Abstract
The leucine rich repeat kinase 2 (LRRK2/dardarin) is implicated in autosomal dominant familial and sporadic Parkinson’s disease (PD); mutations in LRRK2 account for up to 40% of PD cases in some populations. LRRK2 is a large protein with a kinase domain, a GTPase domain, and multiple potential protein interaction domains. As such, delineating the functional pathways for LRRK2 and mechanisms by which PD-linked variants contribute to age-related neurodegeneration could result in pharmaceutically tractable therapies. A growing number of recent studies implicate dysregulation of mitogen activated protein kinases 3 and 1 (also known as ERK1/2) as possible downstream mediators of mutant LRRK2 effects. As these master regulators of growth, differentiation, neuronal plasticity and cell survival have also been implicated in other PD models, a set of common cell biological pathways may contribute to neuronal susceptibility in PD. Here, we review the literature on several major cellular pathways impacted by LRRK2 mutations – autophagy, microtubule/cytoskeletal dynamics, and protein synthesis – in context of potential signaling crosstalk involving the ERK1/2 and Wnt signaling pathways. Emerging implications for calcium homeostasis, mitochondrial biology and synaptic dysregulation are discussed in relation to LRRK2 interactions with other PD gene products. It has been shown that substantia nigra neurons in human PD and Lewy body dementia patients exhibit cytoplasmic accumulations of ERK1/2 in mitochondria, autophagosomes and bundles of intracellular fibrils. Both experimental and human tissue data implicate pathogenic changes in ERK1/2 signaling in sporadic, toxin-based and mutant LRRK2 settings, suggesting engagement of common cell biological pathways by divergent PD etiologies.
1. Introduction
Parkinson’s disease (PD) is the second most common neurodegenerative disorder, affecting approximately 1-2% of the population over the age of 65 [1]. Clinically, PD is characterized by resting tremor, rigidity, bradykinesia, and stooped posture. The majority of these motor impairments arise from the loss of dopaminergic neurons in the substantia nigra pars compacta (SNpc), which results in depletion of dopamine from the nigro-striatal system. Formation of Lewy bodies (LB), intracytoplasmic inclusion bodies primarily composed of α-synuclein, often accompanies this selective neurodegeneration [2]. Though the underlying cause of sporadic PD remains unidentified, exposure to environmental toxins and genetic factors have been implicated in PD pathogenesis.
Higher incidences of sporadic PD have been reported in rural populations associated with agricultural work. This observation suggests that pesticides and herbicides may contribute to PD [3, 4]. Rotenone, a mitochondrial complex I inhibitor, is one of the pesticides that has been linked to PD. It was shown to cause selective degeneration of dopaminergic neurons after systemic administration to rats, and is currently used as a toxin model to study sporadic PD [5]. A toxin previously shown to induce human parkinsonism is MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine). The oxidized product of MPTP, MPP+ is selectively taken up by dopaminergic neurons, accumulates in the mitochondria and inhibits complex I [6]. Both toxins induce mitochondrial damage, implicating a key role for dysregulated mitochondrial homeostasis in sporadic PD.
Though comprising only a fraction of total PD cases, a genetic component to parkinsonian neurodegeneration has become well established (reviewed in [7]). There are at least 16 PARK loci that have been identified and are associated with either autosomal recessive or autosomal dominant PD [8]. Causal mutations have been identified in a number of these loci, and the functional consequence of these mutations on the encoded proteins is an area of intense study. Mutations in the leucine-rich repeat kinase 2 gene (LRRK2) are the most common known cause of familial PD [9-12]. The biochemistry of LRRK2 has been reviewed elsewhere [13, 14]. In this review we discuss major cell biologic processes and signaling pathways that may be regulated by LRRK2, and how mutations in LRRK2 may serve to promote neurodegeneration.
2. LRRK2 and Parkinson’s disease
The LRRK2 locus was first identified in a Japanese family with autosomal dominant parkinsonism by linkage analysis, involving a novel PARK locus on chromosome 12q12 that was named PARK8 [15]. In 2004, two independent groups cloned the responsible gene and identified pathogenic mutations in the PARK8 locus [11, 16].
LRRK2 is a multidomain, 2527 amino acid (~280kDa) protein. The N-terminus consists of an ankyrin-like domain and leucine-rich repeats (LRR). LRRK2 also contains a Ras of complex (ROC) GTPase domain adjacent to a C-terminal of ROC (COR) domain that functions as a linker between the N- and C-terminus of the protein. The kinase and a WD40 domain are present in the C-terminal portion of the protein [17]. The LRRK2 kinase domain shares a close homology with mixed linkage kinases (MLKs), which comprise a sub-class of the mitogen activated protein kinase kinase kinase (MAPKKK) family.
Expression analysis by RT-PCR [11] and Northern Blot [16] revealed that LRRK2 is widely expressed in various tissues and cell types, including the brain, lungs, liver, skeletal muscle and kidneys. Within the brain, it is expressed in multiple regions, with a high level of expression in the putamen, where terminals of the substantia nigra region vulnerable to neurodegeneration in PD project. LRRK2 is predominately localized in the cytoplasm, where it associates with mitochondria [18], endoplasmic reticulum, Golgi, microtubules [10] and vesicular structures such as lysosomes and endosomes [19].
Five pathogenic mutations in LRRK2 have been linked to familial parkinsonisms. These include I2020T [12], G2019S [20-22], R1441G [9], R1441C [11] and Y1699C [16]. The most prevalent mutation is G2019S in the kinase domain which increases LRRK2 kinase activity and is estimated to occur in approximately 6-8% of all familial and 1-2% of sporadic PD cases [23]. LRRK2-G2019S mutants can form inclusion bodies when expressed in cultured cells or neurons, associated with neuronal toxicity [24]. The toxic effects of this mutation can be ameliorated using the selective LRRK2 inhibitor, LRRK2-IN-1 [25].
Most LRRK2 kinase activity studies have been done in vitro using generic substrates, such as myelin basic protein (MBP); identifying in vivo targets of LRRK2 remains an area of active investigation. A number of proteins represent potential substrates, including: Ezrin/Radixin/Moesin (ERM) [26], beta-tubulin [27], 4E-BP1 [28] and alpha-synuclein [29]. In addition to the in vitro analyses of LRRK2 targets, a variety of cell culture and animal studies indicate that LRRK2 may play a role in the regulation of cell death, autophagy, the cytoskeleton, protein translation and cell signaling pathways.
3. The cell biology of LRRK2 and its mutations
3.1 LRRK2 and Autophagy
Autophagy is an important catabolic process in which cytoplasmic components are engulfed into a double membrane vesicle called an autophagosome, after which they are delivered by fusion events to the lysosome for hydrolytic degradation [30]. Cells use autophagy to maintain protein and organellar integrity on an ongoing basis. Autophagy is induced in response to starvation [31, 32] and other cellular stresses implicated in a number of neurodegenerative diseases (reviewed in [33]). In neurons, suppression of basal autophagy results in the accumulation of misfolded proteins and neurodegeneration [34, 35]. Autophagy is a common response to injury, but depending on the context, autophagy induction can be adaptive or maladaptive [36].
A number of studies have emerged that implicate a link between LRRK2 and autophagy regulation, but the exact mechanisms are still poorly understood. Overexpression of pathogenic LRRK2 mutants induces autophagosome accumulation in multiple cell types, including neurons [37, 38], neuronal cell lines [39] and kidney cells [40].
The first study implicating LRRK2 in the (mis)regulation of autophagy utilized retinoic acid differentiated SH-SY5Y cells to show that the G2019S mutant of LRRK2, but not the kinase-deficient mutant, increased autophagosomes in neurites. More mature autophagic vacuoles (multivesicular-like bodies and lysosomes) were observed in the soma [39] suggestive of retrograde trafficking and maturation. RNAi knockdown of components of the autophagic machinery, LC3 and Atg7, indicated that the LRRK2-induced neurite retraction is mediated by autophagy [39], confirming that mutations in LRRK2 regulate the morphology of neuronal processes in PD [41]. Subsequent studies in primary neurons demonstrated that Protein kinase A protects against G2019S- or R1441C-LRRK2-mediated dendrite shortening by phosphorylating and inactivating LC3, the mammalian homolog to the yeast Atg8 autophagy protein [37].
Perturbed calcium homeostasis has been shown to play a role in LRRK2-induced autophagy. Autophagy induced by overexpression of wild type or G2019S LRRK2 has been reported to occur through activation of the Ca2+/CaMKK/AMPK pathway [42]. Treatment with inhibitors of this pathway, such as compound c (AMPK inhibitor), STO-609 (CaMKK-a/b inhibitor), or chelation of intracellular calcium with BAPTA-AM abrogates the increase in autophagosome number induced by LRRK2 overexpression [42]. Additionally, deficiencies in calcium handling lie upstream of dendritic mitophagy, which contributes to dendrite retraction in primary neurons overexpressing G2019S- or R1441C-LRRK2 [38]. Inhibition of calcium influx through L-type voltage gated calcium channels prevents mitochondrial depolarization, mitophagy and dendrite shortening.
It remains controversial whether the autophagosome accumulation induced by LRRK2 is due to an increase in autophagy induction, inhibition of autophagic flux, or both. Overexpression of the LRRK2-R1441G in HEK293 cells caused accumulation of p62 and large autolysosomes, [40]. This study also reported that knockdown of GFP-tagged LRRK2 increased autophagic flux under starvation conditions, enhancing cell survival; the effects under nutrient rich conditions were not investigated. Since bafilomycin inhibits late stages of lysosomal degradation [43], it is unlikely that LRRK2 siRNA would be able to restore flux through the system, suggesting additional mechanisms of cytoprotection against starvation. In a recent study involving a LRRK2 kinase inhibitor (LRRK2-IN-1), LRRK2 activity was placed upstream of autophagy initiation. Though this study highlighted the role of endogenous LRRK2 in autophagic regulation, it did not address how pathogenic mutations associated with PD (LRRK2-G2019S) that are linked to increased kinase activity may induce autophagy [44].
