Abstract
The pharmacology of drugs is often defined by more than one protein target. This property can be exploited to use approved drugs to uncover new targets and signaling pathways in cancer. Towards enabling a rational approach to uncover new targets, we expand a structural protein-ligand interactome (http://www.biodrugscreen.org) by scoring the interaction among 1,000 FDA-approved drugs docked to 2,500 pockets on protein structures of the human genome. This afforded a drug-target network whose properties compared favorably with previous networks constructed with experimental data. Among drugs with highest degree and betweenness two are cancer drugs and one is currently used for treatment of lung cancer. Comparison of predicted cancer and non-cancer targets reveals that the most cancer-specific compounds were also the most selective compounds. Analysis of compound flexibility, hydrophobicity, and size showed that the most selective compounds were low molecular weight fragment-like heterocycles. We use a previously-developed screening approach using the cancer drug erlotinib as a template to screen other approved drugs that mimic its properties. Among the top 12 ranking candidates, four are cancer drugs, two of them kinase inhibitors (like erlotinib). Cellular studies using non-small cell lung cancer (NSCLC) cells revealed that several drugs inhibited lung cancer cell proliferation. We mined patient records at the Regenstrief Medical Record System to explore possible association of exposure to three of these drugs with occurrence of lung cancer. Preliminary in vivo studies using non-small cell lung cancer (NCLSC) xenograft model showed that losartan- and astemizole-treated mice had tumors that weighed 50 (p < 0.01) and 15 (p < 0.01) percent less than vehicle. These results set the stage for further exploration of these drugs and to uncover new drugs for lung cancer.
INTRODUCTION
Genomic and proteomic studies have established that cancer is a systems biology disease that involves a large number of genes spanning multiple signaling pathways as shown in lung,1 pancreatic,2 breast,3 brain4 and colorectal5 cancers. In the case of lung cancer, hundreds of genetic alterations spanning 18 signaling pathways have been found.1, 6 The large number of mutations make it a significant challenge to identify effective treatments for this disease. According to the American Cancer Society, the disease has taken 160,340 lives in the U.S in 2011 alone. Non-small cell lung cancer (NSCLC) is the most prevalent form of the disease (85 percent of all cases). It is characterized by poor prognosis and aggressive behavior. First-line treatment options for the majority of patients include chemotherapeutics that cause significant side effects. New treatments with lower toxicity and greater efficacy are urgently needed.
Studies have shown that approved and experimental drugs as well as chemical probes bind and modulate the function of multiple proteins.7, 8 This property, also known as polypharmacology, offers an opportunity to uncover new targets. Recently, we have explored the possibility of using structure-based docking to generate a protein-compound interactome that can be used as a hypothesis generation tool to uncover new targets for small molecules. We docked more than 1,200 compounds to more than 3,000 pockets from 1,000 proteins. The resulting structural protein-ligand interactome (splinter) is available at http://www.biodrugscreen.org 9. The scoring of protein-compound interactions in this interactome enables the rank-ordering of compounds for individual targets for purposes of hit identification, but also makes it possible to rank-order proteins for a list of potential targets for a compound or drug of interest. In a recent application, we used the interactome to search for compounds that mimicked the binding profile of an existing drug.10 We stipulated that such compounds may exhibit similar pharmacokinetic properties and efficacy to the drug and possibly serve as leads for the development of cancer therapeutics. From this study, several compounds were uncovered with potent anti-cancer properties and in vitro studies suggested suitable pharmacokinetic (PK) properties.10
Here, we extend splinter by docking more than 1,000 FDA-approved drugs to targets in the interactome. The cancer drug erlotinib was used as a template to search for other approved drugs that may possess similar anti-cancer properties. Erlotinib is used in the treatment of non-small cell lung cancer (NSCLC) patients. Twelve drugs are tested for their effect on cell growth in a panel of NSCLC cells. We mined patient records to study the potential association between drug exposure and lung cancer occurrence in patients taking these drugs.11 In vivo preclinical studies using human NSCLC xenografts in NOD-SCID mice were carried out to probe these drugs for their effect in lung cancer.
RESULTS AND DISCUSSION
Docking approved drugs to the human structural proteome
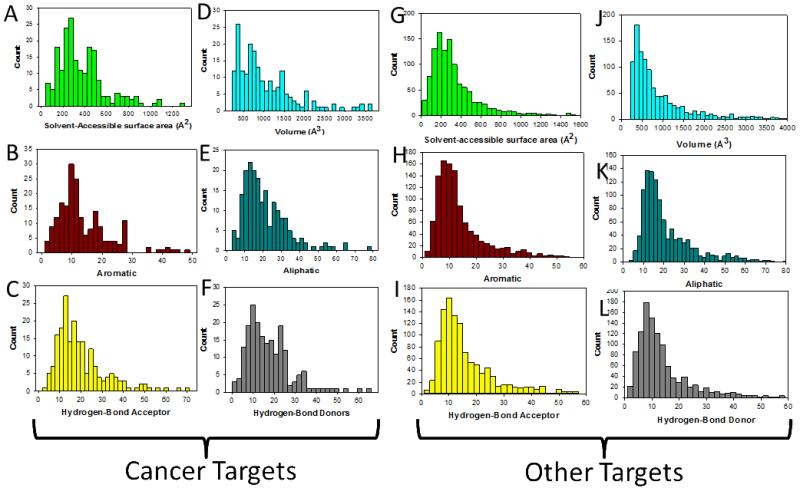
The solvent-accessible surface area (SASA) and volume were determined for each pocket to provide insight into their physico-chemical properties (Fig. 1). The SASA and volume define the shape and size of the pocket. The mean SASA for cancer and non-cancer targets is 367.6 and 353.5 Å2 respectively (Fig. 1A and 1G). To put this number in perspective, a typical SASA for a protein-protein interaction is at least 1000 Å2 while enzyme active site pockets are smaller. More than 90 percent of the pockets fall within 680 Å2. These cavities are located either at protein-protein interaction interfaces, enzyme active sites, or allosteric sites. The mean volume for the cavities is 1029.6 Å3 for cancer targets and 1061.4 Å3 for non-cancer targets (Fig. 1D and 1J). 90 percent of the targets have cavities with volumes that are within 1995 Å3.
Figure 1.
Physico-chemical properties of binding pockets on cancer targets and non-cancer targets. (A, G) Solvent-accessible surface area (SASA). (D, J) Volume of the cavities identified in targets. (B, H) Distribution of aromatic; (E, K) aliphatic; (C, I) hydrogen-bond acceptors; and (F, L) hydrogen bond donors within binding pockets.
