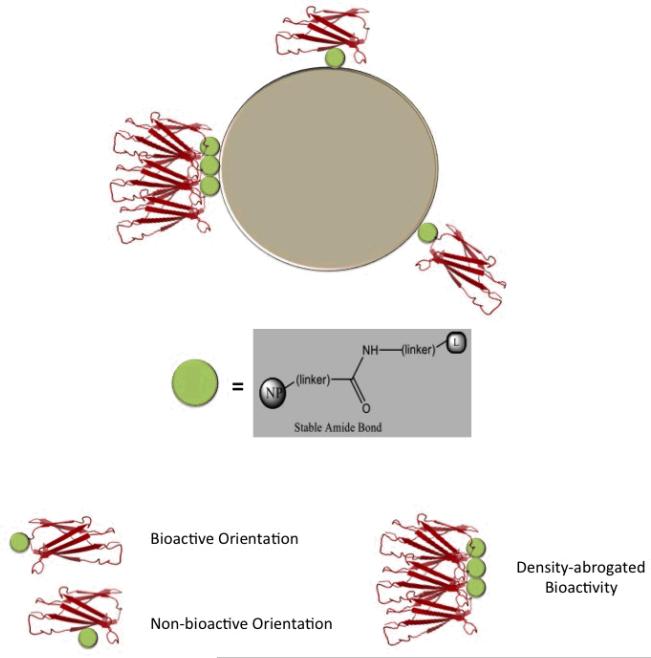
Not all chemistries ensure ligand directed coupling with correct orientation and desired surface density. Conjugation may yield stochastic ligand densities and spatial orientations (random coupling) (Figure 2). Chemistries exist, however, to better control the density and orientation of ligand conjugation via directed coupling. Incorporation of unnatural amino acids into protein-based targeting ligands can site specifically introduce a residue with a desired functional group, by the translational system under in vitro or in vivo conditions [2-4]. Such functional groups, typically not present in natural amino acids, are ideal for site-specific conjugation with nanoparticles or other moieties under bioorthogonal conditions. Suicide substrates, molecules that can covalently tether to specific target proteins, can also direct ligand coupling. The stable and irreversible covalent bond formed between the suicide substrate and the targeting ligand, can allow for the site-specific immobilization of the ligand, facilitating inhibition and labeling while serving as a molecular probe for the target [5-8].
Figure 2. Random coupling can result in various targeting ligand orientations and densities on nanoparticle surfaces.
The targeting ligand is depicted by a protein domain (in red), linked with a stable amide bond (green sphere). Bioactive and non-bioactive orientations, and density-abrogated bioactivity are indicated.
Targeting with Small Molecules
One of the more prevalent targeting ligands conjugated to nanoparticles are small molecules. The major advantages of using a small molecule as targeting ligand is its stability, ease of conjugation with nanoparticles, and the potential low cost, assuming it can be chemically synthesized with high yield. However, there is no systematic approach to develop such ligands, and most small molecule targeting ligands do not bind cell surface receptors with high specificity and affinity. Biotin, also known as vitamin H, has been widely used for facile conjugation with nanoparticles coated with (strept)avidin for in vitro applications. This conjugation method exploits the extremely high affinity (10−14−10−15 M) between biotin and (strept)avidin. Clinical applications of this conjugation system are limited, however, due to the bacterial origin of strept(avidin) and consequent immunogenicity.
Vitamin B9 (folic acid) is a small molecule targeting ligand that has been intensively investigated for clinic applications. Folic acid is a high affinity ligand of endogenous folate receptor, which is frequently up-regulated in many types of human cancers. To date, a wide variety of therapeutic agents have been linked to folic acid for tumor-selective drug delivery, including protein toxins and therapeutics, chemotherapeutic agents, gene therapy vectors, oligonucleotides, radioimaging and radiotherapeutic agents, MRI contrast agents, and drug-loaded liposomes and nanoparticles. It has been demonstrated that nanoparticles or liposomes conjugated with folic acid can be actively internalized via receptor-mediated endocytosis and effectively directed to folate receptor-positive cancer cells [9, 10].
In principle, small molecules that tightly and specifically bind to the extracellular domain of transmembrane cancer biomarkers can be used in ways similar to folate. Sigma receptors are upregulated in many cancer cells. Benzamides (anisamide, in particular), are demonstrated sigma receptor ligands and, therefore, can target nanoparticles to sigma receptor-positive tissues [11, 12].
Carbohydrates, which interact weakly with some cell surface receptors, can also serve as nanoparticle small molecule targeting ligands. Carbohydrates permit nanoparticle glycotargeting, which is based on endogenous lectin interactions with carbohydrates. A disadvantage of this targeting method is that glycotargeting often requires multiple interacting carbohydrates to achieve strong enough binding strength. One known example uses galactose or galactose-mimics as ligands to asialoglycoprotein receptor, an endocytotic cell surface lectin receptor highly expressed on hepatocyte surfaces. DC-SIGN is a C-type lectin receptor preferentially expressed by dendritic cells. Lex and ManLAM carbohydrates, for example, can be used to enhance the binding and uptake of the nanoparticles by dendritic cells, although their targeting features are not as effective as DC-SIGN specific antibodies that are presumably more specific and potent [13].
In general, most target receptors do not have naturally occurring small molecule ligands that tightly and specifically interact with their extracellular domains. One attractive approach to the development of small molecule targeting ligands involves using the natural substrate, a small molecule inhibitor, or the transition state analog of the target receptors as the lead compound for small molecule targeting ligand development. For example, small-molecule PSMA targeting molecules have been developed based on substrate N-acetyl-L-Asp-L-Glu-like analogues in which the central linkage between Asp and Glu is replaced with a phosphonate or urea linkage [14, 15],[16]. Recently, RR-11a, a synthetic enzyme inhibitor of Legumain, an asparaginyl endopeptidase whose cell surface expression is driven by hypoxic stress, was developed for targeted nanoparticle delivery [17]. These ligands can be engineered further to contain a primary amine for conjugation through a flexible linker, such as PEG or other effector molecules, for targeted nanoparticle delivery to cancer cells expressing the biomarkers [18-20]. Development of synthetic small molecule targeting ligands, however, cannot always ensure the creation of small molecules that bind target extracellular domains with high affinity and specificity, presumably due to the much smaller interaction surface areas with target receptors compared with their natural protein ligands. One possible solution is multivalent targeting, often used in nature to significantly increase the binding strength between two molecules.
A series of papers that demonstrate the utilization of small molecules for targeted delivery of nanoparticles (Table 3) show that a variety of small molecule ligands can be used with this approach. This includes carbohydrates, folic acid, as well as synthetic small molecules. There are a range of cell-surface receptors targeted here as well, with a few studies focusing on brain tumor [21, 22] and other cancer cell lines [23].
Table 3.
Recent examples of targeted nanoparticles using small molecules.
| PMID# | Small Molecule | Target | Nanoparticles | Conjugation Chemistry |
|---|---|---|---|---|
| 21871507 | Anisamide | Sigma receptor | LPH | Synthesized as part of DSPE-PEG2000 |
| 22410170 | ManLAM | DC-SIGN | PLGA | Biotin/Streptavidin |
| 22410170 | Lex | DC-SIGN | PLGA | Biotin/Streptavidin |
| 21955528 | Myristic acid | U87 cells | MC-PEI/DNA | Amine/Carboxyl |
| 21882825 | Dimannose | C-type lectin receptors on dendritic cells | Polyanhydride | Amine/Carboxyl |
| 21419870 | RR-11a | Legumain | Liposome | Amine/Carboxyl |
| 22204981 | Folic Acid | HeLa cells | β-Cyclodextrin Micelle | Carbamate/Ester |
Targeting with Polypeptide-based Homing Peptide, Protein Domain, and Antibody Ligands
Polypetide-based targeting ligands, including homing peptides, protein domains, and antibodies, have advantages over other classes of targeting ligands in that they can be systemically developed and generated by using various biological selection and expression systems, respectively. Some major issues of these targeting ligands include immunogenicity, stability, and difficulty for site-specific conjugation with nanoparticles.
Antibodies
The idea of using nanoparticle technology as a drug delivery platform is not new, dating back to the development and investigation of polyalkylcyanoacrylate nanoparticles in the early 1980s [24, 25]. Antibodies were used in this pioneering research because, at this time, the work of Pimm and coworkers in the early 1980s notably predated the development of phage display screening of short peptide libraries [26], yet hybridoma technology had existed for almost a decade [27].
Following Pimm's work, Vaughan and coworkers developed a method of displaying human mAb fragments in a bacteriophage display system [28], which led to the FDA-approved recombinant human mAb Humira for the treatment of rheumatoid arthritis [29]. Another method, described by Russell and Lonberg, created a transgenic mouse whose murine antibody genes were replaced by human versions [30, 31]. Vectibix, a human anti-EGFR that can be used to treat colorectal cancer, was developed using transgenic mice [32]. Other approaches focused on the modification of existing murine mAbs into murine/human chimeras for the purpose of improving their pharmacokinetics [33].
Antibodies, which were considered originally as targeting ligands due to their availability to research and their attributes as specific, in vivo targeting ligands without reliance on tumor enhanced permeability and retention (EPR), gained greater utility as nanoparticle targeting ligands from the aforementioned advances. For in vivo therapeutics, the continued use of antibodies as nanoparticle targeting ligands is due largely to various developments that have overcome the problems of cross-species antibody immunogenicity. Consequently, therapeutic mAb development for the purpose of translation to the clinic remains an active field and, therefore, mAbs persist as major nanoparticle targeting ligands.
