Abstract
Nitric oxide (NO) deficiency is involved in the development of hypertension, a condition that can originate early in life. We examined whether NO deficiency contributed to programmed hypertension in offspring from mothers with calorie-restricted diets and whether melatonin therapy prevented this process. We examined 3-month-old male rat offspring from four maternal groups: untreated controls, 50% calorie-restricted (CR) rats, controls treated with melatonin (0.01% in drinking water), and CR rats treated with melatonin (CR + M). The effect of melatonin on nephrogenesis was analyzed using next-generation sequencing. The CR group developed hypertension associated with elevated plasma asymmetric dimethylarginine (ADMA, a nitric oxide synthase inhibitor), decreased L-arginine, decreased L-arginine-to-ADMA ratio (AAR), and decreased renal NO production. Maternal melatonin treatment prevented these effects. Melatonin prevented CR-induced renin and prorenin receptor expression. Renal angiotensin-converting enzyme 2 protein levels in the M and CR + M groups were also significantly increased by melatonin therapy. Maternal melatonin therapy had long-term epigenetic effects on global gene expression in the kidneys of offspring. Conclusively, we attributed these protective effects of melatonin on CR-induced programmed hypertension to the reduction of plasma ADMA, restoration of plasma AAR, increase of renal NO level, alteration of renin-angiotensin system, and epigenetic changes in numerous genes.
1. Introduction
Hypertension might originate during early life. Maternal malnutrition can impair development, resulting in intrauterine growth restriction (IUGR), permanent structural changes, and disrupted physiological function—a phenomenon called “developmental programming” [1]. In the kidneys of both humans and experimental models, developmental programming reduces nephron numbers, alters the renin-angiotensin system (RAS), and impairs natriuresis, leading to adult kidney disease and hypertension [2–5].
A number of hypotheses have been proposed to explain the developmental programming phenomenon [6]. Oxidative stress is proposed as the underlying link between developmental programing and elevated risks of hypertension and kidney disease in adulthood [7, 8]. Asymmetric dimethylarginine (ADMA), an endogenous inhibitor of NO synthase (NOS), causes oxidative stress and is involved in the development of hypertension [9]. Our recent work demonstrated that an impaired ADMA-NO pathway and low nephron numbers are associated with programmed hypertension in the adult offspring of malnourished or diabetic mothers [10, 11]. Reduced nephron numbers impaired renal tubular sodium reabsorption, and the altered RAS components disrupted sodium retention, ultimately increasing blood pressure (BP) and inducing kidney damage. Histone deacetylases (HDACs) repress gene expression, a mechanism of epigenetic control that is involved in developmental programming. Class I HDACs are critical in nephrogenesis, particularly HDAC1-3 that are highly expressed in nephron precursors [12]. HDACs also play an important role in regulating RAS components during nephrogenesis [13]. These observations suggest that these mechanisms jointly lead to the development of hypertension and kidney disease.
Melatonin, an indoleamine produced from the pineal gland, is an antioxidant and free radical scavenger [14]. Experimental and human studies indicate that melatonin can regulate BP [10, 11]. We recently found that melatonin can prevent oxidative stress and hypertension concurrently in young spontaneously hypertensive rats (SHR) [15]. Emerging evidence supports novel roles of melatonin in epigenetic modulation through the regulation of HDACs [16, 17]. Thus, we examined whether melatonin prevented programmed hypertension in offspring exposed to maternal caloric restriction through reduction of oxidative stress, alteration of the RAS pathway, and modulation of HDACs. Moreover, we identified melatonin-induced gene changes during nephrogenesis and determined whether melatonin treatment induced global changes in biological processes by using next-generation sequencing.