In vivo studies also suggest that altered LRRK2 expression leads to dysregulation of autophagy. Lrrk2−/− mice are viable, physically normal, and have an intact dopaminergic system [45]. However, alpha-synuclein and ubiquitinated protein accumulation, accompanied by dysregulation of the autophagy-lysosomal pathway, has been observed in the kidneys. The observed alterations of autophagic activity are age-dependent and bi-phasic. At 1 month of age there are no observable differences, but autophagic activity is enhanced at 7 months, and then reduced at 20 months [46]. The authors suggest that impaired clearance of proteins observed in aged mice was due to decreased recycling of induced autolysosomes and increased accumulation of autophagic vacuoles.
Transgenic mice expressing LRRK2-G2019S show progressive degeneration of dopaminergic neurons, as demonstrated by shorter neurite length and branching, leading to an autophagy-induced reduction in neurite complexity. This loss of DA neurons, is more pronounced in older G2019S-LRRK2 transgenic mice than younger transgenic or wild type mice, and requires a certain level of LRRK2 overexpression [47]. In Drosophila, overexpression of LRRK2-G2019S mutation causes increased loss of photoreceptors in the retina due to elevated autophagy caused by upregulation of key autophagy gene, Atg5, compared to wild type LRRK2 overexpression [48].
LRRK2 may also be a victim of dysregulated autophagy rather than the perpetrator. Conditional deletion of the essential autophagic gene, Atg7, in DA neurons caused a presynaptic accumulation of alpha-synuclein, p62 and LRRK2 proteins in large intracellular inclusions in the brain [49]. Similarly, there was almost a 4-fold increase in LRRK2 mRNA in Atg5−/− MEFs compared to control cells. These results showed that dysregulation of autophagy could lead to an abnormal elevation in expression of LRRK2 [49].
The role that LRRK2 plays in autophagy regulation remains unclear. However, a large body of evidence suggests that autophagosome accumulation induced by expression of PD-associated mutants contributes to injury in LRRK2 models of PD. Alterations in calcium handling and LRRK2 kinase activity have been shown to contribute to this phenotype, but further investigation is required to unravel the mechanism of autophagy dysregulation in these models.
3.2 LRRK2 and Microtubule dynamics
Cytoskeletal components are important for maintaining the structural support for neurons, vesicular biogenesis, organelle or vesicle transport, and synaptic signaling [50]. Dysfunction in cytoskeleton dynamics is often associated with neurodegenerative diseases [51, 52]. Rotenone, a mitochondrial complex I inhibitor, was shown to induce depolymerization of microtubules leading to disruption in the transport of dopamine vesicles and their accumulation in the soma and finally, causing oxidative stress due to the leakage of dopamine from these vesicles [53].
LRRK2 has been shown to co-localize with β-tubulin [10, 19] and to interact directly with α/β tubulin through the ROC GTPase domain [19, 54]. Wild-type and R1441C LRRK2 had similar affinities for α/β tubulin [19]. Human LRRK2 interacted with β tubulin and preferentially phosphorylated tubulin from bovine brain, which was enhanced three-fold by the G2019S mutation [27]. Interestingly, phosphorylated β-tubulin did not immunoprecipitate with LRRK2 in HEK293 cells despite this interaction. When incubated with microtubule associated protein-rich medium, G2019S-LRRK2 significantly enhanced microtubule assembly/stability. These results suggest that increased phosphorylation of β tubulin by G2019S-LRRK2 mutant results in decreased microtubule dynamics, affecting the structure of the neurons [27]. Recently, multiple PD associated LRRK2 mutants (R1441C, R1441G, Y1699C and I2020T) have been shown to form filamentous structures that associate with microtubules [55]. This association requires kinase function and the WD40 domain. Interestingly, the G2019S mutation did not show any filament formation [55], raising the possibility of different pathogenic mechanisms by the mutation that clearly elevates kinase activity versus the mutations that do not.
The interaction of LRRK2 and tau (a microtubule-associated protein) leads to increased phosphorylation of tau and shorter neurite processes [41]. This phenotype was prevented with the expression of kinase dead (K1906M) fragment of LRRK2, although caution should be taken due to the need for further characterization of this fragment. Interestingly, this interaction and the in vitro phosphorylation of tau by LRRK2 are dependent on tubulin [51]. G2019S- [56] and I2020T- [57], but not R1441C-, LRRK2 mutants phosphorylated more tau than wild-type LRRK2 [58]. Overexpression of the G2019S PD-associated mutant of LRRK2 lead to tau-positive inclusions in neurons and LRRK2 co-localized with tau in these inclusions. In Drosophila, expression of G2019S-LRRK2 induced mislocalization of tau in the dendrites that leads to dendrite degeneration [59]. Tau phosphorylation was dependent on the expression of LRRK2, as increased LRRK2 expression increased tau phosphorylation, which was conversely reduced by RNAi knockdown of LRRK2. The expression level of LRRK2 was inversely correlated with neurite length [57, 58]. Taken together with the autophagy and mitophagy data discussed above, it appears that multiple LRRK2-associated mechanisms may contribute to LRRK2-mediated neurite shortening.
Elongation factor 1A (EF1A), a GTPase that transports aminoacyl-tRNA to the ribosomes during protein translation and is essential for cytoskeletal organization [60, 61], has also shown to interact with LRRK2 [62]. Binding of EF1A significantly reduced the autophosphorylation activity of wild type-, G2019S- and R1441C-LRRK2. The microtubule assembly activity of EF1A was shown to be impaired in the presence of wild-type and G2019S LRRK2 in an in vitro microtubule polymerization assay. Phospho-EF1A could not be detected in this study suggesting that EF1A is not the target of LRRK2 [62].
In dopaminergic neurons, either knockdown of LRRK2 [63] or transgenic mice expressing LRRK2-G2019S mutations [64] led to shortening of neurite extensions. Using the KinasE Substrate Tracking and ELucidation screening (KESTREL) approach, moesin, a protein that anchors the actin cytoskeleton to the plasma membrane, as well as ezrin and radixin were proposed as G2019S-LRRK2 targets [26]. A follow-up study from the same group, however, failed to detect phosphorylation of ERM proteins in HEK293 cells, even after overexpression of LRRK2-wt or LRRK2-G2019S [65]. On the other hand, mutant LRRK2-G2019S did enhance phosphorylation of ERM proteins in another study, which was correlated with retardation of neurite outgrowth, and inhibition of ERM phosphorylation was able to rescue the G2019S-phenotype [64]. Data from NIH3T3 cells suggest that LRRK2 can directly bind F-actin, affecting its polymerization and depolymerization in vitro. These discrepancies suggest that either LRRK2 status modulates ERK phosphorylation indirectly, or that detection of phosphorylated ERM proteins might be dependent on experimental conditions. It will be interesting to see if other groups can detect phospho-ERMs in mammalian neurons.
In summary, current evidence suggests that either too much or too little LRRK2 activity may contribute to neurite shortening, although initial studies indicated that LRRK2 knockdown caused elongated neuronal processes [41]. LRRK2 may interact with and/or phosphorylate or regulate the phosphorylation of several structural and regulatory components of the actin cytoskeleton and microtubule network. Given that neurite shortening is a commonly observed phenotype in neuronal cells expressing pathogenic LRRK2 mutants, understanding these cytoskeletal associations and their possible relationships with autophagy or mitochondrial dynamics may provide insight into the PD pathogenesis.
3.3 LRRK2 and protein translation control
LRRK2 was shown to interact with genes in the TOR/4E-BP pathway in Drosophila [28]. In the same study, human wild type LRRK2 and an I2020T mutant were able to phosphorylate 4E-BP1. Eukaryotic 4E-BP1 binds to initiation factor 4E (eIF4E) and inhibits its function, while phosphorylation alleviates this inhibition. Phosphorylation of 4E-BP1 by LRRK2 decreases survival of Drosophila neurons. In contrast, loss of LRRK2 or gain of 4E-BP1 function rescues pathology in Drosophila Parkin/PINK1 models [66]. Hence, it has been hypothesized that increased LRRK2 expression or kinase activity increases protein translation to a level that overwhelms the cellular degradation machinery, resulting in accumulation of unwanted proteins [28, 66].
However, this interpretation has been challenged by other studies. While phosphorylation of 4E-BP by LRRK2 can be demonstrated in in vitro, it is a relatively poor substrate [67]. Moreover, increased phosphorylation was not observed when LRRK2 was overexpressed in HEK cells. This study suggested that 4E-BP phosphorylation may be an indirect phenomenon related to p38-mediated cell stress [67]. Similar results were obtained in another study in which the phosphorylation status of endogenous 4E-BP1 was not altered in the brains of LRRK2 knockout or mutant LRRK2 transgenic mice, nor were there changes in idiopathic or G2019S PD patient brains, suggesting that 4E-BP1 is not a direct substrate for LRRK2 in mammalian systems [68].
3.4 Mitochondrial pathology in LRRK2 pathogenesis
Mitochondrial dysfunction is widely recognized as a contributor to parkinsonian injury. Common themes observed in multiple genetic or toxin models include increased reactive oxygen species production, altered mitochondrial dynamics, decreases in mitochondrial membrane potential, and impaired calcium homeostasis. The recessive parkinsonian genes PINK1, Parkin and DJ-1 have been particularly implicated in mitochondrial homeostasis, and LRRK2 interacts with these recessive PD genes in Drosophila [69]. Interestingly, recent studies also implicate mitochondrial dysfunction in LRRK2 pathogenic mechanisms.
A loss of mitochondrial membrane potential has been reported in primary mouse cortical neurons, immortalized cell lines, and PD patient fibroblasts expressing mutant LRRK2 [38, 70]. In addition, elevated ROS and diminished ATP levels have been reported in human fibroblasts derived from patients with the G2019S pathogenic mutation [71]. Decreased ΔΨm may initiate mitochondrial fission, and knockdown of PINK1 limits phosphorylation of Drp1 at Ser637, enabling it to be recruited to mitochondria to facilitate fission [72, 73]. Two recent studies suggest that overexpression of WT or mutant LRRK2 also leads to mitochondrial fragmentation in mouse primary cortical neurons, SH-SY5Y, and HeLa cells [74, 75]. Further, a direct interaction between LRRK2 and Drp1 was described in these reports.