A review of papers describing antibody-guided nanoparticles from 2003 to 2012 (Table 4) reveals that most targeting antibodies are monoclonal and mostly murine, though some antibodies from other species, and polyclonals from rabbit, have been effective, as well as some chimeric [34-36] and humanized antibodies [13, 37, 38]. The majority of these antibodies target the extracellular domains (ECDs) of cell surface proteins, which is logical considering their intended application as in vivo targeted nanoparticles, the exception being a diagnostic sensor of NANOG, a transcription factor, composed of a graphite AuNP-coated film [39]. These antibodies have been successfully conjugated to a variety of nanoparticles, from metallic NPs (e.g. AuNPs, SPIONs, 99mTc), polymers (e.g. PLGA, chitosan, HDDP), micelles and liposomes, to silica and quantum dots. However, most of the conjugation techniques employed lack directionality, presumably due to the presence of multiple reactive functional groups on antibodies, yielding heterogeneous antibody orientations on the nanoparticles.
Table 4.
Recent examples of targeted nanoparticles using antibodies.
| PMID# | Antibody | Antibody Type |
Source | Target | Nanoparticles | Conjugation Chemistry |
|---|---|---|---|---|---|---|
| 22394186 | ab76586 | Monoclonal Mouse IgG1 | Abcam | NANOG* | Graphite AuNP-coated film | Amine/Carboxylate |
| 22394186 | ab84231 | Polyclonal Rabbit IgG | Abcam | NANOG* | Graphite AuNP-coated film | Amine/Carboxylate |
| 20825223 | Trastuzumab | Monoclonal Humanized | HER2 ECD | Magnetic nanocrystals | Non-covalent interaction | |
| 18767886 | E2156 | Monoclonal Mouse IgG1 | Sigma | EGFR ECD | AuNPs | Non-covalent interaction |
| 18606202 | mAb62 | Monoclonal Mouse IgG2a | PECAM-1 ECD | Polystyrene | Biotin/Streptavidin | |
| 18606202 | mAb35 | Monoclonal Mouse IgG1 | PECAM-2 ECD | Polystyrene | Biotin/Streptavidin | |
| 18606202 | mAbGi34 | Monoclonal Mouse IgG1 | PECAM-3 ECD | Polystyrene | Biotin/Streptavidin | |
| 18606202 | mAb4G6 | Monoclonal Mouse IgG2b | PECAM-4 ECD | Polystyrene | Biotin/Streptavidin | |
| 18606202 | mAb37 | Monoclonal Mouse IgG1 | PECAM-5 ECD | Polystyrene | Biotin/Streptavidin | |
| 22107797 | ATCC 27660 | Polyclonal Rabbit IgG | ViroStat | Staphylococcus aureus | Polylactide | Non-covalent interaction |
| 21748635 | Trastuzumab | Monoclonal Humanized Mouse IgG1 | HER2 ECD | 99mTc | Biotin/Streptavidin | |
| 22065745 | 29D7 | Monoclonal Mouse IgG1 | Wyeth | TrkB ECD | SPIONs | Rat anti-mouse IgG1 capture |
| 22011314 | Ritux (rituximab) | Monoclonal Chimeric IgG | BC Cancer Agency | CD20 ECD | PEG Lipid NP | Thiol/Maleimide |
| 22114481 | MAB1609 | Monoclonal Mouse IgG1, IgM | Chemicon | Cytokeratin 7/8 ECD | SPIONs | Amine/Carboxylate |
| 22324543 | 7.16.4 | Monoclonal Mouse | UCSF Monoclonal Antibody Core | Rat neu ECD | SPION-chitosan-g-PEG | Thiol/Maleimide |
| 22349096 | C225 (cetuximab, erbitux) | Monoclonal Chimeric IgG1 | Bristol-Myers Squibb | EGFR ECD | AuNPs | Amine/Carboxylate, Au/Thiol |
| 22410170 | Q5/13 | Monoclonal Mouse IgG2a | Beckman Coulter | HLA-DR/DP ECD | PLGA | Biotin/Streptavidin |
| 22410170 | CD83 | Monoclonal Mouse IgG1 | Beckman Coulter | HLA-DR/DP ECD | PLGA | Biotin/Streptavidin |
| 22410170 | CD69 | Monoclonal Mouse IgG1 | BD Biosciences | HLA-DR/DP ECD | PLGA | Biotin/Streptavidin |
| 22410170 | CD80 | Monoclonal Mouse IgG1 | BD Biosciences | HLA-DR/DP ECD | PLGA | Biotin/Streptavidin |
| 22410170 | CD86 | Monoclonal Mouse IgG1 | BD Biosciences | HLA-DR/DP ECD | PLGA | Biotin/Streptavidin |
| 22410170 | CCR7 | Monoclonal Rat IgG2a | BD Biosciences | HLA-DR/DP ECD | PLGA | Biotin/Streptavidin |
| 22410170 | AZN-D1 (αDC-SIGN1) | Monoclonal Mouse hybrid IgG2/IgG4 | Alexion Pharmaceuticals | DC-SIGN ECD | PLGA | Biotin/Streptavidin |
| 22410170 | AZN-D2 | Monoclonal Mouse IgG2 | Alexion Pharmaceuticals | DC-SIGN ECD | PLGA | Biotin/Streptavidin |
| 22410170 | Ab hD1 (αDC-SIGN2) | Monoclonal Humanized IgG2/4 | Alexion Pharmaceuticals | DC-SIGN ECD | PLGA | Biotin/Streptavidin |
| 22410170 | H200 (αDC-SIGN3) | Polyclonal Rabbit IgG | Santa Cruz Biotechnology | DC-SIGN ECD | PLGA | Biotin/Streptavidin |
| 22464249 | Colo205 | Monoclonal | FMMU, China | EpCAM ECD | Silica NPs | NaIO4 oxidation |
| 22464249 | sw480 | Monoclonal | FMMU, China | EpCAM ECD | Silica NPs | NaIO4 oxidation |
| 22464249 | NCM460 | Monoclonal | FMMU, China | EpCAM ECD | Silica NPs | NaIO4 oxidation |
| 22469295 | 9B9 | Monoclonal Rat | ShJU, China | EGFR ECD | PHPA-PEI | Non-covalent interaction |
| 22494888 | Anti-CD44 | Monoclonal | BD Biosciences | CD44 ECD | PEG-Liposome | Thiol/Maleimide |
| 22471719 | C225 (cetuximab, erbitux) | Monoclonal Chimeric | EGFR ECD | PLGA-ZnS:Mn2+ | Amine/Carboxylate | |
| 22515817 | Monoclonal | BD Biosciences | IL-6 | Fe3O4@SiO2 | Glutaraldehyde | |
| 22515817 | Monoclonal | BD Biosciences | IFN-γ | Fe3O4@SiO3 | Glutaraldehyde | |
| 22515817 | Monoclonal | BD Biosciences | AFP (alpha-fetoprotein) | Fe3O4@SiO4 | Glutaraldehyde | |
| 18239128 | FIB504 | Monoclonal Rat IgG2a | β7 integrin ECD | Multilamellar Liposome | Amine/Carboxylate | |
| 14512622 | Polyclonal | PSA ECD | AuNPs | Non-covalent interaction | ||
| 14512622 | Monoclonal | PSA ECD | Iron Oxide NPs | Glutaraldehyde/Amine | ||
| 21838300 | anti-TM34-211 | Monoclonal Rat IgG2a | Murine thrombomodulin ECD | PEO-filomicelles | Biotin/Streptavidin | |
| 21838300 | anti-TM201-411 | Monoclonal Rat IgG2a | Murine thrombomodulin ECD | PEO-filomicelles | Biotin/Streptavidin | |
| 21976974 | Trastuzumab | Monoclonal Humanized Mouse IgG1 | Roche | HER2 ECD | Chitosan NPs | Thiol/Maleimide |
| 21976975 | anti-DR5 | Monoclonal Humanized IgG2b | Santa Cruz Biotechnology | DR5 ECD | PLA | Amine/Carboxylate |
| 22072868 | LCCS Abs | Polyclonal Mouse | liver cancer cell surface-specific (LCCS) | AuNPs | Non-covalent interaction | |
| 22035507 | Anti-Her2 Ab | Monoclonal Mouse IgG1 | Bender MedSystems (eBioscience) | HER2 | Iron Oxide NPs | Amine/Carboxylate |
| 21980236 | Anti-EGFR Ab | Monoclonal Chimera | Beijing Zhong Shan Company | EGFR | Quantum Dots (QD800) | Thiol/Maleimide |
| 22032622 | ScFv-EGFR | ScFv | EGFR | HDDP | Amine/Carboxylate |
Antibody targeting of nanoparticles face several major challenges: antigen binding (the mAb must have high target specificity and affinity and the linker, as well as NPs, must not perturb the desired specificity), conjugation (the Ab-NP linkage must be highly efficient and site-specific), and circulation time (the mAb-NP conjugate linker must be stable during circulation). In addition, immunogenicity and purity are other concerns. The body can perceive antibodies as foreign proteins and clear them, nullifying the action of the targeted NPs. Many conjugation techniques, such as those exploiting lysine side-chain amines and cysteine sulfhydryl groups, yield heterogeneous mixtures of targeted NPs, each with differing Ab:NP molar ratios, conjugation sites, pharmacokinetics, and safety profiles.
Peptides
Much smaller than antibodies but larger than small molecules, short homing peptides offer additional nanoparticle targeting options, and certain advantages over the aforementioned targeting ligands. The design of a small molecule that fits into a usually shallow and hydrophobic binding pocket can be challenging. As a compromise between small molecules and antibodies, short peptides provide smaller size, as well as high specificity and affinity.