2. Material and Methods
2.1. Animal Models
This study was carried out in strict accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of the Kaohsiung Chang Gung Memorial Hospital. Virgin Sprague-Dawley (SD) rats (12–16 weeks old) were obtained from BioLASCO Taiwan Co., Ltd. (Taipei, Taiwan), and were housed and maintained in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. Male SD rats were caged with individual females until mating was confirmed. Calorie-restricted (CR) maternal rats received 11 g/d of a standard chow from day 11 of pregnancy until the day of delivery (day 23) and 20 g/d during the entire lactation period [10]. A subset of CR mothers was treated for the duration of the pregnancy with 0.01% melatonin dissolved in drinking water (CR + M, n = 8). The control group (n = 8) mothers had free access to standard rat chow. As another control, maternal rats were allowed free access to standard rat chow and were treated with 0.01% melatonin in drinking water (M, n = 10). After birth, each litter was left with the mother until weaning; pups were not weighed at birth to prevent maternal rejection. Male offspring, selected at random from each litter, were used in all subsequent experiments. In rats, nephrogenesis occurs predominantly from late gestation to 1-2 weeks postnatum and litters were typically weaned by postnatal week 3. Thus, melatonin was administered to mother rats for a total period of 6 weeks to cover the entire period of nephrogenesis. The dose of melatonin used was based on our previous study [15]. Water bottles were covered with aluminum foil to protect them from light. BP was measured in conscious rats by using an indirect tail-cuff method (BP-2000, Visitech Systems, Inc., Apex, NC, USA) after they had been systematically trained [10]. Three stable consecutive measures were taken and averaged. All offspring were sacrificed at 12 weeks of age and heparinized blood samples were collected. Kidneys were harvested after perfusion with PBS, divided into cortex and medulla regions, and snap-frozen. The activity of dimethylarginine dimethylaminohydrolase (DDAH), an ADMA-metabolizing enzyme, was measured using a colorimetric assay. The assay determined the rate of l-citrulline production and we performed the assay as previously described [18].
2.2. High-Performance Liquid Chromatography (HPLC)
Plasma and kidney l-arginine, l-citrulline, ADMA, and symmetric dimethylarginine (SDMA, a stereoisomer of ADMA) levels were measured using HPLC (HP series 1100, Agilent Technologies, Inc., Santa Clara, CA, USA) with the OPA-3MPA derivatization reagent as we described previously [10]. Standards contained l-arginine, l-citrulline, ADMA, and SDMA in the range of 1–100 μM, 1–100 μM, 0.5–5 μM, and 0.5–5 μM, respectively. The recovery rate was between 90 and 105%. The tissue concentration was factored for protein concentration, which was represented as μmol/mg protein. Plasma and urine creatinine (Cr) levels were analyzed by HPLC as described previously [10]. The creatinine clearance (CCr) was calculated by dividing the total amount of Cr excreted in urine by the Cr concentration in plasma. CCr values were normalized with respect to body weight.
2.3. Electron Paramagnetic Resonance (EPR)
Superoxide production was measured by EPR spectroscopy using a 1-hydroxy-3-carboxypyrrolidine (CPH) hydroxylamine spin probe, as we previously described [11]. The EPR spectra were recorded using an EMX Plus EPR spectrometer (Bruker BioSpin, Rheinstetten, Germany) equipped with an EMX-m40X microwave bridge operating at 9.87 GHz. NO was detected by EPR using N-methyl-D-glucamine dithiocarbamate (MGD) spin probe and FeSO4, as previously described [11]. The EPR spectra were recorded using an EMX Plus EPR spectrometer (Bruker BioSpin) equipped with an EMX-m40X microwave bridge operating at 3.16 GHz.
2.4. Metanephros Organ Culture
Metanephros organ culture was performed as we described previously [11]. Briefly, SD female rats of known mating date were anesthetized and laparotomized. Fetuses were aseptically removed, and metanephroi from fetuses at embryonic day 14 (E14) were collected and freed of exogenous tissue. Explants were placed onto a Steritop filter unit (Millipore, Billerica, MA, USA) floating on a defined serum-free medium and incubated for 6 d in 35 mm Petri dishes at 37°C in a humidified incubator (5% CO2). The defined medium was composed of Eagle's Minimum Essential Medium containing 10% (v/v) fetal calf serum, 100 units/mL penicillin, and 100 μg/mL streptomycin. All of these reagents were obtained from Sigma (St. Louis, MO, USA). The culture medium was changed daily, and no antibiotic or fungicide was present throughout the experiment. Fresh aliquots of each culture medium additive were used for each metanephros culture. The medium was changed daily. Metanephroi were treated with melatonin (1 μM and 1 mM) and harvested after 6 d for real-time polymerase chain reaction.