To get insight into the physico-chemical properties of binding cavities within the cancer and druggable targets, we defined pseudocenters in the binding pockets following the approach of Klebe and coworkers.12 These pseudocenters consisted of aromatic, aliphatic, hydrogen bond donors, and acceptors as shown in Fig. 1. On average, there are 13.8 and 14.6 aromatic pseudocenters in the binding cavities of the proteome for cancer and non-cancer targets (Fig 1B and 1H). We found on average 21.1 and 22.7 aliphatic pseudocenters for cancer and non-cancer targets (Fig. 1E and 1K). Hydrogen bond donor and acceptor reflect hydrogen bonding capacity of residues within the binding cavities. The average number of acceptors was 19.7 and 17.3 for cancer and non-cancer targets (Fig. 1C and 1I). The mean number of donors was 17.3 and 14.6 for cancer non-cancer targets (Fig. 1F and 1L).
Physico-chemical properties and polypharmacology
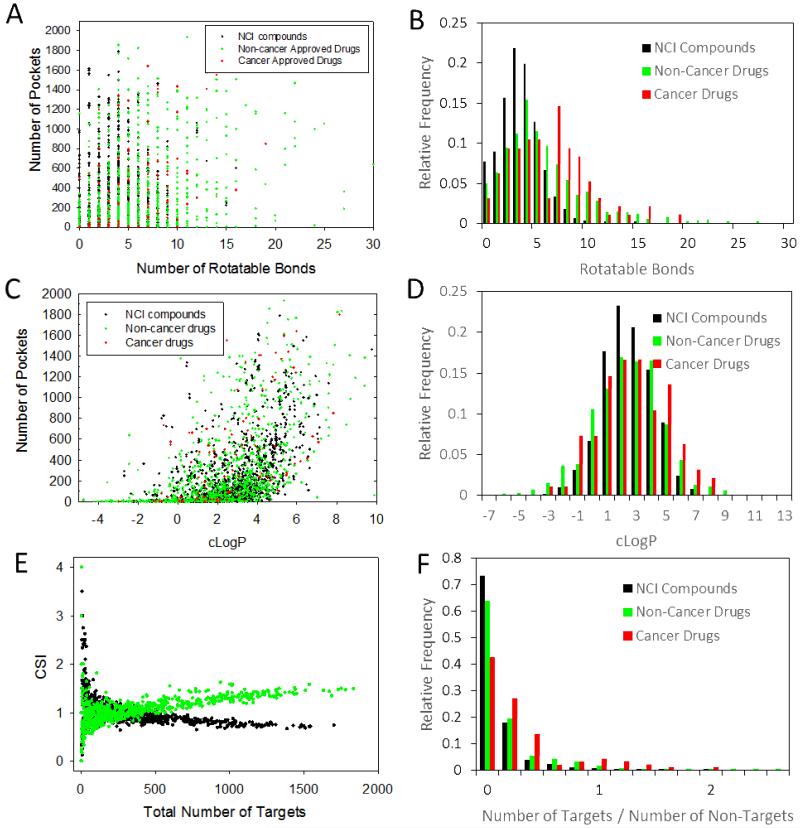
Flexibility and solubility are investigated for approved drugs, approved cancer drugs, and publicly-available NCI compounds. Flexibility is represented by the number of rotatable bonds. Using a threshold value of 0.1 μM, the number of targets for all three classes versus the size of the small molecule is provided in Fig. 2A. The most promiscuous compounds have about 5 rotatable bonds. The most selective compounds had less than 3 rotatable bonds. High promiscuity is predicted even for compounds with more than 10 rotatable bonds (Fig. 2B). In fact, some drugs with 20 rotatable bonds had more than 1,000 predicted targets at the 0.1 μM threshold. Approved cancer drugs followed a similar pattern. Rotatable bonds for NCI compounds, non-cancer drugs and cancer drugs showed different distributions (Fig. 2B). A significantly greater fraction of NCI compounds had 3-5 rotatable bonds compared with drugs and non-cancer drugs. Rotatable bonds were more uniformly distributed among approved drugs. A significant fraction of drugs and approved drugs had more than 7 rotatable bonds in significant contrast to NCI compounds. Cancer drugs were even more likely to have more than 7 rotatable bond than non-cancer drugs.
Figure 2.
Physico-chemical properties of the cancer drugs, non-cancer drugs and non-drug compounds using 0.1 μM binding threshold. (A) Compound rotatable bonds versus number of targets. (B) Distribution of rotatable bonds among NCI compounds, non-cancer and cancer drugs. (C) Logarithm of the partition coefficient (cLogP) vs. number of cavities. (D) Distribution of cLogP among NCI compounds, non-cancer and cancer drugs. (E) Ratio of cancer to non-cancer targets of cancer drugs and non-cancer drugs. (F) Distribution of ratio of targets over non-targets among for NCI compounds, non-cancer drugs, and cancer drugs.
It has been suggested that hydrophobic compounds are more promiscuous.13 Lipophilicity is quantified by the partition coefficient that corresponds to the ratio of the concentration of compound in water versus n-octanol. Several algorithms have been developed to predict the logarithm of the partition coefficient (cLogP).14 A plot of the number of pockets versus cLogP for all three classes of compounds shows a gradual increase in promiscuity for compounds with increasing cLogP (Fig. 2C). This is observed for approved non-cancer drugs, approved cancer drugs, and NCI compounds. The mean cLogP was 2.3, 2.7 and 2.5 for the three classes of compounds, suggesting that cancer drugs had slightly more hydrophobic character than other drugs and compounds. This is illustrated by the distribution in Fig. 2D as a greater proportion of cancer drugs had cLogP values greater than 5. The distribution also shows that compounds from the NCI library were more likely to have a cLogP between 1 and 3.
Drug pharmacology
Compounds that bind selectively to cancer-associated targets are more desirable as they are likely to possess greater efficacy and lower toxicity. To get insight into the selectivity of drugs, the Cancer Selectivity Index (CSI) is defined as the ratio of the number of predicted cancer target proteins from the HCPIN database to predicted non-cancer targets of approved drugs obtained from DrugBank. A plot of CSI versus the total number of predicted targets (HCPIN + DrugBank) for each drug is shown in Fig. 2E. A protein is considered a “target” when the predicted binding affinity from the ChemScore empirical scoring function exceeds a predefined threshold of 0.1 μM. ChemScore has been extensively validated for scoring protein-compound complexes.15, 16 For the majority of drugs, the CSI ratio is in the 0.7 to 1.2 range. This is not completely unexpected since pockets located on cancer targets are similar to those located on non-cancer targets. A close inspection of the data reveals that there were 500 drugs with CSI greater than 1. A significant proportion among them, 64 are approved cancer drugs. Twelve of these have a high degree of preference to cancer targets with CSI values greater than 2 (Table 1). All twelve had a total of less than 18 targets (that exceeded the 0.1 μM threshold). Their chemical structure is provided in Supporting Information Fig. S1. It was notable that the overwhelming majority of these drugs were fragment-like with molecular weights below 300 Da. They consisted of a single heterocyclic or aromatic ring structure with various appendages. This suggests that smaller compounds may be the most effective approach to achieve selective polypharmacology.