In order to maintain healthy mitochondrial networks, damaged mitochondria need to be recycled by mitophagy. Parkin and PINK1 are integrally involved in regulating the selective, autophagic clearance of mitochondria known as mitophagy [76, 77]. We have recently reported that loss of dendritic mitochondria in G2019S or R1441C LRRK2 neurons can be reversed by inhibition of autophagy [38]. Interestingly, perturbed calcium homeostasis was the underlying cause of dendritic mitophagy induced by mutant LRRK2 expression. Consistent with our findings, another group has reported that elevated intracellular calcium levels are necessary for LRRK2 induced autophagy [42]. Delayed calcium clearance has also been observed following PINK1 knockdown and in cells expressing mutant PINK1; thus, impaired calcium buffering is a shared feature of mutant LRRK2 and PINK1-related pathogenic mechanisms [78-80].
Overexpression of PKA protects against injuries induced by either PINK1 knockdown [72] or G2019S-LRRK2 expression [37], providing further evidence that converging signaling cascades contribute to pathogenesis in PINK1 and LRRK2 models. Phosphorylation of Drp1 by PKA appears to be the primary mechanism of protection in PINK1 deficient cells [72], whereas of phosphorylation of LC3 is involved in PKA-mediated protection against injury in cells expressing mutant LRRK2 [37]. PINK also regulates phosphorylation of LC3, albeit through indirect mechanisms, and overexpression of WT-PINK1 prevents autophagy and neurite retraction in cells expressing G2019S-LRRK2 (SJ Cherra & CT Chu, unpublished data).
As with other genetic models of PD, altered mitochondrial dynamics, impaired mitochondrial calcium buffering, elevations in ROS levels, and decreased ΔΨm have been reported in cells expressing PD-associated LRRK2 mutants. The parallels that can be drawn between pathogenic mechanisms in genetic models of PD provide compelling evidence that aberrant regulation of convergent signaling pathways contribute to mitochondrial dysfunction.
3.5 Endosomal/synaptic dysregulation in LRRK2 pathogenesis
The cytoskeletal, autolysosomal and protein synthesis effects of LRRK2 have been shown to affect synaptic vesicle function/dysfunction and the dynamics of recycling endosomes [81]. In Drosophila, loss of LRRK2 causes synaptic overgrowth, while its overexpression has opposite effects [82]. LRRK2 protein targets have been proposed in both the pre- and postsynaptic compartments, namely the microtubule-binding protein Futsch and the protein translation inhibitor 4E-BP, respectively. Presynaptic silencing of LRRK2 causes an increase in vesicular dynamics within the recycling pool [83]. In the postsynaptic compartment, mutant LRRK2 promotes calcium overload through engagement of L-type calcium channels [38] and glutamate receptors (E Plowey & CT Chu, unpublished data). While the molecular mechanisms underlying these synaptic alterations remain unknown, other studies indicate that they may reflect more general problems with RAB7L1-related vesicular protein transport and endosomal, lysosomal and Golgi trafficking [84]. LRRK2 was shown to interact and partially co-localize with Rab5b. Additionally, altering the levels of LRRK2 expression can modulate endocytosis of synaptic vesicles [85]. Interestingly, either overexpression of wild type or PD associated mutants, or knockdown of LRRK2 was shown to slow down the rate of endocytosis. Using yeast as a model system, the defect in endocytosis by LRRK2 was found to be dependent on its GTPase activity [86]. A recent study identified EndoA as a direct substrate for LRRK2, and phosphorylation of EndoA in a Drosophila model system affects the endocytotic process [87]. The increased demand for autophagy generated by protein aggregation and mitochondrial dysfunction, combined with defects in endosomal-lysosomal processes create an imbalance in cellular quality control mechanisms and promotes autophagic stress [88].
In summary, LRRK2 has been shown to play a role in maintaining homeostasis at the synapse through multiple mechanisms. Interactions with known regulators of protein translation, vesicular transport, and autophagy and endocytosis are important for proper synaptic function. As many of the studies have been performed in Drosophila, further investigation is needed to determine the role of these mechanisms in mammalian LRRK2 models of PD.
3.6 Interactions of LRRK2 with the Wnt Signaling Pathway
3.6.1. The Wnt pathway and PD
Wnt (Wingless/Int) signaling pathways have been implicated in the regulation of neurogenesis and function of mature and post mitotic neurons [89]. Wnt ligands are extracellular factors that regulate the differentiation of neuronal stem cells into mature neurons [90]. The Wnt signaling cascade has three major branches; canonical, non-canonical/planar cell polarity and Wnt-Ca2+, of which the canonical pathway has been best described [91]. Activation of this pathway results in stabilization and nuclear recruitment of beta-catenin, which regulates gene expression [92, 93].
The Wnt signaling pathway is important to the development of DA neurons in the midbrain. Wnt1 and Wnt3a were shown to induce specification of committed dopaminergic precursors, whereas, Wnt5a induced their differentiation by promoting the maturation of Nurr+, a nuclear receptor essential for the development and maintenance of DA precursors into DA neurons [94]. Induced expression of Wnt5a in ventral midbrain neural stem cells generated 10 fold more DA neurons, and transplantation of these cells in Parkinson’s mice model resulted in functional recovery [95]. Moreover, compared to single KO mice, Wnt1−/− and Wnt5−/− double knockout mice showed greater loss of DA neurons and Nurr1+ cells [96]. In the absence of Wnt ligand, Nurr1 associates with LEF1 and functions as a co-repressor. The Nurr1 and beta catenin interaction is increased upon Wnt-mediated stabilization of beta-catenin. This disrupts the Nurr1-LEF1 co-repressor complex and induces the expression of beta-catenin responsive genes [97].
Several key components of Wnt-beta-catenin pathway are downregulated in monkeys treated with MPTP [98]. In mice, MPTP treatment of the ventral midbrain increases Wnt1 expression as well as deregulation of FZD-1 and beta-catenin expression, suggesting that the Wnt signaling pathway may contribute to neuroprotection against MPTP toxicity. Aged mice have decreased Wnt1 and loss of beta-catenin, which is correlated with failure to recover from acute MPTP toxicity [99]. Pesticide treatment, which induces sporadic PD, alters expression of Wnt signaling pathway components [100], and inhibits Wnt signaling by Dkk1 that promotes neurodegeneration in mice treated with 6-OHDA [101]. Similarly, Wnt1 has been shown to be protective in human neuroblastoma cell line SH-SY5Y when treated with 6-OHDA through activation of canonical pathway [102].
3.6.2. LRRK2-Wnt interactions
LRRK2 and Wnt signaling were first linked, when mRNA species involved in Wnt signaling pathway cascade was shown to be deregulated by knockdown of LRRK2 in SH-SY5Y cells [103]. Similarly, a systems biology approach revealed that many transcripts in the Wnt signaling cascade are co-regulated with LRRK2 [104].
The LRRK2 ROC-COR tandem domain was reported to interact with Dvl-1, an important mediator in Wnt signaling [105]. This interaction is disrupted by the Y1699C mutation and strengthened by R1441C and R1728 mutations in LRRK2 [105]. Additionally, LRRK2 functions as a scaffold where it interacts with components of beta-catenin destruction complex. Once recruited to plasma membrane, it has been shown to bind the Wnt-corepressor LRP6 upon stimulation. However, overexpression of LRRK2 itself does not stimulate the Wnt pathway. Instead, it requires co-expression of Dvl proteins to be stimulated by Wnt3a. This induction of Wnt signaling is dependent on the kinase and GTPase activity of LRRK2, as inhibition of kinase activity by LRRK2-IN-1 or the inactivating K1347A GTPase mutation prevents Wnt activation [106]. An interaction between LRRK2 and Sgg (the Drosophila homolog of GSK3β), a component of the beta-catenin destruction complex, has been reported in Drosophila [59]. This would imply a role for LRRK2, however, in downregulating canonical Wnt signaling, in contrast to the prior study.
As differential effects among Drosophila and mammalian systems have been observed with other PD-linked genes, it would be interesting to determine if LRRK2 promotes beta-catenin degradation in mammalian cells. As the Wnt signaling pathway is important for neurogenesis, the possible role of LRRK2 in regulation of this pathway, and how it is affected in PD, still needs to be determined. As chemical (kinase specific inhibitors) and molecular tools (knock-out, transgenic animals) become increasingly available, future research linking Wnt signaling and LRRK2 would be interesting.
3.7 Interactions of LRRK2 with MAPK/ERK1/2 Signaling Pathways
3.7.1 ERK1/2 signaling in parkinsonian toxin models
Extracellular-regulated kinase 1 and 2 (ERK1/2) belongs to the mitogen activated protein kinase (MAPK) family, which also includes c-Jun NH2-terminal kinase (JNK) and p38 kinase. These MAPKs are serine/threonine protein kinases that regulate diverse cellular functions such as cell growth, division, differentiation and cell death [107, 108]. LRRK2 has been implicated in regulating multiple branches of the MAPK superfamily, including the dual specificity kinases for the p38 MAPK and the JNK pathways [109, 110].