Targeting homing peptides are typically discovered via phage display, first developed in 1985. Phage display is a screening tool for peptides, allowing selection of peptide sequences with increased affinities to a specific target of choice [40, 41]. The phage display system is a cyclic selection process, where the purified target molecules or specific cell types are incubated with a randomized library of peptide sequences displayed on bacteriophage capsids. Some peptides bind to the target protein, and nonspecific binders are washed away with the specific binders eluted. Binding peptide sequences-bacteriophages are collected, which infect E. Coli and is amplified, followed by additional cycles of selection. Selected peptides have been used as molecular probes for imaging and can be applied as therapeutics as well.
There are numerous publications using short homing peptides to target nanoparticles during the past decade. Studies from 2011-2012 using peptides as nanoparticle targeting ligands (Table 5) predominantly utilized ligands discovered via phage display. Some used natural peptides, such as EGF [42, 43], CANF [44], and Angiopep-2 [45]. About 30% of reviewed papers used cyclic peptides, though this percentage is influenced by the popularity of the RGD peptide as a targeting ligand to αvβ3 integrin [46-51]. All studies targeted cell surface proteins. As with antibodies, the used peptides were successfully conjugated to a variety of nanoparticles, such as metallic NPs (e.g. gadolinium oxide, SPIONs, AuNPs, MBCSPs), micelles and polymers (e.g. chitosan, PLGA, poly(methyl methacrylate)), and dendrimers. Han and coworkers provide an interesting alternative to the typical nanoparticle formulations and conjugation paradigms [42]. They expressed targeting peptides recombinantly fused to the 97 kDa major vault protein (MVP), which self-assembles into Vault Nanoparticles – naturally-occurring nanoparticles present in cell cytoplasm composed of ribonucleoproteins. At times, the orientation of the conjugated peptides in the reviewed studies can be problematic but this is controlled in some applications at the level of peptide synthesis, typically through the additional of a unique functional group to peptide termini, allowing for site-specific conjugation with nanoparticles.
Table 5.
Recent examples of targeted nanoparticles using short homing peptides.
| PMID# | Peptide | Sequence | Cyclic | Target | Nanoparticle | Conjugation Chemistry |
|---|---|---|---|---|---|---|
| 21726134 | ATWLPPR | ATWLPPR | Neuropilin-1 ECD | Gd2O3 in polysiloxane shell | Amine/Carboxylate | |
| 21740042 | EGF | EGFR ECD | Vault nanoparticles | Recombinant fusion | ||
| 21763734 | NGR | NGR | CD13 ECD | ELP Micelle | Non-covalent interaction | |
| 21781994 | S2P | CRTLTVRKC | Stabilin-2 ECD | Chitosan | Amine/Carboxylate, Thiol/Maleimide | |
| 21871505 | EGF | EGFR ECD | GemC18 NPs | Thiol/Maleimide | ||
| 21945679 | I4R | CRKRLDRNC | IL-4R ECD | Hydrophobically modified glycol chitosan | Amine/Carboxylate | |
| 21987727 | AH1 | SPSYVYHQF | MHC Class II ECD | G5-PAMAM dendrimer | Thiol/Maleimide | |
| 21987727 | TRP2180–188 | SVYDFFVWL | MHC Class II ECD | G5-PAMAM dendrimer | Thiol/Maleimide | |
| 21987727 | PAn DR epitope (PADRE) | aKXVAAWTLKAAaZC | MHC Class II ECD | G5-PAMAM dendrimer | Thiol/Maleimide | |
| 21987727 | HA110–120 | SFERFEIFPKEC | MHC Class II ECD | G5-PAMAM dendrimer | Thiol/Maleimide | |
| 22014944 | iRGD | CRGDKGPDC | αvβ3, αvβ5 | PLGA-PLL-PEG | Thiol/Maleimide | |
| 22049461 | CANF | NPR-C ECD | poly (methyl methacrylate)-PEG | Methacrylate/Acetylene | ||
| 22087004 | KLWVLPKGGGCAm | Collagen IV | PLGA PEG | Thiol/Maleimide | ||
| 22093292 | CSK | CSKSSDYQC | Goblet cells | trimethyl chitosan chloride (TMC) | Amine/Carboxylate | |
| 22118776 | TRAIL | hTRAIL(114–281) | DR4, DR5 ECD | HSA NPs | Thiol/Maleimide | |
| 22133551 | Angiopep-2 | TFFYGGSRGKRNNFKTEEY | LRP Receptor ECD | PEG-co-poly(ε-caprolactone) | Thiol/Maleimide | |
| 22179825 | tLyp-1 | CGNKRTR | Yes | Neuropilin-1/2 ECD | Iron oxide nanoworms (NWs) | Biotin/Neutravidin |
| 22196766 | Pep 1 | CHVLWSTRC | Yes | Pancreatic islet capillary endothelial cells | PLGA-b-PEG | Amine/Carboxylate |
| 22197725 | CLL1-L1 | CDLRSAAVC | Yes | CLL1 | PEG nanomicelle telodendrimer | Click Chemistry: Alkyne/Azide |
| 22375916 | RGD | cRGDfK | Yes | αvβ3 integrin | AuNPs | Amine/Carboxylate |
| 22396491 | OA02 | cdG-HoCit-GPQc-Ebes-K-alkyne | α-3 integrin | PEG telodendrimer | Click Chemistry: Alkyne/Azide | |
| 22403681 | Tet-1 | HLNILSTLWKYR | Motor neurons | PLGA | Amine/Carboxylate | |
| 22497548 | RGD | cyclic RGD | Yes | αvβ3 integrin | PEG-PEI | Electrostatic interaction |
| 22533630 | RGD | c(RGDyK) | Yes | αvβ3 integrin | UCNP | Amine/Carboxylate |
| 22559746 | RGD | cRGD | Yes | αvβ3 integrin | CMC | Amine/Carboxylate, Thiol/Maleimide |
| 22560667 | RGD | c(RGDyK) | Yes | αvβ3 integrin | Iron Oxide NPs | Thiol/Maleimide |
| 22561668 | GRGDS | GRGDS | αvβ3 integrin | MBCSP (PLGA-magnetite) | Amine/Carboxylate |
There have been numerous effective in vitro peptides (e.g. targeting protein kinase CK2, glioma, FGF receptor, and many others). Finding peptides that work in an in vivo setting, however, appears more challenging as they are prone to proteolysis, glomerular transit, feature varying toxicities and differential effects on cell signaling, can encourage allergic sensitization, and are not amenable to oral bioavailability [52-54]. In addition, the costs of peptide synthesis can be prohibitive for some special applications [52].
Protein Domains
Targeting ligands based on full-length antibodies have several intrinsic disadvantages compared to ligands with much smaller sizes. First, the large sizes of the full-length antibodies limit the number of antibody molecules that can be accommodated on the surface of nanoparticles. Second, full-length antibodies are composed of multiple light and heavy chains that are linked through disulfide bonds. Such structure complicates its expression level and makes it difficult to achieve site-specific conjugation with nanoparticles. Third, it is both challenging and time-consuming to systematically engineer a full-length antibody with optimized targeting-binding parameters. Considerable effort has been directed to the reduction of the size of antibodies and the development of smaller binding units with antibody-like specificity and affinity. An ideal targeting ligand should be a highly soluble small protein with high stability and minimal aggregation, while it can be highly expressed in bacteria with much lower manufacturing costs. In addition, it should possess functional residues that facilitate conjugation with nanoparticles, preferably in a site-specific manner. The use of antibody fragments represents an interesting compromise in the selection of target specific affinity molecules. The smallest antibody fragments are those based on a single domain, such as naturally occurring heavy-chain antibodies found in camelids (nanobody) and humans (VH domains) [55-66]. These single-domain antibody fragments are well expressed, quite soluble and stable, and yet still able to maintain the specificity and affinity comparable to scFvs. A different approach is to select single domain antibody mimics from partially randomized libraries based on protein scaffolds related or not related to natural antibodies [67]. Scaffolds that have been used to construct single domain protein libraries include immunoglobulin-like β-barrel, zinc fingers, α-helical bundles, Src homology domains, PDZ domains, various repeat proteins, protease inhibitors, and disulfide-bond constrained small toxins [67-74]. Among them, targeting ligands based on FN3 (the tenth type III domain of human fibronectin), Z domain, and DARPins are most promising [1]. Several examples of protein domain based ligands that are suitable for targeted delivery of nanoparticles include FN3-based ligands (monobody) that recognize VEGF receptor and integrin αvβ3, Z domain based ligands (affibody) that recognize EGFR and HER2, and DARPin based ligands that recognizes HER2.
Compared to the number of nanoparticle applications utilizing antibodies and peptides for targeting, relatively few studies from the reviewed period (2007-2012) employed protein domains or non-immunoglobulin antibody mimics (Table 6). These targeting ligands include neurotoxin, transferrin, nanobody, affibody, and other protein domains. Though applications are fewer, protein domain nanoparticle targeting are highly promising, as compared to antibodies and peptides, for targeted delivery of nanoparticles.
Table 6.