Table 1.
Drugs that Exhibit High Selectivity and Preference to Oncology Targets
| Drug Name | Total Cancer targets |
Total Approved Drugs targets |
CSI | Targets |
|---|---|---|---|---|
| Isoetharine | 4 | 1 | 4 | β1 adrenergic receptor |
| Salbutamol | 3 | 1 | 3 | β1,2 adrenergic receptor |
| Guanadrel sulfate |
2 | 1 | 2 | Sodium-dependent noradrenaline transporter |
| Diatrizoate | 4 | 2 | 2 | N/A |
| Rimantadine | 2 | 1 | 2 | Influenza A virus matrix protein 2 |
| Starvudine | 2 | 1 | 2 | HIV1 reverse transcriptase |
| Phensuximide | 2 | 1 | 2 | N/A |
| Diethylpropion | 8 | 4 | 2 | Sodium-dependent noradrenaline and dopamine transporters |
| Bromfenac | 12 | 6 | 2 | COX1, 2 |
| Methyprylon | 2 | 1 | 2 |
γ-aminobutyric acid receptor subunit alpha- 1 |
| Iophendylate | 2 | 1 | 2 | N/A |
| Methsuximide | 10 | 5 | 2 | Voltage-dependent T-type calcium channel |
A measure of the predicted polypharmacology of compounds was defined using the ratio of targets defined by the number of proteins that exceeded the 0.1 μM threshold to the number of proteins below this threshold. The distribution of this ratio is shown in Fig. 2F. The majority of NCI compounds and drugs exhibit a ratio below 0.2, with compounds showing greater selectivity than drugs. Cancer drugs exhibit less selectivity than NCI compounds and non-cancer drugs (Fig. 2F).
A survey of the literature reveals that at least four of these drugs bind to targets that have previously been implicated in cancer. For example, isoetharine and salbutamol are adrenergic β1, β2 agonists and are used for the treatment of bronchospasm, asthma, and chronic obstructive pulmonary disease. They bind and activate the β1 and β2 adrenergic receptors, which are involved in multiple metabolism pathways including calcium signaling, gap junction, salivary secretion, and endocytosis.17 Recent studies suggest these receptors are critical for the development of colorectal cancer.18 The third drug is methyprylon, a sedative of the piperidinedione derivative family and a treatment for insomnia. Up-regulation of microRNA miR-155 inhibits γ-aminobutyric acid A receptor 1 (GABRA1, target of methyprylon) and promotes tumor growth.19 The targets of the fourth drug (bromfenac) are cox1 and cox2, which are well-known to be involved in inflammation, which in turn has been implicated in cancer.20
Drug-target network
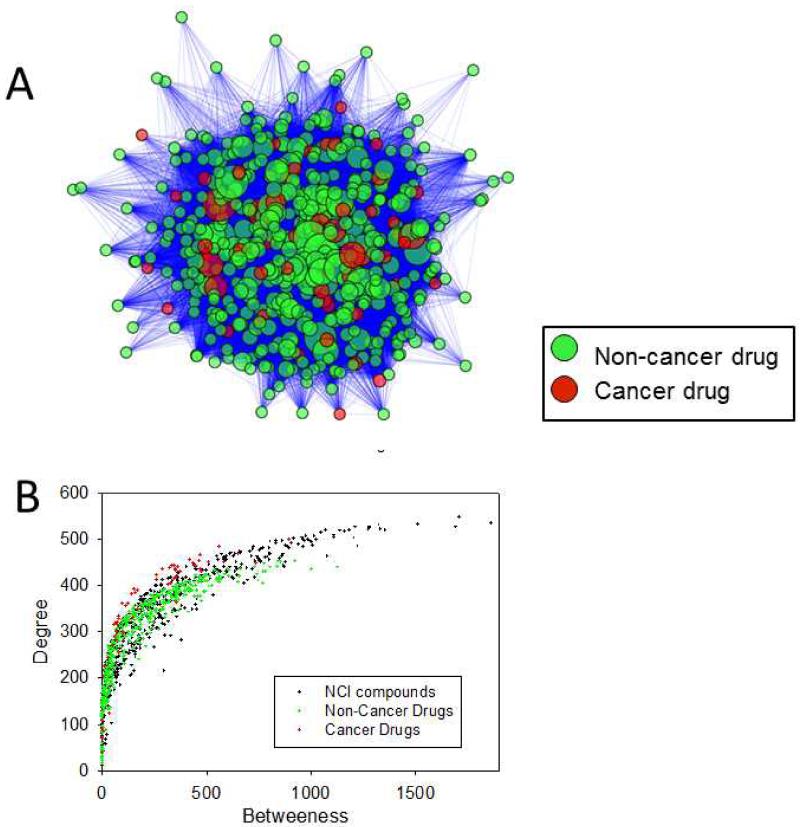
The interaction between small molecules and their targets can be understood within the context of a drug-target network.8, 21 The availability of complete protein-drug or protein-compound interactome affords the construction of a complete drug-target network (Fig. 3). In this network, a node represents a molecule and two nodes are linked if they share a cancer target. A protein is considered a target to a small molecule if its ChemScore predicted affinity is higher than 0.01 μM. We constructed a drug network for cancer and non-cancer FDA-approved drugs (Fig. 3A).
Figure 3.
Drug-target networks for cancer drugs and non-cancer drugs. (A) Overview of drug-target networks. Each node represents a drug and two nodes are linked if they share a target. (B) Degree and betweenness of the drugs. Degree of a node is defined as the number of edges connected to the node. Betweenness of a node is defined as the number of non-redundant shortest pathways going through each node.
A comparison of the two networks reveals a total of 120,314 and 54,632 edges, with 559 and 402 nodes for the NCI compounds and FDA-approved drugs, respectively. To gain insight into the level of interconnections of the nodes, we computed the mean degree for each network (the degree of a node corresponds to the number of edges connected to the node). The NCI compound network exhibited a mean degree of 430, while the FDA-approved drug network showed a mean degree of 272. The number of non-redundant shortest pathways going through each node (betweenness) was also computed for each network. The mean betweenness for the NCI network was 128 while that of the FDA-approved drug network was 131. A plot of the degree versus betweenness is shown in Fig. 3B. The top 10 drugs with highest betweenness and degree are provided in Table 2. It is worth noting that two of the ten compounds are cancer drugs, and one, namely bexarotene, is used in the treatment of lung cancer (Table 2).22 Finally, the mean clustering coefficient was determined to gain insight into the topology of the networks. Clustering coefficient of the NCI compound network is 0.913 while that of the FDA-approved drug network is 0.898. It was found that the NCI compound network was slightly less loosely connected than the FDA-approved drug network. These values are in excellent agreement with a previous study that computed them from a drug-network based on experimental data.23
Table 2.