Activation of ERK1/2 has been shown to be protective as well as cytotoxic in different cell models. Transient, early activation of ERK1/2 is typically neuroprotective [111], whereas delayed and sustained ERK1/2 activation promotes neuronal cell death [111-114]. In the 6-hydroxydopamine (6-OHDA) model, it was found that there is a delayed phase of ERK1/2 activation that is attributable to mitochondrial ROS [115]. Interestingly, activated ERK1/2 is observed in midbrain neurons of human PD and diffuse Lewy body disease patients [116]. A similar increase in activated ERK1/2 was observed in tau associated neuropathology [117], which can be seen in some families with mutant LRRK2 [11]. Ultrastructural and dual immunofluorescence studies have demonstrated that activated ERK1/2 is associated with abnormal mitochondria, autophagosomes, and intracellular filaments [118]. Sustained ERK1/2 activation has also been shown to promote the autophagic clearance of mitochondria in neuronal cells [119], and suppress mitochondrial biogenesis in response to chronic MPP+ toxicity [120]. In addition, ERK1/2 has also been shown to modulate reactive oxygen species (ROS) [121-123], nitric oxide [124] and calcium (reviewed in [125]). Thus, ERK1/2 may be involved in several cellular pathways implicated in LRRK2 pathobiology: autophagy, microtubule dynamics, neurite shortening, altered mitochondrial calcium regulation and mitophagy.
3.7.2. LRRK2: the ERK1/2 link
The most common pathogenic mutation associated with LRRK2 is G2019S, which occurs in the MAPKKK domain of the protein. In the search for possible LRRK2 substrates, leukocytes from patients carrying the G2019S mutation were compared to healthy counterparts. Src, a non-receptor tyrosine kinase that activates both ERK1/2 and p38, was less phosphorylated in G2019S mutants. Similarly, HSP27, a downstream target of p38 was also less phosphorylated. However, there was an increase in phosphorylated ERK1/2 in G2019S leucocytes, suggesting that ERK1/2 could be activated by a different mechanism in these patients [126].
Neurite retraction, a consistent phenotype caused by expression of pathogenic LRRK2 mutations, is driven by ERK1/2-dependent mechanisms, including autophagy [39]. This suggests that ERK1/2 activation may contribute to mutant LRRK2 pathogenicity. In contrast, neuroprotective effects of wild type LRRK2 against hydrogen peroxide stress also seemed to be mediated through ERK1/2 signaling in HEK293 and SH-SY5Y cells [127]. While the G2019S mutant seemed to act through ERK1/2-dependent mechanisms, other studies reveal that the pathogenic Y1699C LRRK2 mutant failed to activate ERK1/2 in SH-SY5Y [127] or SN4741 cell lines [128].
A study using an inducible system confirmed a role for ERK1/2 in LRRK2-associated pathology. Overexpression of wild type, G2019S and R1441C LRRK2 increase phosphorylated ERK1/2 in a time dependent manner, with delayed ERK1/2 activation compared to the wild type in HEK293 cells. Increased phospho-ERK1/2 levels correlates with induction of SNCA, which encodes for α-synuclein. LRRK2 also promoted increased phosphorylation of MEK2, an upstream kinase of ERK1/2 [129]. Indeed, either inhibitors of MEK or dominant negative ERK2 prevents G2019S-mediated autophagic neurite shortening [39], and involvement of ERK1/2 was subsequently confirmed in relation to G2019S LRRK2-induced increases in basal autophagy [130]. In recent study using patient derived iPS cells, genetic correction of LRRK2 G2019S mutation significantly ameliorated PD associated phenotypes, which could also be achieved by inhibition of ERK1/2 [131].
4. Conclusions and Future Direction
Alterations in autophagy, mitophagy, cytoskeletal dynamics, mitochondrial function and the balance of protein synthesis and degradation have been implicated in relation to the normal or pathological function(s) of LRRK2 (Fig. 1), as well as in sporadic or toxin-based parkinsonism. Interestingly, LRRK2 interacts genetically with α-synuclein and with recessive PD genes. In contrast to α-synuclein, whose normal expression is predominantly in axon terminals, it is clear that LRRK2 shows both pre-synaptic and post-synaptic effects in neurons. Moreover, dendrite shortening is a prominent phenotype of neurons expressing the mutant LRRK2 gene. Future studies are needed to address the role of the various cell biological processes impacted by LRRK2 expression, including Wnt-related differentiation, in the context of human neurons affected in PD.
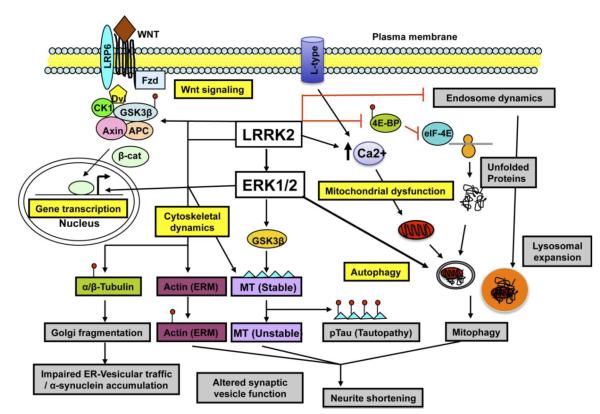
Figure 1. Cell biological pathways implicated in LRRK2-associated pathobiology: intersection with ERK1/2 and Wnt signaling pathways.
Altered endosomal dynamics, increased protein translation by inhibition of repressor protein 4E-BP1, and increased mitophagy contribute to a state of autophagic/lysosomal stress. LRRK2 promotes increased intracellular calcium and activates ERK1/2-dependent autophagy and downregulation of dendritic mitochondria. LRRK2 and ERK1/2 also regulate microtubule dynamics by phosphorylation of tau, actin and α/β-tubulin. LRRK2 also triggers Wnt-β-catenin and ERK1/2-dependent changes in gene transcription. The autophagic, transcriptional and cytoskeletal effects of mutant LRRK2 expression contribute to ERK1/2-dependent neurite shortening, while altered endosomal/vesicular dynamics affect synaptic function. 4E-BP1: translation repressor protein; APC: Adenomatous polyposis coli; CK1:Casein kinase 1; Dvl:Dishevelled; eIF-4E: eukaryotic translation inition factor 4E; ERK1/2:Extra cellular-signal regulated kinase 1/2; ERM: Ezrin-Radixin-Moesin; GSK3β:Glycogen synthase kinase 2beta; LRP6: low density lipoprotein receptor-related protein 6; LRRK2:Leucine-rich repeat kinase 2; MT: Microtubule; β-cat: beta catenin
ERK1/2-related cellular pathways may represent a point of mechanistic convergence between toxin-based models of sporadic PD, and newer studies of pathogenic mutations in LRRK2, which comprise the most common familial forms of PD (Fig. 1). Various studies have emerged linking LRRK2 to the ERK1/2 signaling pathway [39, 129]. Increased mitophagy observed with LRRK2 and associated pathogenic mutation [38, 132] has also been observed when ERK1/2 activity is modulated [114]. Cytoplasmic granules of phosphorylated ERK1/2 are observed in the context of both sporadic and G2019S LRRK2-associated PD patient substantia nigra neurons (Fig. 2) [116, 133]. Most studies have implicated increased kinase activity of LRRK2 in activation of ERK1/2, which could be prevented by MEK inhibitors. These studies would place LRRK2 upstream of ERK1/2. However, as ERK1/2 activation is not universally observed in cells expressing pathogenic LRRK2 mutants, more studies will be required to confirm how LRRK2 interacts with this pathway. A better understanding of mechanisms by which ERK1/2 regulates mitochondrial turnover, protein expression and cytoskeletal dynamics may add insights for sporadic and familial forms of PD linked to LRRK2 as well as several recessive PD genes.
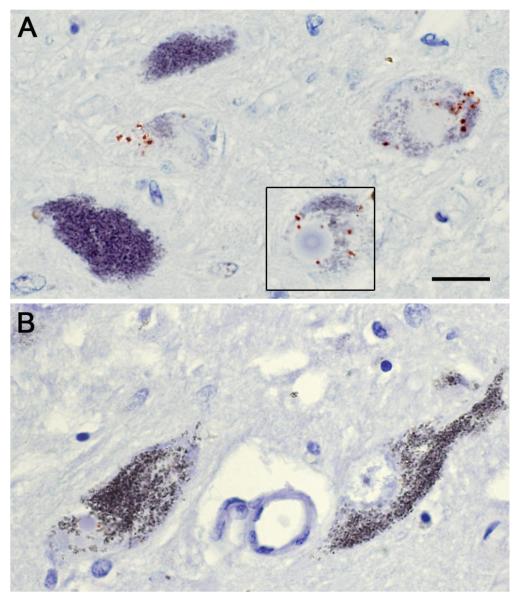
Figure 2. Post-mortem analysis of phosphorylated ERK1/2 in a LRRK2 G2019S PD/transitional Lewy body disease (LBD) patient.
Note the punctate, cytoplasmic staining of phosphorylated ERK1/2 (red chromagen) in pigmented substantia nigra neurons from the G2019S patient (A). These are similar in appearance and numbers to those previously reported in a series of sporadic PD/LBD cases [102], whereas this staining pattern was not seen in the midbrains of control subjects (B). Previous ultrastructural studies revealed phosphorylated ERK1/2 in association with abnormal mitochondria, autophagosomes, and bundles of intracellular filaments in PD/LBD substantia nigra neurons [104]. As reviewed in the text, alterations in mitochondrial homeostasis, autophagy, microtubule dynamics and ERK1/2-dependent protein expression are observed in models of LRRK2-associated pathobiology, suggesting the involvement of common cell biological pathways among sporadic and mutant LRRK2-triggered neurodegeneration. Scale = 20 microns.
Highlights.
Mutations in LRRK2 comprise the most common known cause of Parkinson disease
LRRK2 is implicated in regulating autophagy, cytoskeletal dynamics and protein synthesis
LRRK2 mediates some of these effects through interaction with the ERK1/2 and Wnt signaling pathways
New data implicating LRRK2 in mitochondrial calcium dyshomeostasis, mitophagy and synaptic vesicle dynamics are reviewed.
Common cell biological pathways may underlie sporadic, toxin-based and familial PD.