2.5. Quantitative Real-Time Polymerase Chain Reaction (PCR)
RNA was extracted as described previously [10]. Two-step quantitative real-time PCR was conducted using the QuantiTect SYBR Green PCR Kit (Qiagen, Valencia, CA, USA) and the iCycler iQ Multicolor Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Nephron deficit was assessed by changes in the expression factors known to be involved in branching morphogenesis (BMP4, FGF2, and PAX2) and apoptosis (p53 and Bax). Several components of the RAS were analyzed including renin, prorenin receptor (PRR), angiotensinogen (AGT), angiotensin-converting enzyme (ACE), ACE2, angiotensin II type 1 (AT1R) and 2 receptor (AT2R), and angiotensin (1–7) MAS receptor. Class I HDACs, HDAC-1, -2, -3, and -8, were also examined. We used 18S rRNA (r18S) as a reference. Primers were designed using GeneTool Software (BioTools, Edmonton, Alberta, Canada) (Table 1). All samples were run in duplicate. To quantify the relative gene expression, the comparative threshold cycle (CT) method was employed. For each sample, the average CT value was subtracted from the corresponding average r18S value, calculating the ΔCT. ΔΔCT was calculated by subtracting the average control ΔCT value from the average experimental ΔCT. The fold-increase of the experimental sample relative to the control was calculated using the formula 2−ΔΔCT.
Table 1.
PCR primers sequences.
| Gene | Forward | Reverse |
|---|---|---|
| Bax | 5 ttgctgatggcaacttcaactg 3 | 5 ctttagtgcacagggccttgag 3 |
| P53 | 5 catgagcgttgctctgatg 3 | 5 cagatactcagcatacggatttcc 3 |
| PAX2 | 5 gagactcccagagtggtgtg 3 | 5 cattcccctgttctgatttg 3 |
| FGF2 | 5 ccagttggtatgtggcactg 3 | 5 cagggaagggtttgacaaga 3 |
| BMP4 | 5 gacttcgaggcgacacttctg 3 | 5 agccggtaaagatccctcatg 3 |
| Renin | 5 aacattaccagggcaactttcact 3 | 5 acccccttcatggtgatctg 3 |
| Prorenin receptor | 5 gaggcagtgaccctcaacat 3 | 5 ccctcctcacacaacaaggt 3 |
| Angiotensinogen | 5 gcccaggtcgcgatgat 3 | 5 tgtacaagatgctgagtgaggcaa 3 |
| ACE | 5 caccggcaaggtctgctt 3 | 5 cttggcatagtttcgtgaggaa 3 |
| ACE2 | 5 acccttcttacatcagccctactg 3 | 5 tgtccaaaacctaccccacatat 3 |
| AT1R | 5 gctgggcaacgagtttgtct 3 | 5 cagtccttcagctggatcttca 3 |
| AT2R | 5 caatctggctgtggctgactt 3 | 5 tgcacatcacaggtccaaaga 3 |
| MAS | 5 catctctcctctcggctttgtg 3 | 5 cctcatccggaagcaaagg 3 |
| HDAC-1 | 5 gaactggggacctacggg 3 | 5 gctcttgacaaattccacacac 3 |
| HDAC-2 | 5 agttgcccttgattgtgaga 3 | 5 ccactgttgtccttggatttat 3 |
| HDAC-3 | 5 tgatgaccagagttacaagcac 3 | 5 gggcaacatttc ggacag 3 |
| HDAC-8 | 5 gctacccccggtttatatttacag 3 | 5 ttcgatcagagagtgaaccatactg 3 |
| R18S | 5 gccgcggtaattccagctcca 3 | 5 cccgcccgctcccaagatc 3 |
2.6. Western Blot
Western blot analysis was performed as previously described [10]. We used the following antibodies from Santa Cruz Biotechnology (Santa Cruz, CA, USA): rabbit polyclonal anti-rat PRR (1 : 500, overnight incubation), rabbit anti-rat ACE2 (1 : 1000, overnight incubation), rabbit anti-rat AT1R (1 : 250, overnight incubation), rabbit anti-rat AT2R (1 : 250, overnight incubation), and rabbit anti-rat MAS (1 : 1000, overnight incubation; Santa Cruz Biotechnology). The bands of interest were visualized using enhanced chemiluminescence reagent (PerkinElmer, Waltham, MA, USA) and quantified by densitometry (Quantity One Analysis software, Bio-Rad). Band density was calculated as the integrated optical density (IOD) minus the background value. The density of Ponceau red staining (PonS) was used to correct for variations in total protein loading. Protein abundance was calculated as IOD/PonS.