Recent examples of targeted nanoparticles using protein domains.
| PMID# | Protein | Target | Nanoparticle | Conjugation Chemistry |
|---|---|---|---|---|
| 17964677 | apolipoprotein B-100, LDLR binding domain | LDL Receptor | Micelle | Non-covalent interaction |
| 18076008 | C-termini of clostridium/botulinum neurotoxins (THC, BHC) | GT1b | ABCD NPs | Thiol/Maleimide |
| 19173297 | hepatitis B surface antigen (HBsAg), preS1 domain | HepG2 cell line | Micelle | Ester replacement |
| 20959824 | anti-IGFBP7 | GBM | PEG-Fe3O4 NPs | NHS ester |
| 21306773 | LFA-1, I domain | ICAM-1 | Liposome | Ni/His tag |
| 21609027 | 2Rb18a nanobody | HER2 | AuNPs | Thiol/Maleimide |
| 21609027 | N7 nanobody | PSA | AuNPs | Thiol/Maleimide |
| 21302357 | Affibody-EGFR | EGFR | Au-Silica NPs | Thiol/Maleimide |
| 21508310 | Affibody-EGFR | EGFR | Au-Silica NPs | Thiol/Maleimide |
| 21351748 | Affibody-HER2 | HER2 | Polymeric nanosphere | Amine/Carboxylate |
| 21147502 | Affibody-HER2 | HER2 | NIR QDs and IO NPs | Thiol/Maleimide |
| 20801029 | Affibody-HER2 | HER2 | Bionanocapsules (BNCs) | Genetically displayed |
| 19012296 | Affibody-HER2 | HER2 | PLA-PEG polymeric NPs | Thiol/Maleimide |
| 18937120 | Affibody-HER2 | HER2 | Thermosensitive liposomes | Thiol/Maleimide |
| 21753879 | adiponectin, globular domain (gAd) | Atherosclerotic plaques | Proticle | Non-covalent interaction |
| 21753879 | adiponectin, globular domain (gAd) | Atherosclerotic plaques | Liposome | Thiol/Maleimide |
| 22013169 | anti-IGFBP7 single-domain antibody | IGFBP7 | SPION | Amine/Carboxylate |
| 22037106 | Heptameric Z (EGFR) | EGFR | Ni-lipid NPs | Ni2+/His tag |
| 22118776 | Transferrin | Transferrin Receptor | HSA NPs | Thiol-maleimide |
| 22410170 | gp120 | DC-SIGN | PLGA | Biotin, streptavidin |
Targeting with Aptamers
Since their development in 1990 by the Szostak, Gold, and Joyce groups, aptamers have existed as a separate class of binding molecules [75]. Aptamers are short single-stranded nucleic acids (RNA or DNA) capable of displaying diverse structures with the potential of binding many biochemical targets, from small molecules to large proteins. This ability derives from aptamer sequences; a designed 5’ and 3’ consensus region about 12-20 nucleotides in length flanks a central region of totally or partially randomized nucleotides. The random region determines the diversity of the aptamer pool, which typically achieves 1×1013 to 1×1015 unique sequences. The high sequence and conformational diversity of naïve aptamer pools (not yet selected against a target) makes the discovery of target binding aptamers highly likely. The selection of aptamers capable of binding a target of interest is called ‘Systematic Evolution of Ligands by EXponential enrichment’ (SELEX) [76]. SELEX involves iterative rounds of target binding, partitioning binding from non-binding sequences, and amplification of the enriched binding sequences. Several SELEX variants have been developed since 1990, such as whole cell surface-SELEX (Cell-SELEX) [77, 78], which ensures the selection of aptamers capable of binding the bioactive forms of target proteins on the cell surface. Other conventional SELEX strategies vary in the means of partitioning unbound nucleic acids from target-bound aptamers: cutoff membranes, flow cytometry, EMSA, and capillary electrophoresis.
Aptamers are uniquely suited to nanoparticle targeting. First, it is possible to synthesize aptamers with a specific functional moiety, such as a carboxylate, amino, sulfhydryl or aldehyde, at only one end of the nucleic acid aptamer. This ensures and greatly facilitates site-specific conjugation and prevents the formation of heterogeneous mixtures. Second, aptamers are typically non-immunogenic [79]. Third, many other attributes make aptamers attractive for nanoparticle targeting, such as being non-toxic [79-81] and modifiable for stability in circulation [82]. They can be selected in vitro and in vivo, and be repeatedly and reversibly denatured. Moreover, as aptamers are not dependent on animals or their immune responses, aptamers can be selected against weakly immunogenic targets and toxins. The ability to chemically synthesize aptamers infers little batch variation [83]. They are much smaller than antibodies and can form compact structures, allowing them to bind clefts, binding sites, and enzymatic active sites, which is difficult, if not impossible, for antibodies to achieve [84].
The degradative activity of biologically-abundant nucleases on nucleic acids has been a major barrier to in vivo aptamer-targeted nanoparticle applications. Attempts at translating RNA aptamers for use as therapeutics have focused on replacement of the nuclease-susceptible 2’-hydroxyl RNAs with other moieties. RNAs containing 2’-fluoro and 2’-O-methyl pyrimidines, which can be generated by in vitro transcription with an appropriate T7 RNA polymerase mutant, have known partial resistant to nucleases [85, 86]. The development of aptamers with higher levels of 2’-modification requires lengthy, expensive, and tedious post-selection optimization. The development of the FDA-approved aptamer Macugen (Pegaptanib), for example, involved the selection of an initial anti-VEGFR aptamer (NX1838) bearing 2’-fluoropyrimidines only [87]. NX1838 was then subjected to tedious and time-consuming post-selection modifications involving the selective substitution of purines one-by-one with 2’-O-methyl purines. Systematic testing indicated that all but two natural 2’-hydroxyl purines could be replaced with 2’-O-methyl purines.
Nuclease resistant aptamers that specifically bind to the extracellular domains of transmembrane cancer biomarkers, such as integrin αvβ3, VEGF receptor, EGF receptor, HER2, HER3, MUC1, PSMA, and receptor tyrosine kinase RET, can be used to direct nanoparticles to tumor tissues. Of the reviewed studies (2004-2012) (Table 7) that used aptamers as nanoparticle targeting ligands, all used aptamers to cell surface biomarkers and used either DNA (62%), unmodified RNA (17%), or modified RNA (21%). Compared to numerous RNA nucleases, there are relatively fewer DNases in vivo. DNA aptamers do, however, suffer from characteristics that can complicate their in vitro selection via SELEX, such as the formation of hard to manage G-tetrads. Of those studies using unmodified RNA aptamers, two chose nanoparticles that confer nuclease resistance to the aptamers. Li and coworkers employed AuNPs, which maintained a halocline immediately surrounding the AuNP, yielding a blanketing solution layer of high ionic character that discourages nuclease activity [88, 89]. In another instance, Lee and coworkers found that PSMA-specific RNA A9 aptamer conjugated to dendrimers was nuclease-resistant 24 hours post-exposure [90]. Yu and coworkers intentionally used nucleases as a means of releasing intercalated doxorubicin from A9 aptamers targeting nanoparticles to PSMA [91].
Table 7.
Recent examples of targeted nanoparticles using aptamers.
| PMID# | Aptamer | Aptamer Type | Target | Nanoparticle | Conjugation Chemistry |
|---|---|---|---|---|---|
| 15520166 | A10 | Modified RNA, 2′-F C/U, 3′ inverted dT cap | PSMA | PLA | Amine/Carboxylate |
| 16495043 | A9 | Modified RNA, 2′-F C/U | PSMA | Streptavidin Quantum Dots | Hydrazide |
| 18512972 | A9 | Unmodified RNA | PSMA | AuNPs | Base-pairing hybridization |
| 18978032 | A10 | Modified RNA, 2′-F C/U | PSMA | PLGA-b-PEG | Amine/Carboxylate |
| 19377681 | sgc8c | DNA | CCRF-CEM (T-cell acute lymphoblastic leukemia, T-cell ALL) cells | FCNPs | Amine/Carboxylate |
| 20024341 | sgc8 | DNA | CCRF-CEM (T-cell acute lymphoblastic leukemia, T-cell ALL) cells | Pegylated Liposome | Thiol/Maleimide |
| 20066302 | J18 | Unmodified RNA | EGFR | AuNPs | Base-pairing hybridization |
| 20080797 | TDO5 | DNA | immunoglobin heavy mu chain receptor | Aptamer-PEG-Lipid NPs | Non-covalent interaction |
| 20947949 | GB-10 | DNA | tenascin-c | Dextran Magnetic NPs | Amine/Carboxylate |
| 21233423 | A10 | Modified RNA, 2′-F C/U, 3′ inverted dT cap | PSMA | PLGA-b-PEG | Amine/Carboxylate |
| 21281497 | apt1 | Unmodified RNA, 2′-Ome termini | CD30 | PEI-citrate | Non-covalent interaction |
| 21342659 | MUC1 | DNA | MUC1 | Quantum Dot | Amine/Carboxylate |
| 21530479 | Ky2 | DNA | Kanamycin, kanamycin B, tobramycin | AuNPs | Non-covalent interaction |
| 21641946 | A9 | Unmodified RNA | PSMA | PAMAM dendrimer | Base-pairing hybridization |
| 21648076 | A9 | Unmodified RNA | PSMA | TCL-SPION | Amine/Carboxylate |
| 21732610 | MUC1 | DNA | MUC1 | Three-dimensional (3D) DNA polyhedra | Self-assembly |
| 21788069 | AS1411 | DNA | nucleolin | PEG-PLGA | Amine/Carboxylate |
| 21888350 | sgc8 | DNA | CCRF-CEM cell line | PHMNP | Amine/Carboxylate |
| 21912664 | MUC1 | DNA | MUC1 | PLGA | Amine/Carboxylate |
| 21936502 | A10 | Unmodified RNA | PSMA | QD–PMAT–PEI | Amine/Carboxylate, Thiol/Maleimide |
| 21942498 | sgc8c | DNA | CCRF-CEM cell line | AuNPs | Gold/Thiol |
| 21944470 | AS1411 | DNA | nucleolin | Magnetic Fluorescence NP (MF) | Amine/Carboxylate |
| 22214176 | XEO2 mini | Modified RNA, 2′-Ome C/A/U | PC3, LNCaP | DSPE-PLGA | Thiol/Maleimide |
| 22424140 | sgc8c | DNA | CCRF-CEM cell line | Streptavidin-coated MNPs | Biotin/Streptavidin |
| 22424140 | TDO5 | DNA | Ramos leukemia cell line | Streptavidin-coated MNPs | Biotin/Streptavidin |
| 22424140 | T2-KK1B10 | DNA | K562 leukemia cell line | Streptavidin-coated MNPs | Biotin/Streptavidin |
| 22424140 | KDED2a-3 | DNA | DLD1 colon cell line | Streptavidin-coated MNPs | Biotin/Streptavidin |
| 22424140 | KCHA10 | DNA | HCT116 colon cell line | Streptavidin-coated MNPs | Biotin/Streptavidin |
| 22424140 | TLS11a | DNA | LH86 liver cell line | Streptavidin-coated MNPs | Biotin/Streptavidin |
Across all reviewed studies, aptamers were conjugated to a variety of nanoparticles successfully, as with peptides and antibodies. Unlike peptides and antibodies, however, the maintenance of proper aptamer orientation was rarely a problem. Through the use of capture oligos conjugated first to the nanoparticle, followed by hybridization of the aptamer via a consensus sequence, or through the synthesis of targeting aptamers with a terminal biotin, thiol, or amine, directional conjugation was easily achieved.