Acknowledgments
Supported by the National Institutes of Health (NIH): AG026389 and NS065789 to C.T.C. We thank Dennis Dickson of the Mayo Clinic Jacksonville, supported in part by P50-NS40256 and CurePSP: The Society for PSP Brain Bank, for the unstained sections from the G2019S and control patients.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Lang AE, Lozano AM. Parkinson’s disease. First of two parts. The New England journal of medicine. 1998;339:1044–1053. doi: 10.1056/NEJM199810083391506. [DOI] [PubMed] [Google Scholar]
- [2].Braak H, Ghebremedhin E, Rüb U, Bratzke H, Del Tredici K. Stages in the development of Parkinson’s disease-related pathology. Cell and tissue research. 2004;318:121–134. doi: 10.1007/s00441-004-0956-9. [DOI] [PubMed] [Google Scholar]
- [3].Gorrell JM, DiMonte D, Graham D. The role of the environment in Parkinson’s disease. Environmental health perspectives. 1996;104:652–654. doi: 10.1289/ehp.96104652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Semchuk KM, Love EJ, Lee RG. Parkinson’s disease and exposure to agricultural work and pesticide chemicals. Neurology. 1992;42:1328–1335. doi: 10.1212/wnl.42.7.1328. [DOI] [PubMed] [Google Scholar]
- [5].Betarbet R, Sherer TB, MacKenzie G, Garcia-Osuna M, Panov AV, Greenamyre JT. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nature neuroscience. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- [6].Tipton KF, Singer TP. Advances in our understanding of the mechanisms of the neurotoxicity of MPTP and related compounds. Journal of neurochemistry. 1993;61:1191–1206. doi: 10.1111/j.1471-4159.1993.tb13610.x. [DOI] [PubMed] [Google Scholar]
- [7].Dawson TM, Dawson VL. Rare genetic mutations shed light on the pathogenesis of Parkinson disease. The Journal of clinical investigation. 2003;111:145–151. doi: 10.1172/JCI17575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bekris LM, Mata IF, Zabetian CP. The genetics of Parkinson disease. Journal of geriatric psychiatry and neurology. 2010;23:228–242. doi: 10.1177/0891988710383572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mata IF, Taylor JP, Kachergus J, Hulihan M, Huerta C, Lahoz C, Blazquez M, Guisasola LM, Salvador C, Ribacoba R, Martinez C, Farrer M, Alvarez V. LRRK2 R1441G in Spanish patients with Parkinson’s disease. Neuroscience letters. 2005;382:309–311. doi: 10.1016/j.neulet.2005.03.033. [DOI] [PubMed] [Google Scholar]
- [10].Gloeckner CJ, Kinkl N, Schumacher A, Braun RJ, O’Neill E, Meitinger T, Kolch W, Prokisch H, Ueffing M. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Human molecular genetics. 2006;15:223–232. doi: 10.1093/hmg/ddi439. [DOI] [PubMed] [Google Scholar]
- [11].Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, Stoessl AJ, Pfeiffer RF, Patenge N, Carbajal IC, Vieregge P, Asmus F, Müller-Myhsok B, Dickson DW, Meitinger T, Strom TM, Wszolek ZK, Gasser T. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- [12].Funayama M, Hasegawa K, Ohta E, Kawashima N, Komiyama M, Kowa H, Tsuji S, Obata F. An LRRK2 mutation as a cause for the parkinsonism in the original PARK8 family. Annals of neurology. 2005;57:918–921. doi: 10.1002/ana.20484. [DOI] [PubMed] [Google Scholar]
- [13].Rideout HJ, Stefanis L. The Neurobiology of LRRK2 and its Role in the Pathogenesis of Parkinson’s Disease. Neurochemical research. 2013 doi: 10.1007/s11064-013-1073-5. [DOI] [PubMed] [Google Scholar]
- [14].Greggio E, Cookson MR. Leucine-rich repeat kinase 2 mutations and Parkinson’s disease: three questions. ASN neuro. 2009;1 doi: 10.1042/AN20090007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Funayama M, Hasegawa K, Kowa H, Saito M, Tsuji S, Obata F. A new locus for Parkinson’s disease (PARK8) maps to chromosome 12p11.2-q13.1. Annals of neurology. 2002;51:296–301. doi: 10.1002/ana.10113. [DOI] [PubMed] [Google Scholar]
- [16].Paisán-Ruíz C, Jain S, Evans EW, Gilks WP, Simón J, van der Brug M, López de Munain A, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Martí-Massó JF, Pérez-Tur J, Wood NW, Singleton AB. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- [17].Mata IF, Wedemeyer WJ, Farrer MJ, Taylor JP, Gallo KA. LRRK2 in Parkinson’s disease: protein domains and functional insights. Trends in neurosciences. 2006;29:286–293. doi: 10.1016/j.tins.2006.03.006. [DOI] [PubMed] [Google Scholar]
- [18].West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Biskup S, Moore DJ, Celsi F, Higashi S, West AB, Andrabi SA, Kurkinen K, Yu S-W, Savitt JM, Waldvogel HJ, Faull RLM, Emson PC, Torp R, Ottersen OP, Dawson TM, Dawson VL. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Annals of neurology. 2006;60:557–569. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- [20].Gilks WP, Abou-Sleiman PM, Gandhi S, Jain S, Singleton A, Lees AJ, Shaw K, Bhatia KP, Bonifati V, Quinn NP, Lynch J, Healy DG, Holton JL, Revesz T, Wood NW. A common LRRK2 mutation in idiopathic Parkinson’s disease. Lancet. 2005;365:415–416. doi: 10.1016/S0140-6736(05)17830-1. [DOI] [PubMed] [Google Scholar]
- [21].Ishihara L, Gibson RA, Warren L, Amouri R, Lyons K, Wielinski C, Hunter C, Swartz JE, Elango R, Akkari PA, Leppert D, Surh L, Reeves KH, Thomas S, Ragone L, Hattori N, Pahwa R, Jankovic J, Nance M, Freeman A, Gouider-Khouja N, Kefi M, Zouari M, Ben Sassi S, Ben Yahmed S, El Euch-Fayeche G, Middleton L, Burn DJ, Watts RL, Hentati F. Screening for Lrrk2 G2019S and clinical comparison of Tunisian and North American Caucasian Parkinson’s disease families. Movement disorders : official journal of the Movement Disorder Society. 2007;22:55–61. doi: 10.1002/mds.21180. [DOI] [PubMed] [Google Scholar]
- [22].Munhoz RP, Wakutani Y, Marras C, Teive HA, Raskin S, Werneck LC, Moreno D, Sato C, Lang AE, Rogaeva E. The G2019S LRRK2 mutation in Brazilian patients with Parkinson’s disease: phenotype in monozygotic twins. Movement disorders : official journal of the Movement Disorder Society. 2008;23:290–294. doi: 10.1002/mds.21832. [DOI] [PubMed] [Google Scholar]
- [23].Correia Guedes L, Ferreira JJ, Rosa MM, Coelho M, Bonifati V, Sampaio C. Worldwide frequency of G2019S LRRK2 mutation in Parkinson’s disease: a systematic review. Parkinsonism & related disorders. 2010;16:237–242. doi: 10.1016/j.parkreldis.2009.11.004. [DOI] [PubMed] [Google Scholar]
- [24].Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, Ahmad R, Miller DW, Kesavapany S, Singleton A, Lees A, Harvey RJ, Harvey K, Cookson MR. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiology of disease. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- [25].Deng X, Dzamko N, Prescott A, Davies P, Liu Q, Yang Q, Lee J-D, Patricelli MP, Nomanbhoy TK, Alessi DR, Gray NS. Characterization of a selective inhibitor of the Parkinson’s disease kinase LRRK2. Nature chemical biology. 2011;7:203–205. doi: 10.1038/nchembio.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jaleel M, Nichols RJ, Deak M, Campbell DG, Gillardon F, Knebel A, Alessi DR. LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson’s disease mutants affect kinase activity. The Biochemical journal. 2007;405:307–317. doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gillardon F. Leucine-rich repeat kinase 2 phosphorylates brain tubulin-beta isoforms and modulates microtubule stability--a point of convergence in parkinsonian neurodegeneration? Journal of neurochemistry. 2009;110:1514–1522. doi: 10.1111/j.1471-4159.2009.06235.x. [DOI] [PubMed] [Google Scholar]
- [28].Imai Y, Gehrke S, Wang H-Q, Takahashi R, Hasegawa K, Oota E, Lu B. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. The EMBO journal. 2008;27:2432–2443. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Qing H, Zhang Y, Deng Y, McGeer EG, McGeer PL. Lrrk2 interaction with alpha-synuclein in diffuse Lewy body disease. Biochemical and biophysical research communications. 2009;390:1229–1234. doi: 10.1016/j.bbrc.2009.10.126. [DOI] [PubMed] [Google Scholar]