Top FDA-Approved Drugs with the Highest Betweenness and Degree Obtained from a Network Analysis of the Data
| Name | Betweenness | Degree | Indication |
|---|---|---|---|
| Tacrine | 2027 | 348 | Alzheimer’s |
| Sulfisoxazole | 1896 | 310 | Antibacterial |
| Adapalene | 905 | 376 | Acne |
| Flurbiprofen | 786 | 293 | Inflammation and Pain |
| Naftifine | 651 | 367 | Antifungal |
| Conjugated Estrogens | 622 | 360 | - |
| Nilotinib | 593 | 355 | leukemia |
| Proflavine | 590 | 359 | bacteriostatic |
| Bexarotene | 571 | 374 | Cancer including lung |
| Tolcapone | 531 | 349 | Parkinson |
A search for new cancer drugs using compound polypharmacology
Previously, we had used predicted off-targets of erlotinib to identify compounds that mimicked the pharmacokinetic and anti-cancer properties of the cancer drug. Here, we extend this concept to identify FDA-approved drugs that could be potentially used in the treatment of lung cancer. Following the same approach a fingerprint is defined based on erlotinib’s predicted off-targets.10 We selected the top 12 drugs with highest similarity to the binding profile of erlotinib as measured by a Tanimoto coefficient that was determined by comparing fingerprints (Table 3). It was notable that among these 12 drugs, 4 are either currently used for the treatment of cancer, or have been considered in clinical trials as candidates for the treatment of cancer; these include lapatinib, dasatinib, bexarotene, and podofilox. Lapatinib is approved for advanced or metastatic breast cancer. Dasatinib is approved BCR-ABL positive chronic myelogenous leukemia. Bexarotene is approved for treatment of T-cell lymphoma and is being studied in a Phase II lung cancer clinical trial. All of these drugs were tested in clinical trials for treatment of lung cancer. Podophyillotoxin (podofilox) is approved to treat external genital warts; due to its tubulin modulation property and antimitotic function, podophyillotoxin and its derivatives may have anticancer properties.24 Another significant outcome of the ranking by fingerprint using erlotinib is the fact that two of the anti-cancer drugs, namely lapatinib and dasatinib, are kinase inhibitors just like erlotinib. The most significant aspect of this observation is that the fingerprint approach can be used to identify other drugs that target the same protein as the template drug without the use of chemical structure. It is interesting to note that none of the drugs shared any structural similarity with erlotinib. Hence, our fingerprint approach obviates the need to use chemical structure to identify pairs of small molecules that share similar targets.
Table 3.
Top FDA-Approved Drugs Identified using Erlotinib Binding Profile
| Name | Indication (DrugBank) |
|---|---|
| Ergotamine | Migraine headaches |
| Treprostinil | Pulmonary Arterial Hypertension |
| Bexarotene | Cutaneous T-cell lymphoma |
| Astemizole | Seasonal allergic rhinitis |
| Podofilox | External genital warts (Condyloma acuminatum) |
| Forasartan | Hypertension |
| Acenocoumarol | Thromboembolic diseases |
| Desoxycorticoserone Pivalate |
Adrenocortical insufficiency |
| Dihydroergotamine | Migraine headaches |
| Latanoprost | Glaucoma or occular hypertension |
| Lapatinib | Advanced or metastatic breast cancer |
| Dasatinib | Chronic, accelerated, or myeloid or lymphoid blast phase chronic myeloid leukemia |
In an effort to assess the effect of each drug on cancer cell growth, we performed an MTT study for each drug in three NSCLC cancer cell lines, namely H1299, A459, H460, and one non-cancer WI38 fibroblast cell line. All 32 MTT curves are provided in the Supporting Information Fig. S1 to S4. EC50 that were obtained from these curves are provided in Table 4. The most cytotoxic drug was podophyillotoxin with EC50 in the nanomolar concentrations (Table 4). This was not a surprising finding since this drug is a derivative of etoposide a well-known chemotherapeutic. The second most potent drug was dasatinib. In A549 and WI38, the compound inhibited proliferation at sub-micromolar EC50. Astemizole was the next most cytotoxic drug, EC50 of 12, 9, 10, and 8 μM for H1299, H460, A549 and WI38 cells, respectively. Lapatinib, another kinase inhibitor, showed significantly less inhibition of cell proliferation in all three cell lines with EC50 values between 30 to 40 μM. Bexarotene, which was previously tested in lung cancer clinical trials revealed weaker anti-proliferative effect (EC50 = ~50 μM), showed weaker effect on WI38 proliferation. Ergotamine, an analog of dihydroergotamine, had higher potency with an EC50 ~ 25 μM in H1299 and H460, and even greater potency in A549 cells (13 μM). What sets this compound apart from the others is that it had significantly less effect on WI38, providing a potential therapeutic window. Losartan, a drug used mainly to treat high blood pressure, showed very little cytotoxicity even at concentrations up to 100 μM.
Table 4.
EC50 Values for 8 Drugs on Lung Cancer Cells
| Drug Name | EC50 (μM) |
|||
|---|---|---|---|---|
| H1299 | H460 | A549 | WI38 | |
| Dihydroergotamine | 70±9 | 67±1 | 43±2 | 80±24 |
| Astemizole | 12±1 | 9±1 | 10±1 | 8±1 |
| Podophyllotoxin | 0.003±0.032 | 0.0002±0.0007 | 0.024±0.002 | - |
| Ergotamine | 252±3 | 26±1 | 14±1 | 57±4 |
| Losartan | - | - | 63±7 | 9±3 |
| Lapatinib | 41±2 | 32±1 | 67±11 | 32±1 |
| Dasatinib | - | - | 0.1±0.04 | 0.7±0.4 |
| Bexarotene | 44+2 | 52±2 | 58±4 | 76±10 |
Mining and statistical analysis of clinical drug exposure and disease occurrence
Patient cohorts were defined based on different drugs. For losartan, patient cohort was constructed as co-occurrence of hypertension prior to lung cancer plus mono-occurrence of hypertension without any kind of cancer. For ergotamine, patient cohort was constructed as co-occurrence of migraine pain prior to 12 major types of cancer (Supporting Information Table S1) plus mono-occurrence of migraine pain without any kind of cancer. All cohorts contained patients who had first diagnosis of hypertension or migraine pain at 30 years of age or older. We extracted 67,109 patients in the losartan/hypertension cohort, among which 65,411 patients had not been exposed to losartan and 1,698 patients had been exposed to losartan before first diagnosis of lung cancer or last visit date; and among which 1,574 patients were diagnosed with lung cancer sometime after first hypertension diagnosis and 65,535 patients were not diagnosed with any cancer before last visit date (Table 5). For ergotamine/migraine pain cohort, we extracted 44,721 patients in the ergotamine/migraine pain cohort, among which 44,509 patients had not been to ergotamine and 212 patients had been treated with ergotamine before first diagnosis of any of major cancer types; and among which 1,171 patients were diagnosed with any of 12 major cancers after first migraine pain diagnosis and 43,550 patients were not diagnosed with any cancer before last visit date (Table 6).