Chimeras, Multifunctionalization, and other unconventional approaches
Attempts to improve nanoparticle performance, either therapeutically or diagnostically, have increasingly turned to multifunctional and, more recently, chimeric targeting systems. Multifunctional targeting involves the conjugation of various ligands within the same class (e.g. aptamers, peptides, etc.) but with different individual targets (e.g. HER3, tenascin-C, PSMA), whereas chimeric targeting uses targeting ligands across classes (e.g. an aptamer with a peptide). The installation of a multifunctional or chimeric targeting system into a nanoparticle-payload technology attempts to extend one or more key characteristics: cell uptake, target specificity, utilization of multiple targeting strategies, and attribute exploitation of multiple targeting ligands classifications. Recent examples of multifunctionalized nanoparticles are listed in Table 8, including those developed by Bhattacharyya and coworkers using anti-EGFR and MOV18 anti-folate receptor α antibodies [92], and those by Kluza and coworkers using Anx and RGD peptides [93]. Ko and coworkers actually used three targeting ligands to produce a nanoparticle with both multifunctional and chimeric features (DNA aptamer AS1411, DNA aptamer TTA1, and peptide RGD) [94].
Table 8.
Recent examples of targeted nanoparticles using multifunctional and chimeric approaches.
| PMID# | Ligand 1 |
Ligand 1 Class (Sequence) |
Ligand 1 Target |
Ligand 2 |
Ligand 2 Class (Sequence) |
Ligand 2 Target |
Ligand 3 |
Ligand 3 Class (Sequence) |
Ligand 3 Target |
Nanoparticle | Conjugation Chemistry |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 20155973 | Anti- HER2 |
Antibody; Monoclonal |
HER2 | S6 | Aptamer, RNA |
HER2 | AuNPs | Ab: Amine/Carboxylate; Aptamer: Thiol/Maleimide |
|||
| 21071077 | AS1411 | Aptamer, DNA | Nucleolin | TTA1 | Aptamer, DNA |
Tenascin-C | RGD | Peptide (cRGDfk) |
αvβ3 integrin |
MNPs | Amine/Carboxylate |
| 21147500 | A10 | Aptamer, RNA | PSMA | DUP-1 | Aptamer, Peptide |
PSMA (-) cells |
TCL- SPION |
Amine/Carboxylate | |||
| 21971980 | C225 (cetuximab, erbitux) |
Antibody; Monoclonal, Chimeric |
EGFR | MOV18 | Antibody; Monoclonal, Mouse IgG1 |
Folate Receptor α |
AuNPs | Non-covalent interaction |
|||
| 22079810 | Anginex (Anx) |
Peptide (ANIKLSVQMKLFKRHLKWKIIVKLNDGRELSLD) |
galectin-1 | RGD | Peptide (cRGDfk) |
αvβ3 integrin |
Liposomes | Thiol/Maleimide | |||
| 22118775 | RGD | Peptide (cRGDfk) |
αvβ3 integrin |
Transferrin | Glycoprotein | Transferrin Receptor |
HPAE-co-PLA/DPPE | Thiol/Maleimide | |||
| 22342711 | RGD | Peptide (RGD) |
αvβ3 integrin |
DEVD | Peptide (DEVD) |
Caspase 3 |
Au-Iron Oxide |
Gold/Thiol |
The majority of nanoparticle targeting research aims at the specific delivery of nanoparticles via targeting ligands directed against endogenous differences between normal and pathologic tissues. Many other avenues of nanoparticle targeting research, however, are also under investigation. These include the ex vivo induction of molecular physiological changes, exploiting unique characteristics inherent in the pathologic environment, autologous harnessing of the immune system as an active participant in nanoparticle-based therapy, and the use of targeted bacteriophages, to name a few. Hariri and coworkers investigated the use of radiation to guide FePt nanoparticles to tumor sites using a short peptide that targets TIP-1 receptor, a receptor upregulated on endothelial cells in response to radiation-induced injury [94]. Basel and coworkers demonstrated that targeting ligands may not always be necessary for effective nanoparticle targeting via the exploitation of high concentrations of cancer-associated protease (CAP), such as urokinase plasminogen activator (uPA), matrix metalloproteases (MMPs), and some cathepsins, in dysplastic tissues [95]. In a radical shift of nanoparticle targeting strategy, Choi and Kennedy demonstrated that macrophages and human T cells could be loaded with gold nanoparticles (AuNPs) and used to deliver those AuNPs to tumor sites [96, 97]. Building off early bacteriophage work by Smith [98], many studies have used bacteriophages as nanoparticle platforms presenting weakly immunogenic targets for the purpose of provoking immune responses [99-102]. Most recently, Lee et al. combined bacteriophage, AuNPs, and magnetic beads for combined colorimetric protein detection and identification [103].
Conclusion
One of the most challenging problems in the targeted delivery of nanoparticles is to develop high-quality targeting ligands that can give rise to more specific accumulation of nanoparticles in tumors than in other tissues. Such smart molecules can be systematically developed through affinity selection from combinatorial libraries displaying small molecules, short peptides, antibodies and antibody fragments, engineered protein domains, and nucleic acid aptamers. The availability of these types of targeting ligands and their successful conjugation with nanoparticles will have significant applications in targeted imaging, diagnosis, and treatment of malignant tumors and other diseases that are based on nanotechnology.
Acknowledgments
The targeting ligand development work in the Liu lab was supported by National Institutes of Health Grants CA119343, CA151652 and CA157738 (to R.L.).