- [30].Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. The Journal of pathology. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell metabolism. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Rabinowitz JD, White E. Autophagy and metabolism. Science (New York, N.Y.) 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cherra SJ, 3rd, Dagda RK, Chu CT. Review: autophagy and neurodegeneration: survival at a cost? Neuropathology and applied neurobiology. 2010;36:125–132. doi: 10.1111/j.1365-2990.2010.01062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- [35].Komatsu M, Waguri S, Chiba T, Murata S, Iwata J.-i., Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- [36].Oczypok EA, Oury TD, Chu CT. It’s a cell-eat-cell world: autophagy and phagocytosis. The American journal of pathology. 2013;182:612–622. doi: 10.1016/j.ajpath.2012.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cherra SJ, Kulich SM, Uechi G, Balasubramani M, Mountzouris J, Day BW, Chu CT. Regulation of the autophagy protein LC3 by phosphorylation. The Journal of cell biology. 2010;190:533–539. doi: 10.1083/jcb.201002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cherra SJ, Steer E, Gusdon AM, Kiselyov K, Chu CT. Mutant LRRK2 elicits calcium imbalance and depletion of dendritic mitochondria in neurons. The American journal of pathology. 2013;182:474–484. doi: 10.1016/j.ajpath.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Plowey ED, Cherra SJ, Liu Y-J, Chu CT. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. Journal of neurochemistry. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Alegre-Abarrategui J, Christian H, Lufino MMP, Mutihac R, Venda LL, Ansorge O, Wade-Martins R. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Human molecular genetics. 2009;18:4022–4034. doi: 10.1093/hmg/ddp346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].MacLeod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The familial Parkinsonism gene LRRK2 regulates neurite process morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- [42].Gómez-Suaga P, Luzón-Toro B, Churamani D, Zhang L, Bloor-Young D, Patel S, Woodman PG, Churchill GC, Hilfiker S. Leucine-rich repeat kinase 2 regulates autophagy through a calcium-dependent pathway involving NAADP. Human molecular genetics. 2012;21:511–525. doi: 10.1093/hmg/ddr481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fass E, Shvets E, Degani I, Hirschberg K, Elazar Z. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. The Journal of biological chemistry. 2006;281:36303–36316. doi: 10.1074/jbc.M607031200. [DOI] [PubMed] [Google Scholar]
- [44].Manzoni C, Mamais A, Dihanich S, Abeti R, Soutar MP, Plun-Favreau H, Giunti P, Tooze SA, Bandopadhyay R, Lewis PA. Inhibition of LRRK2 kinase activity stimulates macroautophagy. Biochimica et biophysica acta. 2013;1833:2900–2910. doi: 10.1016/j.bbamcr.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tong Y, Yamaguchi H, Giaime E, Boyle S, Kopan R, Kelleher RJ, Shen J. Loss of leucine-rich repeat kinase 2 causes impairment of protein degradation pathways, accumulation of alpha-synuclein, and apoptotic cell death in aged mice. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9879–9884. doi: 10.1073/pnas.1004676107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tong Y, Giaime E, Yamaguchi H, Ichimura T, Liu Y, Si H, Cai H, Bonventre JV, Shen J. Loss of leucine-rich repeat kinase 2 causes age-dependent bi-phasic alterations of the autophagy pathway. Molecular neurodegeneration. 2012;7 doi: 10.1186/1750-1326-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ramonet D, Daher JPL, Lin BM, Stafa K, Kim J, Banerjee R, Westerlund M, Pletnikova O, Glauser L, Yang L, Liu Y, Swing DA, Beal MF, Troncoso JC, McCaffery JM, Jenkins NA, Copeland NG, Galter D, Thomas B, Lee MK, Dawson TM, Dawson VL, Moore DJ. Dopaminergic neuronal loss, reduced neurite complexity and autophagic abnormalities in transgenic mice expressing G2019S mutant LRRK2. PloS one. 2011;6:e18568. doi: 10.1371/journal.pone.0018568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hindle S, Afsari F, Stark M, Middleton CA, Evans GJO, Sweeney ST, Elliott CJH. Dopaminergic expression of the Parkinsonian gene LRRK2-G2019S leads to non-autonomous visual neurodegeneration, accelerated by increased neural demands for energy. Human molecular genetics. 2013;22:2129–2140. doi: 10.1093/hmg/ddt061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Friedman LG, Lachenmayer ML, Wang J, He L, Poulose SM, Komatsu M, Holstein GR, Yue Z. Disrupted autophagy leads to dopaminergic axon and dendrite degeneration and promotes presynaptic accumulation of α-synuclein and LRRK2 in the brain. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:7585–7593. doi: 10.1523/JNEUROSCI.5809-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kirschner M, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- [51].Cairns NJ, Lee VM-Y, Trojanowski JQ. The cytoskeleton in neurodegenerative diseases. The Journal of pathology. 2004;204:438–449. doi: 10.1002/path.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].McMurray CT. Neurodegeneration: diseases of the cytoskeleton? Cell death and differentiation. 2000;7:861–865. doi: 10.1038/sj.cdd.4400764. [DOI] [PubMed] [Google Scholar]
- [53].Ren Y, Liu W, Jiang H, Jiang Q, Feng J. Selective vulnerability of dopaminergic neurons to microtubule depolymerization. The Journal of biological chemistry. 2005;280:34105–34112. doi: 10.1074/jbc.M503483200. [DOI] [PubMed] [Google Scholar]
- [54].Gandhi PN, Wang X, Zhu X, Chen SG, Wilson-Delfosse AL. The Roc domain of leucine-rich repeat kinase 2 is sufficient for interaction with microtubules. Journal of neuroscience research. 2008;86:1711–1720. doi: 10.1002/jnr.21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kett LR, Boassa D, Ho CC-Y, Rideout HJ, Hu J, Terada M, Ellisman M, Dauer WT. LRRK2 Parkinson disease mutations enhance its microtubule association. Human molecular genetics. 2012;21:890–899. doi: 10.1093/hmg/ddr526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Melrose HL, Dachsel JC, Behrouz B, Lincoln SJ, Yue M, Hinkle KM, Kent CB, Korvatska E, Taylor JP, Witten L, Liang YQ, Beevers JE, Boules M, Dugger BN, Serna VA, Gaukhman A, Yu X, Castanedes-Casey M, Braithwaite AT, Ogholikhan S, Yu N, Bass D, Tyndall G, Schellenberg GD, Dickson DW, Janus C, Farrer MJ. Impaired dopaminergic neurotransmission and microtubule-associated protein tau alterations in human LRRK2 transgenic mice. Neurobiol Dis. 2010;40:503–517. doi: 10.1016/j.nbd.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ujiie S, Hatano T, Kubo S-I, Imai S, Sato S, Uchihara T, Yagishita S, Hasegawa K, Kowa H, Sakai F, Hattori N. LRRK2 I2020T mutation is associated with tau pathology. Parkinsonism & related disorders. 2012;18:819–823. doi: 10.1016/j.parkreldis.2012.03.024. [DOI] [PubMed] [Google Scholar]
- [58].Kawakami F, Yabata T, Ohta E, Maekawa T, Shimada N, Suzuki M, Maruyama H, Ichikawa T, Obata F. LRRK2 phosphorylates tubulin-associated tau but not the free molecule: LRRK2-mediated regulation of the tau-tubulin association and neurite outgrowth. PloS one. 2012;7:e30834. doi: 10.1371/journal.pone.0030834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lin C-H, Tsai P-I, Wu R-M, Chien C-T. LRRK2 G2019S mutation induces dendrite degeneration through mislocalization and phosphorylation of tau by recruiting autoactivated GSK3ß. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:13138–13149. doi: 10.1523/JNEUROSCI.1737-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gross SR, Kinzy TG. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nature structural & molecular biology. 2005;12:772–778. doi: 10.1038/nsmb979. [DOI] [PubMed] [Google Scholar]
- [61].Shiina N, Gotoh Y, Kubomura N, Iwamatsu A, Nishida E. Microtubule severing by elongation factor 1 alpha. Science (New York, N.Y.) 1994;266:282–285. doi: 10.1126/science.7939665. [DOI] [PubMed] [Google Scholar]
- [62].Gillardon F. Interaction of elongation factor 1-alpha with leucine-rich repeat kinase 2 impairs kinase activity and microtubule bundling in vitro. Neuroscience. 2009;163:533–539. doi: 10.1016/j.neuroscience.2009.06.051. [DOI] [PubMed] [Google Scholar]
- [63].Meixner A, Boldt K, Van Troys M, Askenazi M, Gloeckner CJ, Bauer M, Marto JA, Ampe C, Kinkl N, Ueffing M. A QUICK screen for Lrrk2 interaction partners--leucine-rich repeat kinase 2 is involved in actin cytoskeleton dynamics. Molecular & cellular proteomics : MCP. 2011;10:M110.001172. doi: 10.1074/mcp.M110.001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Parisiadou L, Xie C, Cho HJ, Lin X, Gu X-L, Long C-X, Lobbestael E, Baekelandt V, Taymans J-M, Sun L, Cai H. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:13971–13980. doi: 10.1523/JNEUROSCI.3799-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Nichols RJ, Dzamko N, Hutti JE, Cantley LC, Deak M, Moran J, Bamborough P, Reith AD, Alessi DR. Substrate specificity and inhibitors of LRRK2, a protein kinase mutated in Parkinson’s disease. The Biochemical journal. 2009;424:47–60. doi: 10.1042/BJ20091035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Tain LS, Mortiboys H, Tao RN, Ziviani E, Bandmann O, Whitworth AJ. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nature neuroscience. 2009;12:1129–1135. doi: 10.1038/nn.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kumar A, Greggio E, Beilina A, Kaganovich A, Chan D, Taymans J-M, Wolozin B, Cookson MR. The Parkinson’s disease associated LRRK2 exhibits weaker in vitro phosphorylation of 4E-BP compared to autophosphorylation. PloS one. 2010;5:e8730. doi: 10.1371/journal.pone.0008730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Trancikova A, Mamais A, Webber PJ, Stafa K, Tsika E, Glauser L, West AB, Bandopadhyay R, Moore DJ. Phosphorylation of 4E-BP1 in the mammalian brain is not altered by LRRK2 expression or pathogenic mutations. PloS one. 2012;7:e47784. doi: 10.1371/journal.pone.0047784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Venderova K, Kabbach G, Abdel-Messih E, Zhang Y, Parks RJ, Imai Y, Gehrke S, Ngsee J, Lavoie MJ, Slack RS, Rao Y, Zhang Z, Lu B, Haque ME, Park DS. Leucine-Rich Repeat Kinase 2 interacts with Parkin, DJ-1 and PINK-1 in a Drosophila melanogaster model of Parkinson’s disease. Human molecular genetics. 2009;18:4390–4404. doi: 10.1093/hmg/ddp394. [DOI] [PubMed] [Google Scholar]