2.7. Next-Generation Sequencing and Analysis
In rats, nephrogenesis occurs predominantly from late gestation to 7–10 days postnatum. Thus, offspring from the control and M groups were sacrificed at 1 week of age. Kidneys were isolated and snap-frozen for whole-genome RNA next-generation sequencing (RNA-seq), performed by Welgene Biotech Co., Ltd. (Taipei, Taiwan). Purified RNA was quantified at 260 nm (OD600) by using ND-1000 spectrophotometer (Nanodrop Technology, Wilmington, DE, USA) and analyzed using a Bioanalyzer 2100 (Agilent Technology) with RNA 6000 LabChip kit (Agilent Technologies). All procedures were performed according to the Illumina protocol. For all samples, library construction was performed using the TruSeq RNA Sample Prep Kit v2 for ∼160 bp (single-end) sequencing and the Solexa platform. The sequence was directly determined by sequencing-by-synthesis technology using the TruSeq SBS Kit. Raw sequences were obtained using the Illumina GA Pipeline software CASAVA v1.8, which was expected to generate 10 million reads per sample. Quantification for gene expression was calculated as reads per kilobase of exon per million mapped reads [19]. For differential expression analysis, Cufflink v 2.1.1 and CummeRbund v 2.0.0 were used to perform statistical analyses of the gene expression profiles. The reference genome and gene annotations were retrieved from the Ensembl database (http://asia.ensembl.org/index.html). Gene ontology analysis for significant genes was performed using KEGG (http://www.genome.jp/kegg/) and NIH DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/) to identify regulated biological themes.
2.8. Statistical Analysis
TheShapiro-Wilk normality test was used to determine which data were normally distributed. Normally distributed data are given as mean ± standard error of the mean. For most parameters, statistical analysis was performed using one-way analysis of variance (ANOVA) and Tukey's post hoc test for multiple comparisons. BP was analyzed by two-way repeated-measures ANOVA and Tukey's post hoc test. A P value < 0.05 was considered statistically significant. Analyses were performed using the Statistical Package for the Social Sciences (SPSS) software (Chicago, IL, USA).
3. Results
3.1. The Effects of Melatonin on Morphological and Biochemical Values in CR Rats
Litter sizes were not significantly altered by caloric restriction in the mother rat or by melatonin treatment. The amounts of water intake and urine output were not significantly different in the control and CR groups. Male pup mortality rates did not differ between the four groups analyzed. As shown in Table 2, the CR and M groups had lower and higher body weight (BW) than the control at 12 weeks of age, respectively, whereas the CR + M group had an intermediate BW. Kidney weight and kidney weight-to-BW ratio did not differ between the control and CR groups. Melatonin significantly increased kidney weight and kidney weight-to-BW ratio in the M and CR + M groups. Although heart weight was not different between control and CR groups, the heart weight-to-BW ratio was greater in the CR group. Melatonin caused increased heart weight and heart weight-to-BW ratio in the M group, but not in the CR + M group. CR increased systolic and diastolic BP and mean arterial pressure at 12 weeks of age. Melatonin therapy prevented these effects of CR. In addition, melatonin therapy reduced diastolic BP and mean arterial pressure in the M group compared to the control. As shown in Figure 1, mean arterial pressure was similar in the four groups at 4 weeks of age. By 8 weeks of age, mean arterial pressure had increased in the CR group relative to controls. A significant reduction in mean arterial pressure was measured in the M and CR + M groups versus the control at 8 and 12 weeks of age. In contrast, plasma creatinine level did not differ between the four groups. These data demonstrated that CR induced programmed hypertension but had no effect on renal function on 12-week-old offspring.
Table 2.
Morphological and biochemical values in different experimental groups.
| Control | CR | M | CR + M | |
|---|---|---|---|---|
| n = 8 | n = 8 | n = 10 | n = 8 | |
| Mortality | 10% | 0% | 0% | 0% |
| Body weight (BW) (g) | 435 ± 14 | 356 ± 4* | 489 ± 8∗# | 370 ± 9∗$ |
| Left kidney weight (g) | 1.22 ± 0.06 | 1.01 ± 0.02 | 1.97 ± 0.05∗# | 1.48 ± 0.03#$ |
| Left kidney weight/100g BW | 0.28 ± 0.01 | 0.28 ± 0.01 | 0.4 ± 0.01∗# | 0.4 ± 0.01∗# |
| Heart weight (g) | 1.23 ± 0.05 | 1.24 ± 0.02 | 1.63 ± 0.01∗# | 1.16 ± 0.05$ |
| Heart weight/100 g BW | 0.28 ± 0.01 | 0.35 ± 0.01* | 0.35 ± 0.01* | 0.31 ± 0.01 |
| Systolic blood pressure (mmHg) | 162 ± 2 | 180 ± 2* | 155 ± 1# | 166 ± 1$ |
| Diastolic blood pressure (mmHg) | 122 ± 2 | 134 ± 3* | 108 ± 2∗# | 113 ± 1∗# |
| Mean arterial pressure (mmHg) | 135 ± 2 | 149 ± 2* | 124 ± 1∗# | 131 ± 1#$ |
| CCr, mL·min−1·kg body weight−1 | 9.12 ± 3.45 | 8.5 ± 3.0 | 7.34 ± 2.32 | 7.81 ± 2.76 |
CCr: clearance of creatinine; *P < 0.05 versus control; # P < 0.05 versus CR; $ P < 0.05 versus M.