Table 5.
Patient Records Data to Assess Effect of Losartan in Cancer
| Lung cancer | No cancer | Total | |
|---|---|---|---|
| Losartan | 26 | 1672 | 1698 |
| No losartan | 1548 | 63863 | 65411 |
| Total | 1574 | 65535 | 67109 |
Table 6.
Patient Records Data to Assess Effect of Ergotamine in Cancer
| Any cancer | No cancer | Total | |
|---|---|---|---|
| Ergotamine | 13 | 199 | 212 |
| No ergotamine | 1158 | 43351 | 44509 |
| Total | 1171 | 43550 | 44721 |
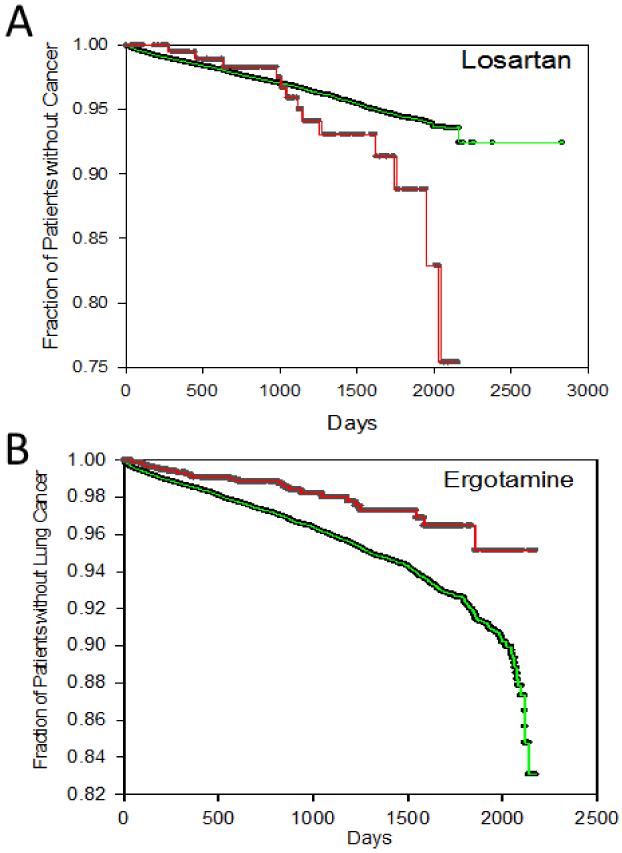
Survival statistical analysis was conducted for the association of drug exposure and risk of cancer (Fig. 4). Time to occurrence of cancer by drug exposure status was analyzed using the Kaplan-Meier method and log-rank test. Survival time (time to occurrence of cancer) was defined as the time from the date of first diagnosis of hypertension (for losartan) or migraine pain (for ergotamine) until date of first diagnosis of lung cancer (for losartan) or any of the 12 major cancer types (for ergotamine). Drug exposure status was considered positive if the patient was prescribed the drug before first diagnosis of cancer or last visit date. Patients who did not have cancer were censored until last visit date. The y-axis corresponds to the fraction of patients who had not been diagnosed with cancer. The x-axis corresponds to survival time in days from first diagnosis of hypertension until first diagnosis of lung cancer (for losartan group) (Fig. 4A), and days from first diagnosis of migraine pain until first diagnosis of any of 12 major cancers in the case of ergotamine (Fig. 4B). For losartan (Fig. 4A green curve), survival time was longer than without losartan (Fig. 4A red curve) at any cancer percentage in range. While with ergotamine (Fig. 4B green curve), survival time was shorter than without ergotamine (Fig. 4B red curve) at most cancer percentage in range. These conditions suggested further statistical confirmation on association of exposure of losartan and astemizole with reduced and enhanced cancer risk respectively.25
Figure 4.
Kaplan-Meier time to cancer occurrence curves of patients with (green curve) or without (red curve) losartan (A) and ergotamine (B). Time to cancer occurrence was measured from occurrence of 1st hypertension diagnosis to 1st diagnosis of lung cancer (A) and from occurrence of 1st migraine pain diagnosis to 1st diagnosis of any of the major cancer types (B).
In vivo studies in mouse xenograft models
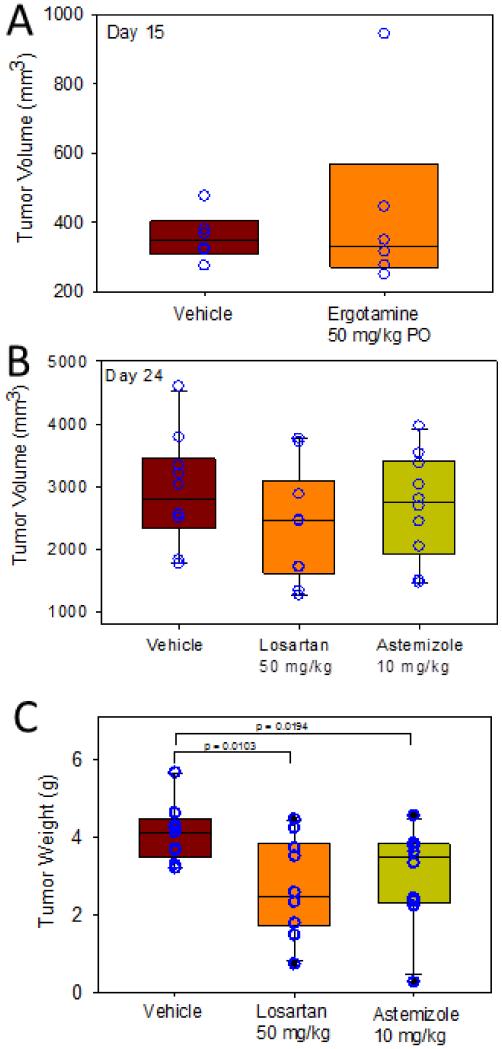
Astemizole, losartan, and ergotamine were evaluated for their effect in vivo on tumor growth using an H460 NSCLC human xenograft model. Two studies were carried out. The first study was done by orally dosing mice with ergotamine at 50 mg/kg orally (n = 7) (Fig. 5A). Vehicle mice (n = 8) were dosed with the methylcellulose. The study was terminated at day 21. While differences in tumor volume in vehicle versus compound-treated mice were not statistically significant, there were some trends worth noting in this early exploratory study. At day 15, tumor volume ranged from 249 to 944 mm3 for ergotamine-treated mice, and 274 to 743 mm3 for vehicle.