- [70].Papkovskaia TD, Chau K-Y, Inesta-Vaquera F, Papkovsky DB, Healy DG, Nishio K, Staddon J, Duchen MR, Hardy J, Schapira AHV, Cooper JM. G2019S leucine-rich repeat kinase 2 causes uncoupling protein-mediated mitochondrial depolarization. Human molecular genetics. 2012;21:4201–4213. doi: 10.1093/hmg/dds244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Mortiboys H, Johansen KK, Aasly JO, Bandmann O. Mitochondrial impairment in patients with Parkinson disease with the G2019S mutation in LRRK2. Neurology. 2010;75:2017–2020. doi: 10.1212/WNL.0b013e3181ff9685. [DOI] [PubMed] [Google Scholar]
- [72].Dagda RK, Gusdon AM, Pien I, Strack S, Green S, Li C, Van Houten B, Cherra SJ, Chu CT. Mitochondrially localized PKA reverses mitochondrial pathology and dysfunction in a cellular model of Parkinson’s disease. Cell death and differentiation. 2011;18:1914–1923. doi: 10.1038/cdd.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Sandebring A, Thomas KJ, Beilina A, van der Brug M, Cleland MM, Ahmad R, Miller DW, Zambrano I, Cowburn RF, Behbahani H, Cedazo-Mínguez A, Cookson MR. Mitochondrial alterations in PINK1 deficient cells are influenced by calcineurin-dependent dephosphorylation of dynamin-related protein 1. PloS one. 2009;4:e5701. doi: 10.1371/journal.pone.0005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Niu J, Yu M, Wang C, Xu Z. Leucine-rich repeat kinase 2 disturbs mitochondrial dynamics via Dynamin-like protein. Journal of neurochemistry. 2012;122:650–658. doi: 10.1111/j.1471-4159.2012.07809.x. [DOI] [PubMed] [Google Scholar]
- [75].Wang X, Yan MH, Fujioka H, Liu J, Wilson-Delfosse A, Chen SG, Perry G, Casadesus G, Zhu X. LRRK2 regulates mitochondrial dynamics and function through direct interaction with DLP1. Human molecular genetics. 2012;21:1931–1944. doi: 10.1093/hmg/dds003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Dagda RK, Cherra SJ, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. The Journal of biological chemistry. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Narendra DP, Jin SM, Tanaka A, Suen D-F, Gautier CA, Shen J, Cookson MR, Youle RJ. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS biology. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Gandhi S, Wood-Kaczmar A, Yao Z, Plun-Favreau H, Deas E, Klupsch K, Downward J, Latchman DS, Tabrizi SJ, Wood NW, Duchen MR, Abramov AY. PINK1-associated Parkinson’s disease is caused by neuronal vulnerability to calcium-induced cell death. Molecular cell. 2009;33:627–638. doi: 10.1016/j.molcel.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Heeman B, Van den Haute C, Aelvoet S-A, Valsecchi F, Rodenburg RJ, Reumers V, Debyser Z, Callewaert G, Koopman WJH, Willems PHGM, Baekelandt V. Depletion of PINK1 affects mitochondrial metabolism, calcium homeostasis and energy maintenance. Journal of cell science. 2011;124:1115–1125. doi: 10.1242/jcs.078303. [DOI] [PubMed] [Google Scholar]
- [80].Marongiu R, Spencer B, Crews L, Adame A, Patrick C, Trejo M, Dallapiccola B, Valente EM, Masliah E. Mutant Pink1 induces mitochondrial dysfunction in a neuronal cell model of Parkinson’s disease by disturbing calcium flux. Journal of neurochemistry. 2009;108:1561–1574. doi: 10.1111/j.1471-4159.2009.05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Plowey ED, Chu CT. Synaptic dysfunction in genetic models of Parkinson’s disease: A role for autophagy? Neurobiol Dis. 2010 doi: 10.1016/j.nbd.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Lee S, Liu HP, Lin WY, Guo H, Lu B. LRRK2 kinase regulates synaptic morphology through distinct substrates at the presynaptic and postsynaptic compartments of the Drosophila neuromuscular junction. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:16959–16969. doi: 10.1523/JNEUROSCI.1807-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Piccoli G, Condliffe SB, Bauer M, Giesert F, Boldt K, De Astis S, Meixner A, Sarioglu H, Vogt-Weisenhorn DM, Wurst W, Gloeckner CJ, Matteoli M, Sala C, Ueffing M. LRRK2 controls synaptic vesicle storage and mobilization within the recycling pool. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:2225–2237. doi: 10.1523/JNEUROSCI.3730-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].MacLeod DA, Rhinn H, Kuwahara T, Zolin A, Di Paolo G, McCabe BD, Marder KS, Honig LS, Clark LN, Small SA, Abeliovich A. RAB7L1 interacts with LRRK2 to modify intraneuronal protein sorting and Parkinson’s disease risk. Neuron. 2013;77:425–439. doi: 10.1016/j.neuron.2012.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Shin N, Jeong H, Kwon J, Heo HY, Kwon JJ, Yun HJ, Kim CH, Han BS, Tong Y, Shen J, Hatano T, Hattori N, Kim KS, Chang S, Seol W. LRRK2 regulates synaptic vesicle endocytosis. Experimental cell research. 2008;314:2055–2065. doi: 10.1016/j.yexcr.2008.02.015. [DOI] [PubMed] [Google Scholar]
- [86].Xiong Y, Coombes CE, Kilaru A, Li X, Gitler AD, Bowers WJ, Dawson VL, Dawson TM, Moore DJ. GTPase activity plays a key role in the pathobiology of LRRK2. PLoS genetics. 2010;6:e1000902. doi: 10.1371/journal.pgen.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Matta S, Van Kolen K, da Cunha R, van den Bogaart G, Mandemakers W, Miskiewicz K, De Bock PJ, Morais VA, Vilain S, Haddad D, Delbroek L, Swerts J, Chavez-Gutierrez L, Esposito G, Daneels G, Karran E, Holt M, Gevaert K, Moechars DW, De Strooper B, Verstreken P. LRRK2 controls an EndoA phosphorylation cycle in synaptic endocytosis. Neuron. 2012;75:1008–1021. doi: 10.1016/j.neuron.2012.08.022. [DOI] [PubMed] [Google Scholar]
- [88].Chu CT. Autophagic stress in neuronal injury and disease. Journal of neuropathology and experimental neurology. 2006;65:423–432. doi: 10.1097/01.jnen.0000229233.75253.be. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Li G, Pleasure SJ. Morphogenesis of the dentate gyrus: what we are learning from mouse mutants. Developmental neuroscience. 2005;27:93–99. doi: 10.1159/000085980. [DOI] [PubMed] [Google Scholar]
- [90].Ciani L, Salinas PC. WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nature reviews. Neuroscience. 2005;6:351–362. doi: 10.1038/nrn1665. [DOI] [PubMed] [Google Scholar]
- [91].Buechling T, Boutros M. Wnt signaling signaling at and above the receptor level. Current topics in developmental biology. 2011;97:21–53. doi: 10.1016/B978-0-12-385975-4.00008-5. [DOI] [PubMed] [Google Scholar]
- [92].Cadigan KM, Nusse R. Wnt signaling: a common theme in animal development. Genes & development. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- [93].Polakis P. Wnt signaling and cancer. Genes & development. 2000;14:1837–1851. [PubMed] [Google Scholar]
- [94].Castelo-Branco G, Wagner J, Rodriguez FJ, Kele J, Sousa K, Rawal N, Pasolli HA, Fuchs E, Kitajewski J, Arenas E. Differential regulation of midbrain dopaminergic neuron development by Wnt-1, Wnt-3a, and Wnt-5a. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:12747–12752. doi: 10.1073/pnas.1534900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Parish CL, Castelo-Branco G, Rawal N, Tonnesen J, Sorensen AT, Salto C, Kokaia M, Lindvall O, Arenas E. Wnt5a-treated midbrain neural stem cells improve dopamine cell replacement therapy in parkinsonian mice. The Journal of clinical investigation. 2008;118:149–160. doi: 10.1172/JCI32273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Andersson ER, Saltó C, Villaescusa JC, Cajanek L, Yang S, Bryjova L, Nagy II, Vainio SJ, Ramirez C, Bryja V, Arenas E. Wnt5a cooperates with canonical Wnts to generate midbrain dopaminergic neurons in vivo and in stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E602–610. doi: 10.1073/pnas.1208524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Kitagawa H, Ray WJ, Glantschnig H, Nantermet PV, Yu Y, Leu C-T, Harada S.-i., Kato S, Freedman LP. A regulatory circuit mediating convergence between Nurr1 transcriptional regulation and Wnt signaling. Molecular and cellular biology. 2007;27:7486–7496. doi: 10.1128/MCB.00409-07. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [98].Ohnuki T, Nakamura A, Okuyama S, Nakamura S. Gene expression profiling in progressively MPTP-lesioned macaques reveals molecular pathways associated with sporadic Parkinson’s disease. Brain research. 2010;1346:26–42. doi: 10.1016/j.brainres.2010.05.066. [DOI] [PubMed] [Google Scholar]
- [99].L’Episcopo F, Tirolo C, Testa N, Caniglia S, Morale MC, Cossetti C, D’Adamo P, Zardini E, Andreoni L, Ihekwaba AEC, Serra PA, Franciotta D, Martino G, Pluchino S, Marchetti B. Reactive astrocytes and Wnt/β-catenin signaling link nigrostriatal injury to repair in 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine model of Parkinson’s disease. Neurobiology of disease. 2011;41:508–527. doi: 10.1016/j.nbd.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Gollamudi S, Johri A, Calingasan NY, Yang L, Elemento O, Beal MF. Concordant signaling pathways produced by pesticide exposure in mice correspond to pathways identified in human Parkinson’s disease. PloS one. 2012;7:e36191. doi: 10.1371/journal.pone.0036191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Dun Y, Li G, Yang Y, Xiong Z, Feng M, Wang M, Zhang Y, Xiang J, Ma R. Inhibition of the canonical Wnt pathway by Dickkopf-1 contributes to the neurodegeneration in 6-OHDA-lesioned rats. Neuroscience letters. 2012;525:83–88. doi: 10.1016/j.neulet.2012.07.030. [DOI] [PubMed] [Google Scholar]