References
- 1.Liu R, Kay BK, Jiang S, Chen S. Nanoparticle Delivery: Targeting and Nonspecific Binding. MRS Bulletin. 2009;34(6):432–40. [Google Scholar]
- 2.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(1):19–24. doi: 10.1073/pnas.012583299. Epub 2001/12/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson JA, Lu YY, Van Deventer JA, Tirrell DA. Residue-specific incorporation of non-canonical amino acids into proteins: recent developments and applications. Current opinion in chemical biology. 2010;14(6):774–80. doi: 10.1016/j.cbpa.2010.09.013. Epub 2010/11/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu CC, Mack AV, Tsao ML, Mills JH, Lee HS, Choe H, et al. Protein evolution with an expanded genetic code. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(46):17688–93. doi: 10.1073/pnas.0809543105. Epub 2008/11/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Algar WR, Prasuhn DE, Stewart MH, Jennings TL, Blanco-Canosa JB, Dawson PE, et al. The controlled display of biomolecules on nanoparticles: a challenge suited to bioorthogonal chemistry. Bioconjugate chemistry. 2011;22(5):825–58. doi: 10.1021/bc200065z. Epub 2011/05/19. [DOI] [PubMed] [Google Scholar]
- 6.Los GV, Wood K. The HaloTag: a novel technology for cell imaging and protein analysis. Methods Mol Biol. 2007;356:195–208. doi: 10.1385/1-59745-217-3:195. Epub 2006/09/22. [DOI] [PubMed] [Google Scholar]
- 7.Hodneland CD, Lee YS, Min DH, Mrksich M. Selective immobilization of proteins to self-assembled monolayers presenting active site-directed capture ligands. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(8):5048–52. doi: 10.1073/pnas.072685299. Epub 2002/04/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jongsma MA, Litjens RH. Self-assembling protein arrays on DNA chips by auto-labeling fusion proteins with a single DNA address. Proteomics. 2006;6(9):2650–5. doi: 10.1002/pmic.200500654. Epub 2006/04/06. [DOI] [PubMed] [Google Scholar]
- 9.Low Ps, Henne Wa, Doorneweerd Dd. Discovery and development of folic-acid-based receptor targeting for imaging and therapy of cancer and inflammatory diseases. Acc Chem Res. 2008;41(1):120–9. doi: 10.1021/ar7000815. Epub 2007 Jul 27. [DOI] [PubMed] [Google Scholar]
- 10.Zhao X, Li H, Lee Rj. Targeted drug delivery via folate receptors. Expert Opin Drug Deliv. 2008;5(3):309–19. doi: 10.1517/17425247.5.3.309. [DOI] [PubMed] [Google Scholar]
- 11.Mach RH, Huang Y, Freeman RA, Wu L, Vangveravong S, Luedtke RR. Conformationally-flexible benzamide analogues as dopamine D3 and sigma 2 receptor ligands. Bioorganic & medicinal chemistry letters. 2004;14(1):195–202. doi: 10.1016/j.bmcl.2003.09.083. Epub 2003/12/20. [DOI] [PubMed] [Google Scholar]
- 12.Banerjee R, Tyagi P, Li S, Huang L. Anisamide-targeted stealth liposomes: a potent carrier for targeting doxorubicin to human prostate cancer cells. Int J Cancer. 2004;112(4):693–700. doi: 10.1002/ijc.20452. Epub 2004/09/24. [DOI] [PubMed] [Google Scholar]
- 13.Cruz LJ, Tacken PJ, Pots JM, Torensma R, Buschow SI, Figdor CG. Comparison of antibodies and carbohydrates to target vaccines to human dendritic cells via DC-SIGN. Biomaterials. 2012;33(16):4229–39. doi: 10.1016/j.biomaterials.2012.02.036. Epub 2012/03/14. [DOI] [PubMed] [Google Scholar]
- 14.Kozikowski Ap, Nan F, Conti P, Zhang J, Ramadan E, Bzdega T, et al. Design of remarkably simple, yet potent urea-based inhibitors of glutamate carboxypeptidase II (NAALADase). J Med Chem. 2001;44(3):298–301. doi: 10.1021/jm000406m. [DOI] [PubMed] [Google Scholar]
- 15.Kozikowski Ap, Zhang J, Nan F, Petukhov Pa, Grajkowska E, Wroblewski Jt, et al. Synthesis of urea- based inhibitors as active site probes of glutamate carboxypeptidase II: efficacy as analgesic agents. J Med Chem. 2004;47(7):1729–38. doi: 10.1021/jm0306226. [DOI] [PubMed] [Google Scholar]
- 16.Low PS, Kularatne SA. Folate-targeted therapeutic and imaging agents for cancer. Curr Opin Chem Biol. 2009;13(3):256–62. doi: 10.1016/j.cbpa.2009.03.022. Epub 2009/05/08. [DOI] [PubMed] [Google Scholar]
- 17.Liao D, Liu Z, Wrasidlo W, Chen T, Luo Y, Xiang R, et al. Synthetic enzyme inhibitor: a novel targeting ligand for nanotherapeutic drug delivery inhibiting tumor growth without systemic toxicity. Nanomedicine : nanotechnology, biology, and medicine. 2011;7(6):665–73. doi: 10.1016/j.nano.2011.03.001. Epub 2011/03/23. [DOI] [PubMed] [Google Scholar]
- 18.Chandran Ss, Banerjee Sr, Mease Rc, Pomper Mg, Denmeade Sr. Characterization of a targeted nanoparticle functionalized with a urea-based inhibitor of prostate-specific membrane antigen (PSMA). Cancer Biol Ther. 2008;7(6):974–82. doi: 10.4161/cbt.7.6.5968. Epub 2008 Mar 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee Sr, Foss Ca, Castanares M, Mease Rc, Byun Y, Fox Jj, et al. Synthesis and evaluation of technetium-99m- and rhenium-labeled inhibitors of the prostate-specific membrane antigen (PSMA). J Med Chem. 2008;51(15):4504–17. doi: 10.1021/jm800111u. Epub 2008 Jul 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humblet V, Misra P, Bhushan Kr, Nasr K, Ko Ys, Tsukamoto T, et al. Multivalent Scaffolds for Affinity Maturation of Small Molecule Cell Surface Binders and Their Application to Prostate Tumor Targeting. J Med Chem. 2008;24:24. doi: 10.1021/jm801033c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Gu B, Meng Q, Yan Z, Gao H, Chen X, et al. The use of myristic acid as a ligand of polyethylenimine/DNA nanoparticles for targeted gene therapy of glioblastoma. Nanotechnology. 2011;22(43):435101. doi: 10.1088/0957-4484/22/43/435101. Epub 2011/10/01. [DOI] [PubMed] [Google Scholar]
- 22.Nie G, Hah HJ, Kim G, Lee YE, Qin M, Ratani TS, et al. Hydrogel nanoparticles with covalently linked coomassie blue for brain tumor delineation visible to the surgeon. Small. 2012;8(6):884–91. doi: 10.1002/smll.201101607. Epub 2012/01/11. [DOI] [PubMed] [Google Scholar]
- 23.Liu T, Li X, Qian Y, Hu X, Liu S. Multifunctional pH-disintegrable micellar nanoparticles of asymmetrically functionalized beta-cyclodextrin-based star copolymer covalently conjugated with doxorubicin and DOTA-Gd moieties. Biomaterials. 2012;33(8):2521–31. doi: 10.1016/j.biomaterials.2011.12.013. Epub 2011/12/30. [DOI] [PubMed] [Google Scholar]
- 24.Couvreur P, Kante B, Lenaerts V, Scailteur V, Roland M, Speiser P. Tissue distribution of antitumor drugs associated with polyalkylcyanoacrylate nanoparticles. Journal of pharmaceutical sciences. 1980;69(2):199–202. doi: 10.1002/jps.2600690222. Epub 1980/02/01. [DOI] [PubMed] [Google Scholar]
- 25.Kreuter J, Hartmann HR. Comparative study on the cytostatic effects and the tissue distribution of 5- fluorouracil in a free form and bound to polybutylcyanoacrylate nanoparticles in sarcoma 180-bearing mice. Oncology. 1983;40(5):363–6. doi: 10.1159/000225763. Epub 1983/01/01. [DOI] [PubMed] [Google Scholar]
- 26.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228(4705):1315–7. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 27.Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256(5517):495–7. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 28.Vaughan TJ, Williams AJ, Pritchard K, Osbourn JK, Pope AR, Earnshaw JC, et al. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat Biotechnol. 1996;14(3):309–14. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
- 29.Weinblatt ME, Keystone EC, Furst DE, Moreland LW, Weisman MH, Birbara CA, et al. Adalimumab, a fully human anti-tumor necrosis factor alpha monoclonal antibody, for the treatment of rheumatoid arthritis in patients taking concomitant methotrexate: the ARMADA trial. Arthritis and rheumatism. 2003;48(1):35–45. doi: 10.1002/art.10697. Epub 2003/01/16. [DOI] [PubMed] [Google Scholar]
- 30.Russell ND, Corvalan JR, Gallo ML, Davis CG, Pirofski L. Production of protective human antipneumococcal antibodies by transgenic mice with human immunoglobulin loci. Infection and immunity. 2000;68(4):1820–6. doi: 10.1128/iai.68.4.1820-1826.2000. Epub 2000/03/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lonberg N. Human antibodies from transgenic animals. Nat Biotechnol. 2005;23(9):1117–25. doi: 10.1038/nbt1135. Epub 2005/09/10. [DOI] [PubMed] [Google Scholar]
- 32.Chua YJ, Cunningham D. Panitumumab. Drugs Today (Barc) 2006;42(11):711–9. doi: 10.1358/dot.2006.42.11.1032061. Epub 2006/12/16. [DOI] [PubMed] [Google Scholar]
- 33.Reichert JM, Valge-Archer VE. Development trends for monoclonal antibody cancer therapeutics. Nat Rev Drug Discov. 2007;6(5):349–56. doi: 10.1038/nrd2241. Epub 2007/04/14. [DOI] [PubMed] [Google Scholar]
- 34.Popov J, Kapanen AI, Turner C, Ng R, Tucker C, Chiu G, et al. Multivalent rituximab lipid nanoparticles as improved lymphoma therapies: indirect mechanisms of action and in vivo activity. Nanomedicine (Lond) 2011;6(9):1575–91. doi: 10.2217/nnm.11.50. Epub 2011/10/21. [DOI] [PubMed] [Google Scholar]