Figure 5.
Effects of losartan and astemizole on tumor growth in an H460 NSCLC xenograft mouse model.
Another study was carried out with losartan and astemizole. These drugs were administered i.p. at a dose of 50 and 10 mg/kg daily, respectively (n = 10 for losartan and vehicle; n = 9 for astemizole). Another difference is that mice were dosed with drug for a period of 7 days before tumors were implanted. At day 24, tumor size was measured (Fig. 5B). Tumor volume ranged from 1,770 to 4,600 mm3 for vehicle mice. For treated mice, tumor size ranged from 1271 to 3,773 for losartan, and 1,470 to 3,969 mm3 for astemizole, respectively. The median tumor volume was 2,800, 2,463, and 2,810 for vehicle, losartan, and astemizole, respectively. Tumor weights, which measured on the last day of the study were 50 percent smaller for losartan-treated mice (p<0.01), and 15 percent for mice treated with astemizole (p<0.01) (Fig. 5C). Four of the losartan-treated mice developed tumors that weight less than 2.5 g, compared with none of the vehicle (smallest tumors for vehicle was 3.2 g). Three astemizole treated mice developed tumors that weighed less than 3.2 g. During this study, the animal’s body weight was monitored (Supporting Information Fig S5) and no significant alteration was found.
Histopathology studies were performed on the resected lung tumors to evaluate the cell cycle arrest of NSCLC cells. The mitotic index (MI) was measured, which is defined as the ratio of mitotic cell to non-mitotic cells for tumor tissues treated with vehicle, losartan, and astemizole. The results were 7, 18, and 8 respectively. These data seem to suggest that losartan has a significant propensity to cause G2M arrest in the cell cycle, which may lead to apoptosis, similar to mechanism of Paclitaxel, a microtubule stabilizer and a well-known cancer drug.26, 27
CONCLUSION
We extend our protein-compound interactome splinter by docking FDA-approved drugs to a large set of proteins within the dataset. The scoring of these protein-compound structures using ChemScore led to a predicted binding affinity for each protein-compound pair. The resulting matrix of predicted binding affinities can be used to rank proteins for each drug to identify the most likely targets for that drug, or to rank drugs for individual proteins to identify potential hit compounds. A protein is defined as a target for a drug if its predicted binding affinity exceeds a pre-defined threshold value. This matrix was instrumental to enable us to get deeper insight into the pharmacology of these drugs particularly in cancer. Since our interactome consists of cancer and non-cancer proteins, it was possible to identify drugs that exhibited greater selectivity to cancer targets. The data revealed that selectivity for cancer targets can only be achieved only for compounds with fewer predicted targets overall. In addition, it was possible to study the predicted polypharmacology of compounds and drugs. In general compounds from chemical libraries had greater promiscuity than drugs, but cancer drugs exhibited more promiscuity than non-cancer drugs. In addition, physico-chemical properties of compounds and drugs led to significant differences predicted polypharmacology. The data also revealed that smaller fragment-like compounds exhibited greater selectivity. Finally, protein-compound scores enabled a network analysis and led to the discovery of highly interconnected hubs that may yield new cancer therapeutics among existing FDA-approved drugs. Interestingly, the parameters of these networks based on predicted binding affinity were in good agreement with previous network constructed on experimentally-determined interactions.
Beyond a deeper understanding of compound pharmacology, the protein-compound score matrix provided an opportunity to extend on previous work that revealed that binding profiles can be used effectively to identify compounds that share similar pharmacology.28 The binding profile of compounds was encapsulated into a fingerprint. We defined these fingerprints as bits of 0 and 1 that correspond to whether the compounds exceeded a pre-defined threshold. In our previous application we used a drug to search commercial libraries for compounds that mimic the properties of that drug.28 Here, we extend this approach to FDA-approved drugs that we have docked to all proteins within our interactome. As we have done previously, we use the lung cancer drug erlotinib as a template and use its fingerprint to search for other approved drugs that share a similar fingerprint with the expectation that these drugs will possess similar pharmacology to erlotinib. The fingerprints are compared using a Tanimoto coefficient as we have done previously.28 From this analysis, the top 12 drugs that possessed the most similar fingerprints as erlotinib were further analyzed. It was interesting that three of these drugs are already in use for treatment of lung and other cancers. Among the remaining nine drugs, cellular studies revealed that except for one case, these drugs were micromolar inhibitors of NSCLC proliferation in a panel of NSCLC that include A549, H1299 and H460.
We selected two drugs (losartan and ergotamine) that are commonly prescribed in the clinic and for which there is extensive clinical data at the Regenstrief Institute database. We were interested in evaluating whether patients that take these drugs are less likely to develop cancer than those that do not. Mining patient records at the Regenstrief Institute, our preliminary results indicate that ergotamine may hasten the onset of cancer; while losartan had the opposite effect. Further statistical analyses and controls are needed in future studies to make a definite link between these drugs and lung cancer in patients. Three drugs were tested in a sub-cutaneous model of NSCLC in NOD-SCID mice. Mice treated with losartan and astemizole had tumors that weighed 50% and 15% less than vehicle, respectively. In histopathological analysis of resected lung tumors, losartan induced more significant G2M arrest in the cell cycle.
MATERIALS AND METHODS
Docking approved drugs structures
Previously, we had docked 1592 compounds from the NCI diversity set to 1918 binding pockets that were found at the surface of protein structures that have been previously implicated in cancer.9, 10 In this work, an additional 1084 FDA-approved small molecule drugs obtained from DrugBank 29 were docked to 2546 cavities on 1738 proteins following the same process that we described previously.9 The strength of the interaction between drug and target was determined using the ChemScore empirical scoring function.