Figure 1.
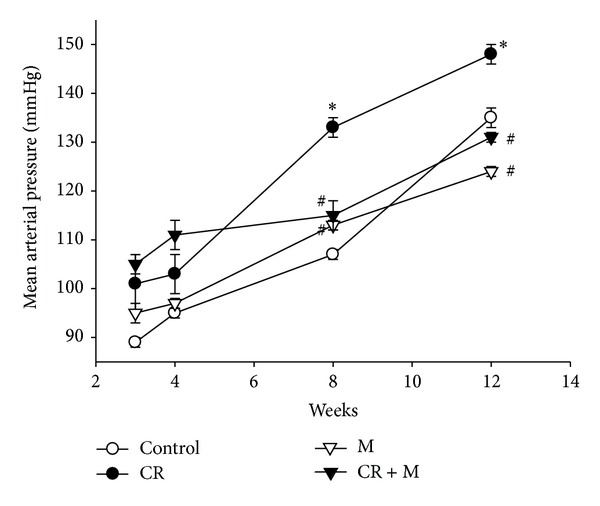
Effect of melatonin and caloric restriction (CR) on mean arterial pressure in male offspring at 12 weeks of age. *P < 0.05 versus control; # P < 0.05 versus CR.
3.2. The Effects of Melatonin on l-Arginine, l-Citrulline, and Dimethylarginine Levels
As shown in Table 3, plasma levels of ADMA and SDMA were elevated nearly 70% and 150% following maternal CR, respectively. In contrast, the l-arginine levels and l-arginine-to-ADMA ratio were decreased by 30% and 55%, respectively. Melatonin treatment significantly increased l-arginine levels and l-arginine-to-ADMA ratio, but decreased ADMA and SDMA levels in the CR + M group. In the kidney, levels of l-citrulline, l-arginine, ADMA, and SDMA did not differ between the four groups. However, renal l-arginine-to-ADMA ratio was higher in the CR + M group versus the M group. We next analyzed superoxide and NO production in the kidney by using EPR. We found no difference in renal superoxide level among the four groups (control: 745 ± 28, CR: 823 ± 107, M: 665 ± 35, CR + M: 757 ± 42 arbitrary units; P > 0.05). CR significantly reduced renal NO levels, but not in the presence of melatonin (control: 412 ± 43, CR: 284 ± 18, M: 308 ± 34, CR + M: 414 ± 55 arbitrary units; control versus CR, P < 0.05; CR versus CR + M, P < 0.05).
Table 3.
L-Citrulline, L-arginine, and dimethylarginine levels in the plasma and kidney.
| Control | CR | M | CR + M | |
|---|---|---|---|---|
| Plasma (μmol) | ||||
| L-Citrulline | 50 ± 4.1 | 61 ± 3.6 | 59.3 ± 5.1 | 55.8 ± 6.9 |
| L-Arginine | 121.1 ± 14 | 84.4 ± 2.4* | 113.6 ± 8.7# | 112.8 ± 13.6# |
| ADMA | 1.31 ± 0.1 | 2.21 ± 0.18* | 1.18 ± 0.06# | 1.08 ± 0.12# |
| SDMA | 0.66 ± 0.04 | 1.62 ± 0.27* | 0.97 ± 0.09# | 0.92 ± 0.08# |
| L-Arginine-to-ADMA ratio | 92 ± 8 | 40 ± 4* | 98 ± 10# | 105 ± 6# |
| Kidney (μmol/mg protein) | ||||
| L-Citrulline | 52.5 ± 8.6 | 53.1 ± 4.6 | 97.6 ± 8.4 | 68.8 ± 12.4 |
| L-Arginine | 425 ± 62.3 | 552.9 ± 58.9 | 522.8 ± 61.6 | 488.1 ± 56 |
| ADMA | 5.09 ± 0.88 | 6.33 ± 0.71 | 6.72 ± 1.03 | 4.84 ± 0.61 |
| SDMA | 4.3 ± 0.65 | 5.3 ± 0.51 | 5.57 ± 0.79 | 4.59 ± 0.73 |
| L-Arginine-to-ADMA ratio | 86 ± 4 | 89 ± 5 | 80 ± 4 | 103 ± 8$ |
*P < 0.05 versus control; # P < 0.05 versus CR; $ P < 0.05 versus M.