- [102].Wei L, Sun C, Lei M, Li G, Yi L, Luo F, Li Y, Ding L, Liu Z, Li S, Xu P. Activation of Wnt/β-catenin pathway by exogenous Wnt1 protects SH-SY5Y cells against 6-hydroxydopamine toxicity. Journal of molecular neuroscience : MN. 2013;49:105–115. doi: 10.1007/s12031-012-9900-8. [DOI] [PubMed] [Google Scholar]
- [103].Häbig K, Walter M, Poths S, Riess O, Bonin M. RNA interference of LRRK2-microarray expression analysis of a Parkinson’s disease key player. Neurogenetics. 2008;9:83–94. doi: 10.1007/s10048-007-0114-0. [DOI] [PubMed] [Google Scholar]
- [104].Ferree A, Guillily M, Li H, Smith K, Takashima A, Squillace R, Weigele M, Collins JJ, Wolozin B. Regulation of physiologic actions of LRRK2: focus on autophagy. Neuro-degenerative diseases. 2012;10:238–241. doi: 10.1159/000332599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Sancho RM, Law BMH, Harvey K. Mutations in the LRRK2 Roc-COR tandem domain link Parkinson’s disease to Wnt signalling pathways. Human molecular genetics. 2009;18:3955–3968. doi: 10.1093/hmg/ddp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Berwick DC, Harvey K. LRRK2 functions as a Wnt signaling scaffold, bridging cytosolic proteins and membrane-localized LRP6. Human molecular genetics. 2012;21:4966–4979. doi: 10.1093/hmg/dds342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Luttrell LM. ’Location, location, location’: activation and targeting of MAP kinases by G protein-coupled receptors. Journal of molecular endocrinology. 2003;30:117–126. doi: 10.1677/jme.0.0300117. [DOI] [PubMed] [Google Scholar]
- [108].Xia Z, Dickens M, Raingeaud J, Davis RJ, Greenberg ME. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science (New York, N.Y.) 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- [109].Hsu CH, Chan D, Greggio E, Saha S, Guillily MD, Ferree A, Raghavan K, Shen GC, Segal L, Ryu H, Cookson MR, Wolozin B. MKK6 binds and regulates expression of Parkinson’s disease-related protein LRRK2. J Neurochem. 2010;112:1593–1604. doi: 10.1111/j.1471-4159.2010.06568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Gloeckner CJ, Schumacher A, Boldt K, Ueffing M. The Parkinson disease-associated protein kinase LRRK2 exhibits MAPKKK activity and phosphorylates MKK3/6 and MKK4/7, in vitro. Journal of neurochemistry. 2009;109:959–968. doi: 10.1111/j.1471-4159.2009.06024.x. [DOI] [PubMed] [Google Scholar]
- [111].Luo Y, DeFranco DB. Opposing roles for ERK1/2 in neuronal oxidative toxicity: distinct mechanisms of ERK1/2 action at early versus late phases of oxidative stress. The Journal of biological chemistry. 2006;281:16436–16442. doi: 10.1074/jbc.M512430200. [DOI] [PubMed] [Google Scholar]
- [112].Gómez-Santos C, Ferrer I, Reiriz J, Viñals F, Barrachina M, Ambrosio S. MPP+ increases alpha-synuclein expression and ERK/MAP-kinase phosphorylation in human neuroblastoma SH-SY5Y cells. Brain research. 2002;935:32–39. doi: 10.1016/s0006-8993(02)02422-8. [DOI] [PubMed] [Google Scholar]
- [113].Kulich SM, Chu CT. Sustained extracellular signal-regulated kinase activation by 6-hydroxydopamine: implications for Parkinson’s disease. Journal of neurochemistry. 2001;77:1058–1066. doi: 10.1046/j.1471-4159.2001.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Zhu J-H, Horbinski C, Guo F, Watkins S, Uchiyama Y, Chu CT. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. The American journal of pathology. 2007;170:75–86. doi: 10.2353/ajpath.2007.060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Kulich SM, Horbinski C, Patel M, Chu CT. 6-Hydroxydopamine induces mitochondrial ERK activation. Free radical biology & medicine. 2007;43:372–383. doi: 10.1016/j.freeradbiomed.2007.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Zhu J-H, Kulich SM, Oury TD, Chu CT. Cytoplasmic aggregates of phosphorylated extracellular signal-regulated protein kinases in Lewy body diseases. The American journal of pathology. 2002;161:2087–2098. doi: 10.1016/S0002-9440(10)64487-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Ferrer I, Barrachina M, Tolnay M, Rey MJ, Vidal N, Carmona M, Blanco R, Puig B. Phosphorylated protein kinases associated with neuronal and glial tau deposits in argyrophilic grain disease. Brain pathology (Zurich, Switzerland) 2003;13:62–78. doi: 10.1111/j.1750-3639.2003.tb00007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Zhu J-H, Guo F, Shelburne J, Watkins S, Chu CT. Localization of phosphorylated ERK/MAP kinases to mitochondria and autophagosomes in Lewy body diseases. Brain pathology (Zurich, Switzerland) 2003;13:473–481. doi: 10.1111/j.1750-3639.2003.tb00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Dagda RK, Zhu J, Kulich SM, Chu CT. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson’s disease. Autophagy. 2008;4:770–782. doi: 10.4161/auto.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Zhu JH, Gusdon AM, Cimen H, Van Houten B, Koc E, Chu CT. Impaired mitochondrial biogenesis contributes to depletion of functional mitochondria in chronic MPP+ toxicity: dual roles for ERK1/2. Cell death & disease. 2012;3:e312. doi: 10.1038/cddis.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Chu CT, Levinthal DJ, Kulich SM, Chalovich EM, DeFranco DB. Oxidative neuronal injury. The dark side of ERK1/2. European journal of biochemistry / FEBS. 2004;271:2060–2066. doi: 10.1111/j.1432-1033.2004.04132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].de Bernardo S, Canals S, Casarejos MJ, Solano RM, Menendez J, Mena MA. Role of extracellular signal-regulated protein kinase in neuronal cell death induced by glutathione depletion in neuron/glia mesencephalic cultures. Journal of neurochemistry. 2004;91:667–682. doi: 10.1111/j.1471-4159.2004.02744.x. [DOI] [PubMed] [Google Scholar]
- [123].Kulich SM, Chu CT. Role of reactive oxygen species in extracellular signal-regulated protein kinase phosphorylation and 6-hydroxydopamine cytotoxicity. Journal of biosciences. 2003;28:83–89. doi: 10.1007/BF02970136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Canals S, Casarejos MJ, de Bernardo S, Rodríguez-Martín E, Mena MA. Nitric oxide triggers the toxicity due to glutathione depletion in midbrain cultures through 12-lipoxygenase. The Journal of biological chemistry. 2003;278:21542–21549. doi: 10.1074/jbc.M213174200. [DOI] [PubMed] [Google Scholar]
- [125].Wiegert JS, Bading H. Activity-dependent calcium signaling and ERK-MAP kinases in neurons: a link to structural plasticity of the nucleus and gene transcription regulation. Cell calcium. 2011;49:296–305. doi: 10.1016/j.ceca.2010.11.009. [DOI] [PubMed] [Google Scholar]
- [126].White LR, Toft M, Kvam SN, Farrer MJ, Aasly JO. MAPK-pathway activity, Lrrk2 G2019S, and Parkinson’s disease. Journal of neuroscience research. 2007;85:1288–1294. doi: 10.1002/jnr.21240. [DOI] [PubMed] [Google Scholar]
- [127].Liou AKF, Leak RK, Li L, Zigmond MJ. Wild-type LRRK2 but not its mutant attenuates stress-induced cell death via ERK pathway. Neurobiology of disease. 2008;32:116–124. doi: 10.1016/j.nbd.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Heo HY, Park J-M, Kim C-H, Han BS, Kim K-S, Seol W. LRRK2 enhances oxidative stress-induced neurotoxicity via its kinase activity. Experimental cell research. 2010;316:649–656. doi: 10.1016/j.yexcr.2009.09.014. [DOI] [PubMed] [Google Scholar]
- [129].Carballo-Carbajal I, Weber-Endress S, Rovelli G, Chan D, Wolozin B, Klein CL, Patenge N, Gasser T, Kahle PJ. Leucine-rich repeat kinase 2 induces alpha-synuclein expression via the extracellular signal-regulated kinase pathway. Cellular signalling. 2010;22:821–827. doi: 10.1016/j.cellsig.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Bravo-San Pedro JM, Niso-Santano M, Gómez-Sánchez R, Pizarro-Estrella E, Aiastui-Pujana A, Gorostidi A, Climent V, López de Maturana R, Sanchez-Pernaute R, López de Munain A, Fuentes JM, González-Polo RA. The LRRK2 G2019S mutant exacerbates basal autophagy through activation of the MEK/ERK pathway. Cellular and molecular life sciences : CMLS. 2013;70:121–136. doi: 10.1007/s00018-012-1061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Reinhardt P, Schmid B, Burbulla LF, Schöndorf DC, Wagner L, Glatza M, Höing S, Hargus G, Heck SA, Dhingra A, Wu G, Müller S, Brockmann K, Kluba T, Maisel M, Krüger R, Berg D, Tsytsyura Y, Thiel CS, Psathaki O-E, Klingauf J, Kuhlmann T, Klewin M, Müller H, Gasser T, Schöler HR, Sterneckert J. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell stem cell. 2013;12:354–367. doi: 10.1016/j.stem.2013.01.008. [DOI] [PubMed] [Google Scholar]
- [132].Chu CT. Diversity in the regulation of autophagy and mitophagy: lessons from Parkinson’s disease. Parkinson’s disease. 2011;2011:789431. doi: 10.4061/2011/789431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Ferrer I, Blanco R, Carmona M, Puig B, Barrachina M, Gómez C, Ambrosio S. Active, phosphorylation-dependent mitogen-activated protein kinase (MAPK/ERK), stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK), and p38 kinase expression in Parkinson’s disease and Dementia with Lewy bodies. Journal of neural transmission (Vienna, Austria : 1996) 2001;108:1383–1396. doi: 10.1007/s007020100015. [DOI] [PubMed] [Google Scholar]