Calculation of physico-chemical properties
The protein targets were collected from two sources: HCPIN30 and DrugBank29, 31 databases. The first HCPIN release contained structures up to February of 2006. We created a local updated version of the database. We obtained sequence information for all HCPIN targets at the UniProt Web site (http://www.uniprot.org) using the SwissProt name provided by the HCPIN Web site. Proteins without a SwissProt name were not included. DrugBank provided sequence information for all targets of existing approved drugs obtained directly from the DrugBank Web site. In total, we collected 3,155 human sequences, 1,147 and 2,241 corresponded to DrugBank and HCPIN proteins, respectively. Among them, 233 were overlapped between the two databases. A blast search was carried against RCSB Protein Data Bank (PDB) proteins to map sequence to structures. The crystal structures were obtained from the PDB. 572 and 1065 PDB structures were identified for DrugBank and HCPIN sequences respectively. Solvents, ligands and binding partners were removed from the crystal structures. The Reduce program 32 was used to add hydrogen atoms to proteins and optimize some of the residue orientations. The MGLTools (v1.5.2) 33 was used to assign Gasteiger charges to the protein and generate a structural file for docking. The structural files were then processed with Relibase+ 34, 35 to detect binding pockets and compute pocket physico-chemical properties, including volume, aromatic pseudocenters, aliphatic pseudocenters, hydrogen bond donor, hydrogen bond acceptor and donor/acceptor. A probe radius of 1.4 Å was used to compute the pocket solvent accessible surface area (SASA). The physico-chemical properties for drugs and compounds, including cLogP and number of rotatable bonds, were computed with the QikProp program in the Schrödinger package. The mol2-formatted coordinate files of drugs and NCI diversity compounds were downloaded from DrugBank29 and ZINC.36
Erlotinib binding profile calculation
We used the crystal complex of elotinib bound to EGFR as the reference structure and identified 11 potential targets1 including EGFR from HCPIN by docking elotinib to HCPIN target crystal structures if available. The targets were selected using a consensus scoring function consisting of ChemScore37 and GoldScore38 implemented in SYBYL program. Complex scored more favorable than the reference structures was considered as a potential target and set an ON bit in the binding profile fingerprint. Hence there are 11 ON bits in case of elotinib, which constitutes the fingerprint of binding. The binding profile of any other FDA-approved small molecules was compared to the elotinib. The similarity was measured by Tanimoto coefficient.39
Cell culture
Human NSCLC cell lines H1299 and H460 cells were cultured in RPMI-1640 medium (Cellgro, Manassas, VA). Human epithelial cell line A549 was cultured in Dulbecco’s Modified Eagle Medium (Cellgro, Manassas, VA). Each medium was supplemented with 10% FBS and 1% penicillin/streptomycin in a 5% CO2 atmosphere at 37 °C.
Proliferation assay
The procedure consisted of culturing cells in 10% FBS-DMEM or RPMI-1640 medium containing various amounts of compounds. 20 mM drug stock in 100% DMSO was serially diluted and added into each well of a 96-well plate. Cells were treated and incubated for 3 days. Viable cells were quantified by MTT assay at absorbance of 570 and 630 nm.
Mining and statistical analysis of clinical drug exposure and disease occurrence data
Retrospective, observational clinical studies were performed with patient data in the Indiana Network of Patient Care (INPC) database formatted to Common Data Model (CDM) of the Observational Medicine Outcomes Partnership (OMOP), which is an NIH-funded public-private partnership for drug safety surveillance.40, 41 INPC is a local health information infrastructure, which is maintained at the Regenstrief Institute. It includes most of the Regenstrief Medical Record System (RMRS) clinical data (660 million separate results) from five major hospital systems (fifteen separate hospitals) of central Indiana, county and state public health departments, Indiana Medicaid, and RxHub.42 After INPC was formatted to the CDM format of OMOP, the database contained records of 2002480 distinct persons spanning from January 1, 2003 to December 31, 2009. The data structure of OMOP CDM 43 allowed us to retrieve patient data such as demographic data, starting/ending date of multiple episodes of drug exposure, starting date of disease diagnosis, and last visit date with database query language SQL. The extraction of diseases including hypertension, migraine pain and cancers was based on codes in Ninth Revision of International Classification of Disease (ICD-9) adopted by WHO in 1975 (Supporting Information Table S1). We focused on 12 major types of cancer which are among top ten deadly cancers in the U.S. either in male, female, or both for the year of 2008. Statistical analysis and graphing were performed with SAS (9.2), IBM SPSS Statistics 19 and SigmaPlot (11.0).
In vivo xenograft studies
NOD/SCID mice were obtained from the on-site breeding colony maintained by the In Vivo Therapeutics Core at the Indiana University Simon Cancer Center (IUSM, Indianapolis, IN). H460 cells (2 × 106) were injected subcutaneously into the right flank of 8-10 week old NOD-SCID mice. These cells were obtained directly from ATCC (Manassas, VA) and used at low passage (<10). Mice were randomized to treatment group based on average tumor volume (mm3). Two different studies were conducted. The first consisted of PO dosing of mice with ergotamine (n =7) at a dose of 50 mg/kg or a PBS solvent control (n = 8). Mice were dosed once a day for 14 days. The second study involved three groups: Astemizole, losartan, and PBS control. For this study, prior to tumor implant, the animals were pre-treated with drugs daily for a period of 7 days. Following subcutaneous tumor implantation animals were treated with astemizole and losartan that were administered intraperitoneally at 10 mg/kg, once a day for 28 days, respectively. Tumor growth was measured over time via electronic caliper and volume calculated as Length * Width2/2 in millimeters. After four weeks mice were euthanized, the lungs were resected, fixed in formalin solution, sectioned, and stained with hematoxylin and eosin (H&E) for analysis.
Supplementary Material
ACKNOWLEDGMENTS
The research was supported by the NIH (CA135380 and AA0197461) and the INGEN grant from the Lilly Endowment, Inc. (SOM). We acknowledge Indiana University School of Medicine Lungs for Life fellowship to LL. XP is a recipient of National Library of Medicine Biomedical Informatics Fellowship (LM007117-14). Computer time on the Big Red supercomputer at Indiana University is funded by the National Science Foundation and by Shared University Research grants from IBM, Inc. to Indiana University. We wish to acknowledge Tony Sinn, Jayne Silver and Kacie Peterman and the In Vivo Therapeutics Core for their expert technical assistance with the in vivo studies.