- 35.Deepagan VG, Sarmento B, Menon D, Nascimento A, Jayasree A, Sreeranganathan M, et al. In vitro targeted imaging and delivery of camptothecin using cetuximab-conjugated multifunctional PLGA-ZnS nanoparticles. Nanomedicine (Lond) 2012;7(4):507–19. doi: 10.2217/nnm.11.139. Epub 2012/04/05. [DOI] [PubMed] [Google Scholar]
- 36.Yang K, Zhang FJ, Tang H, Zhao C, Cao YA, Lv XQ, et al. In-vivo imaging of oral squamous cell carcinoma by EGFR monoclonal antibody conjugated near-infrared quantum dots in mice. International journal of nanomedicine. 2011;6:1739–45. doi: 10.2147/IJN.S23348. Epub 2011/10/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X, Wang Y, Hnatowich DJ. A nanoparticle for tumor targeted delivery of oligomers. Methods Mol Biol. 2011;764:91–105. doi: 10.1007/978-1-61779-188-8_6. Epub 2011/07/13. [DOI] [PubMed] [Google Scholar]
- 38.Ding B, Wu X, Fan W, Wu Z, Gao J, Zhang W, et al. Anti-DR5 monoclonal antibody-mediated DTIC- loaded nanoparticles combining chemotherapy and immunotherapy for malignant melanoma: target formulation development and in vitro anticancer activity. International journal of nanomedicine. 2011;6:1991–2005. doi: 10.2147/IJN.S24094. Epub 2011/10/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chikkaveeraiah BV, Solda A, Choudhary D, Maran F, Rusling JF. Ultrasensitive nanostructured immunosensor for stem and carcinoma cell pluripotency gatekeeper protein NANOG. Nanomedicine (Lond) 2012;7(7):957–65. doi: 10.2217/nnm.11.178. Epub 2012/03/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deutscher SL. Phage display in molecular imaging and diagnosis of cancer. Chem Rev. 2010;110(5):3196–211. doi: 10.1021/cr900317f. Epub 2010/02/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kehoe JW, Kay BK. Filamentous phage display in the new millennium. Chem Rev. 2005;105(11):4056–72. doi: 10.1021/cr000261r. Epub 2005/11/10. [DOI] [PubMed] [Google Scholar]
- 42.Han M, Kickhoefer VA, Nemerow GR, Rome LH. Targeted vault nanoparticles engineered with an endosomolytic peptide deliver biomolecules to the cytoplasm. ACS nano. 2011;5(8):6128–37. doi: 10.1021/nn2014613. Epub 2011/07/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandoval MA, Sloat BR, Lansakara PD, Kumar A, Rodriguez BL, Kiguchi K, et al. EGFR-targeted stearoyl gemcitabine nanoparticles show enhanced anti-tumor activity. Journal of controlled release : official journal of the Controlled Release Society. 2012;157(2):287–96. doi: 10.1016/j.jconrel.2011.08.015. Epub 2011/08/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Pressly ED, Abendschein DR, Hawker CJ, Woodard GE, Woodard PK, et al. Targeting angiogenesis using a C-type atrial natriuretic factor-conjugated nanoprobe and PET. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2011;52(12):1956–63. doi: 10.2967/jnumed.111.089581. Epub 2011/11/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xin H, Sha X, Jiang X, Chen L, Law K, Gu J, et al. The brain targeting mechanism of Angiopep conjugated poly(ethylene glycol)-co-poly(epsilon-caprolactone) nanoparticles. Biomaterials. 2012;33(5):1673–81. doi: 10.1016/j.biomaterials.2011.11.018. Epub 2011/12/03. [DOI] [PubMed] [Google Scholar]
- 46.Scari G, Porta F, Fascio U, Avvakumova S, Dal Santo V, De Simone M, et al. Gold nanoparticles capped by a GC-containing peptide functionalized with an RGD motif for integrin targeting. Bioconjugate chemistry. 2012;23(3):340–9. doi: 10.1021/bc200143d. Epub 2012/03/02. [DOI] [PubMed] [Google Scholar]
- 47.Ming X, Feng L. Targeted delivery of a splice-switching oligonucleotide by cationic polyplexes of RGD-oligonucleotide conjugate. Molecular pharmaceutics. 2012;9(5):1502–10. doi: 10.1021/mp300113c. Epub 2012/04/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou A, Wei Y, Wu B, Chen Q, Xing D. Pyropheophorbide A and c(RGDyK) comodified chitosan- wrapped upconversion nanoparticle for targeted near-infrared photodynamic therapy. Molecular pharmaceutics. 2012;9(6):1580–9. doi: 10.1021/mp200590y. Epub 2012/04/27. [DOI] [PubMed] [Google Scholar]
- 49.Lv PP, Ma YF, Yu R, Yue H, Ni DZ, Wei W, et al. Targeted delivery of insoluble cargo (paclitaxel) by PEGylated chitosan nanoparticles grafted with Arg-Gly-Asp (RGD). Molecular pharmaceutics. 2012;9(6):1736–47. doi: 10.1021/mp300051h. Epub 2012/05/09. [DOI] [PubMed] [Google Scholar]
- 50.Zhang F, Huang X, Zhu L, Guo N, Niu G, Swierczewska M, et al. Noninvasive monitoring of orthotopic glioblastoma therapy response using RGD-conjugated iron oxide nanoparticles. Biomaterials. 2012;33(21):5414–22. doi: 10.1016/j.biomaterials.2012.04.032. Epub 2012/05/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wadajkar AS, Bhavsar Z, Ko CY, Koppolu B, Cui W, Tang L, et al. Multifunctional particles for melanoma-targeted drug delivery. Acta biomaterialia. 2012;8(8):2996–3004. doi: 10.1016/j.actbio.2012.04.042. Epub 2012/05/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koczulla AR, Bals R. Antimicrobial peptides: current status and therapeutic potential. Drugs. 2003;63(4):389–406. doi: 10.2165/00003495-200363040-00005. Epub 2003/02/01. [DOI] [PubMed] [Google Scholar]
- 53.Bradshaw J. Cationic antimicrobial peptides : issues for potential clinical use. BioDrugs : clinical immunotherapeutics, biopharmaceuticals and gene therapy. 2003;17(4):233–40. doi: 10.2165/00063030-200317040-00002. Epub 2003/08/06. [DOI] [PubMed] [Google Scholar]
- 54.Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacological reviews. 2003;55(1):27–55. doi: 10.1124/pr.55.1.2. Epub 2003/03/05. [DOI] [PubMed] [Google Scholar]
- 55.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363(6428):446–8. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 56.Muyldermans S, Atarhouch T, Saldanha J, Barbosa Ja, Hamers R. Sequence and structure of VH domain from naturally occurring camel heavy chain immunoglobulins lacking light chains. Protein Eng. 1994;7(9):1129–35. doi: 10.1093/protein/7.9.1129. [DOI] [PubMed] [Google Scholar]
- 57.Desmyter A, Transue TR, Ghahroudi MA, Thi MH, Poortmans F, Hamers R, et al. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat Struct Biol. 1996;3(9):803–11. doi: 10.1038/nsb0996-803. [DOI] [PubMed] [Google Scholar]
- 58.Martin F, Volpari C, Steinkuhler C, Dimasi N, Brunetti M, Biasiol G, et al. Affinity selection of a camelized V(H) domain antibody inhibitor of hepatitis C virus NS3 protease. Protein Eng. 1997;10(5):607–14. doi: 10.1093/protein/10.5.607. [DOI] [PubMed] [Google Scholar]
- 59.Reiter Y, Schuck P, Boyd LF, Plaksin D. An antibody single-domain phage display library of a native heavy chain variable region: isolation of functional single-domain VH molecules with a unique interface. J Mol Biol. 1999;290(3):685–98. doi: 10.1006/jmbi.1999.2923. [DOI] [PubMed] [Google Scholar]
- 60.Riechmann L, Muyldermans S. Single domain antibodies: comparison of camel VH and camelised human VH domains. J Immunol Methods. 1999;231(1-2):25–38. doi: 10.1016/s0022-1759(99)00138-6. [DOI] [PubMed] [Google Scholar]
- 61.Muyldermans S. Single domain camel antibodies: current status. J Biotechnol. 2001;74(4):277–302. doi: 10.1016/s1389-0352(01)00021-6. [DOI] [PubMed] [Google Scholar]
- 62.Conrath Ke, Lauwereys M, Galleni M, Matagne A, Frere Jm, Kinne J, et al. Beta-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob Agents Chemother. 2001;45(10):2807–12. doi: 10.1128/AAC.45.10.2807-2812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Desmyter A, Decanniere K, Muyldermans S, Wyns L. Antigen specificity and high affinity binding provided by one single loop of a camel single-domain antibody. J Biol Chem. 2001;276(28):26285–90. doi: 10.1074/jbc.M102107200. [DOI] [PubMed] [Google Scholar]
- 64.Holt Lj, Herring C, Jespers Ls, Woolven Bp, Tomlinson Im. Domain antibodies: proteins for therapy. Trends Biotechnol. 2003;21(11):484–90. doi: 10.1016/j.tibtech.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 65.Chen W, Zhu Z, Feng Y, Xiao X, Dimitrov Ds. Construction of a large phage-displayed human antibody domain library with a scaffold based on a newly identified highly soluble, stable heavy chain variable domain. J Mol Biol. 2008;382(3):779–89. doi: 10.1016/j.jmb.2008.07.054. Epub 2008 Jul 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen W, Zhu Z, Feng Y, Dimitrov Ds. Human domain antibodies to conserved sterically restricted regions on gp120 as exceptionally potent cross-reactive HIV-1 neutralizers. Proc Natl Acad Sci U S A. 2008;105(44):17121–6. doi: 10.1073/pnas.0805297105. Epub 2008 Oct 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nygren PA, Uhlen M. Scaffolds for engineering novel binding sites in proteins. Curr Opin Struct Biol. 1997;7(4):463–9. doi: 10.1016/s0959-440x(97)80108-x. [DOI] [PubMed] [Google Scholar]
- 68.Pessi A, Bianchi E, Crameri A, Venturini S, Tramontano A, Sollazzo M. A designed metal-binding protein with a novel fold. Nature. 1993;362(6418):367–9. doi: 10.1038/362367a0. [DOI] [PubMed] [Google Scholar]
- 69.Martin F, Toniatti C, Salvati AL, Venturini S, Ciliberto G, Cortese R, et al. The affinity-selection of a minibody polypeptide inhibitor of human interleukin-6. Embo J. 1994;13(22):5303–9. doi: 10.1002/j.1460-2075.1994.tb06864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McConnell SJ, Hoess RH. Tendamistat as a scaffold for conformationally constrained phage peptide libraries. J Mol Biol. 1995;250(4):460–70. doi: 10.1006/jmbi.1995.0390. [DOI] [PubMed] [Google Scholar]
- 71.Skerra A. Engineered protein scaffolds for molecular recognition. J Mol Recognit. 2000;13(4):167–87. doi: 10.1002/1099-1352(200007/08)13:4<167::AID-JMR502>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 72.Binz Hk, Pluckthun A. Engineered proteins as specific binding reagents. Curr Opin Biotechnol. 2005;16(4):459–69. doi: 10.1016/j.copbio.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 73.Binz Hk, Amstutz P, Pluckthun A. Engineering novel binding proteins from nonimmunoglobulin domains. Nat Biotechnol. 2005;23(10):1257–68. doi: 10.1038/nbt1127. [DOI] [PubMed] [Google Scholar]
- 74.Skerra A. Alternative non-antibody scaffolds for molecular recognition. Curr Opin Biotechnol. 2007;18(4):295–304. doi: 10.1016/j.copbio.2007.04.010. Epub 2007 Jul 20. [DOI] [PubMed] [Google Scholar]
- 75.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu Rev Biochem. 1999;68:611–47. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 76.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–10. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 77.Daniels Da, Chen H, Hicke Bj, Swiderek Km, Gold L. A tenascin-C aptamer identified by tumor cell SELEX: systematic evolution of ligands by exponential enrichment. Proc Natl Acad Sci U S A. 2003;100(26):15416–21. doi: 10.1073/pnas.2136683100. Epub 2003 Dec 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shangguan D, Li Y, Tang Z, Cao Zc, Chen Hw, Mallikaratchy P, et al. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc Natl Acad Sci U S A. 2006;103(32):11838–43. doi: 10.1073/pnas.0602615103. Epub 2006 Jul 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Campos WRL, Coopusamy D, Morris L, Mayosi BM, Khati M. Cytotoxicological Analysis of a gp120 Binding Aptamer with Cross-Clade Human Immunodeficiency Virus Type 1 Entry Inhibition Properties: Comparison to Conventional Antiretrovirals. Antimicrob Agents Chemother. 2009;53(7):3056–64. doi: 10.1128/AAC.01502-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Group TES. Preclinical and phase 1A clinical evaluation of an anti-VEGF pegylated aptamer (EYE001) for the treatment of exudative age-related macular degeneration. Retina. 2002;22(2):143–52. doi: 10.1097/00006982-200204000-00002. [DOI] [PubMed] [Google Scholar]
- 81.Group TES. Anti-vascular endothelial growth factor therapy for subfoveal choroidal neovascularization secondary to age-related macular degeneration: Phase II study results. Ophthalmology. 2003;110(5):979–86. doi: 10.1016/S0161-6420(03)00085-X. [DOI] [PubMed] [Google Scholar]
- 82.Pagratis NC, Bell C, Chang Y-F, Jennings S, Fitzwater T, Jellinek D, et al. Potent 2[prime]-amino-, and 2[prime]-fluoro-2[prime]- deoxyribonucleotide RNA inhibitors of keratinocyte growth factor. 1997;15(1):68–73. doi: 10.1038/nbt0197-68. [DOI] [PubMed] [Google Scholar]
- 83.Jayasena SD. Aptamers: An Emerging Class of Molecules That Rival Antibodies in Diagnostics. Clin Chem. 1999;45(9):1628–50. [PubMed] [Google Scholar]
- 84.Khati M. The future of aptamers in medicine. Journal of Clinical Pathology. 2010;63(6):480–7. doi: 10.1136/jcp.2008.062786. [DOI] [PubMed] [Google Scholar]
- 85.White RR, Shan S, Rusconi CP, Shetty G, Dewhirst MW, Kontos CD, et al. Inhibition of rat corneal angiogenesis by a nuclease-resistant RNA aptamer specific for angiopoietin-2. Proceedings of the National Academy of Sciences. 2003;100(9):5028–33. doi: 10.1073/pnas.0831159100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pieken W, Olsen D, Benseler F, Aurup H, Eckstein F. Kinetic characterization of ribonuclease-resistant 2'-modified hammerhead ribozymes. Science. 1991;253(5017):314–7. doi: 10.1126/science.1857967. [DOI] [PubMed] [Google Scholar]
- 87.Bell C, Lynam E, Landfair D, Janjic N, Wiles M. Oligonucleotide NX1838 inhibits VEGF165-mediated cellular responses in vitro. In Vitro Cellular & Developmental Biology - Animal. 1999;35(9):533–42. doi: 10.1007/s11626-999-0064-y. [DOI] [PubMed] [Google Scholar]
- 88.Seferos DS, Prigodich AE, Giljohann DA, Patel PC, Mirkin CA. Polyvalent DNA nanoparticle conjugates stabilize nucleic acids. Nano letters. 2009;9(1):308–11. doi: 10.1021/nl802958f. Epub 2008/12/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Giljohann DA, Seferos DS, Prigodich AE, Patel PC, Mirkin CA. Gene regulation with polyvalent siRNA-nanoparticle conjugates. J Am Chem Soc. 2009;131(6):2072–3. doi: 10.1021/ja808719p. Epub 2009/01/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee IH, An S, Yu MK, Kwon HK, Im SH, Jon S. Targeted chemoimmunotherapy using drug-loaded aptamer-dendrimer bioconjugates. Journal of controlled release : official journal of the Controlled Release Society. 2011;155(3):435–41. doi: 10.1016/j.jconrel.2011.05.025. Epub 2011/06/07. [DOI] [PubMed] [Google Scholar]
- 91.Yu MK, Kim D, Lee IH, So JS, Jeong YY, Jon S. Image-guided prostate cancer therapy using aptamer-functionalized thermally cross-linked superparamagnetic iron oxide nanoparticles. Small. 2011;7(15):2241–9. doi: 10.1002/smll.201100472. Epub 2011/06/08. [DOI] [PubMed] [Google Scholar]
- 92.Bhattacharyya S, Khan JA, Curran GL, Robertson JD, Bhattacharya R, Mukherjee P. Efficient delivery of gold nanoparticles by dual receptor targeting. Adv Mater. 2011;23(43):5034–8. doi: 10.1002/adma.201102287. Epub 2011/10/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kluza E, Jacobs I, Hectors SJ, Mayo KH, Griffioen AW, Strijkers GJ, et al. Dual-targeting of alphavbeta3 and galectin-1 improves the specificity of paramagnetic/fluorescent liposomes to tumor endothelium in vivo. Journal of controlled release : official journal of the Controlled Release Society. 2012;158(2):207–14. doi: 10.1016/j.jconrel.2011.10.032. Epub 2011/11/15. [DOI] [PubMed] [Google Scholar]
- 94.Ko HY, Choi KJ, Lee CH, Kim S. A multimodal nanoparticle-based cancer imaging probe simultaneously targeting nucleolin, integrin alphavbeta3 and tenascin-C proteins. Biomaterials. 2011;32(4):1130–8. doi: 10.1016/j.biomaterials.2010.10.034. Epub 2010/11/13. [DOI] [PubMed] [Google Scholar]
- 95.Basel MT, Shrestha TB, Troyer DL, Bossmann SH. Protease-sensitive, polymer-caged liposomes: a method for making highly targeted liposomes using triggered release. ACS nano. 2011;5(3):2162–75. doi: 10.1021/nn103362n. Epub 2011/02/15. [DOI] [PubMed] [Google Scholar]
- 96.Kennedy LC, Bear AS, Young JK, Lewinski NA, Kim J, Foster AE, et al. T cells enhance gold nanoparticle delivery to tumors in vivo. Nanoscale research letters. 2011;6(1):283. doi: 10.1186/1556-276X-6-283. Epub 2011/06/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cho HR, Choi SH, Lee N, Hyeon T, Kim H, Moon WK. Macrophages homing to metastatic lymph nodes can be monitored with ultrasensitive ferromagnetic iron-oxide nanocubes and a 1.5T clinical MR scanner. PloS one. 2012;7(1):e29575. doi: 10.1371/journal.pone.0029575. Epub 2012/01/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228(4705):1315–7. doi: 10.1126/science.4001944. Epub 1985/06/14. [DOI] [PubMed] [Google Scholar]
- 99.Lindqvist BH, Naderi S. Peptide presentation by bacteriophage P4. FEMS microbiology reviews. 1995;17(1-2):33–9. doi: 10.1111/j.1574-6976.1995.tb00185.x. Epub 1995/08/01. [DOI] [PubMed] [Google Scholar]
- 100.Sternberg N, Hoess RH. Display of peptides and proteins on the surface of bacteriophage lambda. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(5):1609–13. doi: 10.1073/pnas.92.5.1609. Epub 1995/02/28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Santini C, Brennan D, Mennuni C, Hoess RH, Nicosia A, Cortese R, et al. Efficient display of an HCV cDNA expression library as C-terminal fusion to the capsid protein D of bacteriophage lambda. Journal of molecular biology. 1998;282(1):125–35. doi: 10.1006/jmbi.1998.1986. Epub 1998/09/12. [DOI] [PubMed] [Google Scholar]
- 102.Hashemi H, Pouyanfard S, Bandehpour M, Noroozbabaei Z, Kazemi B, Saelens X, et al. Immunization with M2e-displaying T7 bacteriophage nanoparticles protects against influenza A virus challenge. PloS one. 2012;7(9):e45765. doi: 10.1371/journal.pone.0045765. Epub 2012/10/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lee JH, Domaille DW, Cha JN. Amplified protein detection and identification through DNA-conjugated M13 bacteriophage. ACS nano. 2012;6(6):5621–6. doi: 10.1021/nn301565e. Epub 2012/05/17. [DOI] [PubMed] [Google Scholar]