REFERENCES
- 1.Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, Yue P, Zhang Y, Pant KP, Bhatt D, Ha C, Johnson S, Kennemer MI, Mohan S, Nazarenko I, Watanabe C, Sparks AB, Shames DS, Gentleman R, de Sauvage FJ, Stern H, Pandita A, Ballinger DG, Drmanac R, Modrusan Z, Seshagiri S, Zhang Z. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 2.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 4.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Jr., Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JK, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PV, Ballinger DG, Sparks AB, Hartigan J, Smith DR, Suh E, Papadopoulos N, Buckhaults P, Markowitz SD, Parmigiani G, Kinzler KW, Velculescu VE, Vogelstein B. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 6.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, Fulton L, Fulton RS, Zhang Q, Wendl MC, Lawrence MS, Larson DE, Chen K, Dooling DJ, Sabo A, Hawes AC, Shen H, Jhangiani SN, Lewis LR, Hall O, Zhu Y, Mathew T, Ren Y, Yao J, Scherer SE, Clerc K, Metcalf GA, Ng B, Milosavljevic A, Gonzalez-Garay ML, Osborne JR, Meyer R, Shi X, Tang Y, Koboldt DC, Lin L, Abbott R, Miner TL, Pohl C, Fewell G, Haipek C, Schmidt H, Dunford-Shore BH, Kraja A, Crosby SD, Sawyer CS, Vickery T, Sander S, Robinson J, Winckler W, Baldwin J, Chirieac LR, Dutt A, Fennell T, Hanna M, Johnson BE, Onofrio RC, Thomas RK, Tonon G, Weir BA, Zhao X, Ziaugra L, Zody MC, Giordano T, Orringer MB, Roth JA, Spitz MR, Wistuba, Ozenberger B, Good PJ, Chang AC, Beer DG, Watson MA, Ladanyi M, Broderick S, Yoshizawa A, Travis WD, Pao W, Province MA, Weinstock GM, Varmus HE, Gabriel SB, Lander ES, Gibbs RA, Meyerson M, Wilson RK. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hopkins AL. Nat Chem Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 8.Paolini GV, Shapland RH, van Hoorn WP, Mason JS, Hopkins AL. Nat Biotechnol. 2006;24:805–815. doi: 10.1038/nbt1228. [DOI] [PubMed] [Google Scholar]
- 9.Li L, Bum-Erdene K, Baenziger PH, Rosen JJ, Hemmert JR, Nellis JA, Pierce ME, Meroueh SO. Nucleic acids research. 2010;38:D765–773. doi: 10.1093/nar/gkp852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Li J, Khanna M, Jo I, Baird JP, Meroueh SO. ACS Med Chem Lett. 2010;1:229–233. doi: 10.1021/ml100031a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R. New Engl J Med. 2005;353:123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 12.Schmitt S, Kuhn D, Klebe G. Journal of molecular biology. 2002;323:387–406. doi: 10.1016/s0022-2836(02)00811-2. [DOI] [PubMed] [Google Scholar]
- 13.Hopkins AL, Mason JS, Overington JP. Curr Opin Struct Biol. 2006;16:127–136. doi: 10.1016/j.sbi.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 14.Sangster J. European Journal of Medicinal Chemistry. 1997;32:842–842. [Google Scholar]
- 15.Wang R, Lu Y, Fang X, Wang S. J Chem Inf Comp Sci. 2004;44:2114–2125. doi: 10.1021/ci049733j. [DOI] [PubMed] [Google Scholar]
- 16.Ferrara P, Gohlke H, Price DJ, Klebe G, Brooks CL. J. Med. Chem. 2004;47:3032–3047. doi: 10.1021/jm030489h. [DOI] [PubMed] [Google Scholar]
- 17.Brodde OE. Pharmacology & therapeutics. 2008;117:1–29. doi: 10.1016/j.pharmthera.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Wong HPS, Ho JWC, Koo MWL, Yu L, Wu WKK, Lam EKY, Tai EKK, Ko JKS, Shin VY, Chu KM. Life sciences. 2011;88:1108–1112. doi: 10.1016/j.lfs.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 19.D’URSO PI, D’URSO OF, Storelli C, Mallardo M, DAMIANO GIANFREDA C, Montinaro A, Cimmino A, Pietro C, Marsigliante S. International journal of oncology. 2012;41:228–234. doi: 10.3892/ijo.2012.1420. [DOI] [PubMed] [Google Scholar]
- 20.Cross JT, Poole EM, Ulrich CM. The pharmacogenomics journal. 2008;8:237–247. doi: 10.1038/sj.tpj.6500487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Nature Biotechnology. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 22.Dragnev KH, Petty WJ, Shah SJ, Lewis LD, Black CC, Memoli V, Nugent WC, Hermann T, Negro-Vilar A, Rigas JR, Dmitrovsky E. Clin Cancer Res. 2007;13:1794–1800. doi: 10.1158/1078-0432.CCR-06-1836. [DOI] [PubMed] [Google Scholar]
- 23.Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Nat Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- 24.Clark P, Cottier B. The activity of 10-, 14-, and 21-day schedules of single-agent etoposide in previously untreated patients with extensive small cell lung cancer. 1992 [PubMed] [Google Scholar]
- 25.Azoulay L, Assimes TL, Yin H, Bartels DB, Schiffrin EL, Suissa S. PloS one. 2012;7:e50893. doi: 10.1371/journal.pone.0050893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmed W, Rahmani M, Dent P, Grant S. Cell Cycle. 2004;3:1305–1311. doi: 10.4161/cc.3.10.1161. [DOI] [PubMed] [Google Scholar]
- 27.Pasquier E, Honore S, Pourroy B, Jordan MA, Lehmann M, Briand C, Braguer D. Cancer Res. 2005;65:2433–2440. doi: 10.1158/0008-5472.CAN-04-2624. [DOI] [PubMed] [Google Scholar]
- 28.Li L, Li J, Khanna M, Jo I, Baird JP, Meroueh SO. ACS. Med. Chem. Lett. 2010;1:229–233. doi: 10.1021/ml100031a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. Nucleic acids research. 2008;36:D901–906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang YJ, Hang D, Lu LJ, Tong L, Gerstein MB, Montelione GT. Mol Cell Proteomics. 2008;7:2048–2060. doi: 10.1074/mcp.M700550-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. Nucleic acids research. 2006;34:D668–672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Word JM, Lovell SC, Richardson JS, Richardson DC. J Mol Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 33.Sanner MF. J Mol Graph Model. 1999;17:57–61. [PubMed] [Google Scholar]
- 34.Hendlich M, Bergner A, Gunther J, Klebe G. J. Mol. Biol. 2003;326:607–620. doi: 10.1016/s0022-2836(02)01408-0. [DOI] [PubMed] [Google Scholar]
- 35.Bergner A, Gunther J, Hendlich M, Klebe G, Verdonk M. Biopolymers. 2001;61:99–110. doi: 10.1002/1097-0282(2001/2002)61:2<99::AID-BIP10075>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 36.Irwin JJ, Shoichet BK. J Chem Inf Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. J Comput Aid Mol Des. 1997;11:425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- 38.Jones G, Willett P. Curr Opin Biotechnol. 1995;6:652–656. doi: 10.1016/0958-1669(95)80107-3. [DOI] [PubMed] [Google Scholar]
- 39.Flower DR. Journal of molecular graphics & modelling. 1998;16:239–253. 264. doi: 10.1016/s1093-3263(98)80008-9. [DOI] [PubMed] [Google Scholar]
- 40.Overhage JM, Ryan PB, Reich CG, Hartzema AG, Stang PE. Journal of the American Medical Informatics Association. 2012;19:54–60. doi: 10.1136/amiajnl-2011-000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stang PE, Ryan PB, Racoosin JA, Overhage JM, Hartzema AG, Reich C, Welebob E, Scarnecchia T, Woodcock J. Annals of internal medicine. 2010;153:600. doi: 10.7326/0003-4819-153-9-201011020-00010. [DOI] [PubMed] [Google Scholar]
- 42.McDonald CJ, Overhage JM, Barnes M, Schadow G, Blevins L, Dexter PR, Mamlin B. Health Affairs. 2005;24:1214–1220. doi: 10.1377/hlthaff.24.5.1214. [DOI] [PubMed] [Google Scholar]
- 43.Ryan P, Griffin D, Reich C. 2009 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.