3.3. The Effects of Melatonin on the ADMA Pathway
Next, we examined the expression/activity of proteins involved in the ADMA pathway. We found that renal level of protein arginine N-methyltransferase 1 (PRMT-1), an ADMA-synthesizing enzyme, was significantly lower in the M and CR + M groups than that in control and CR groups (Figure 2(b)). However, protein levels of DDAH-1 and -2, ADMA-metabolizing enzymes, in the kidney were not different between the four groups (Figures 2(c) and 2(d)). We found that renal DDAH activity did not differ between control and CR groups (Figure 2(e)). However, melatonin increased renal DDAH activity in both the M and CR + M groups. Thus, we speculate that the increase of systemic ADMA observed with CR is due to excessive synthesis or decreased metabolism in extrarenal tissues. On the other hand, the reduced plasma ADMA levels in response to melatonin might be due to decreased ADMA synthesis and increased ADMA breakdown in the kidney.
Figure 2.
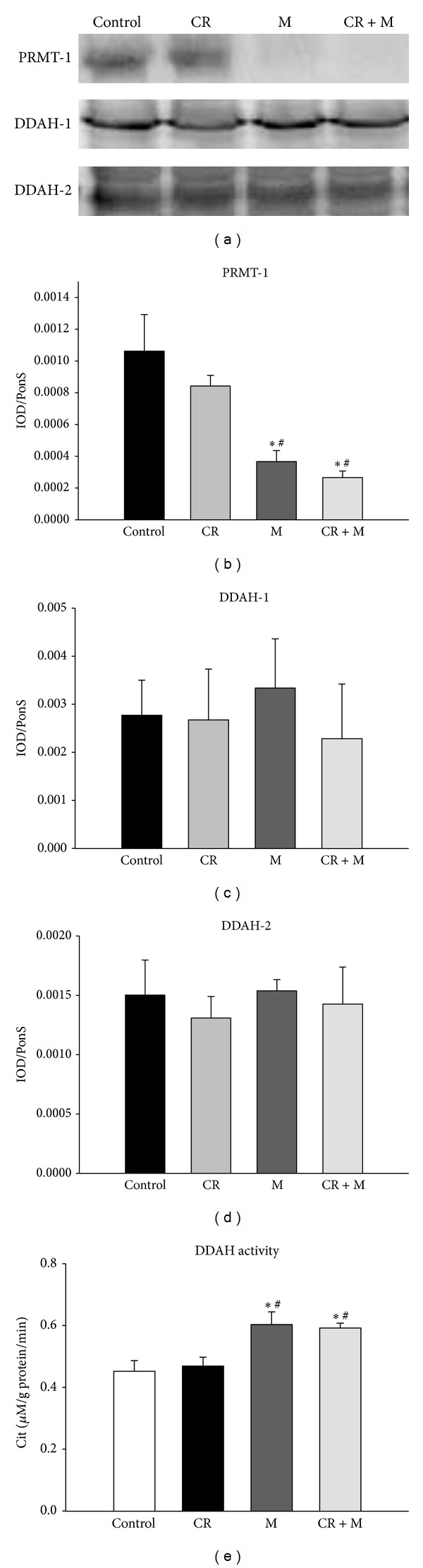
Representative western blots (a) showing protein arginine methyltransferase 1 (PRMT-1; ∼42 kDa), dimethylarginine dimethylaminohydrolase 1 (DDAH-1; ∼34 kDa), and DDAH-2 (∼30 kDa) bands in CR offspring at 12 weeks of age. Relative abundance of renal cortical (b) PRMT-1, (c) DDAH-1, and (d) DDAH-2. (e) Effect of melatonin and CR on renal DDAH activity in male offspring at 12 weeks of age. *P < 0.05 versus control; # P < 0.05 versus CR.
3.4. The Effects of Melatonin on Nephrogenesis
We investigated whether changes in nephrogenesis- or apoptosis-related gene expression were associated with CR-induced reduced nephron numbers, as we found previously [10]. Consistent with our previous report [10], renal expression of p53 and the proapoptotic factor Bax did not differ between the control and CR groups (Figure 3(a)). Similarly, growth factors BMP4 and FGF2 were unaltered by CR or melatonin in the kidney. However, melatonin significantly increased the expression of the transcriptional activator PAX2 in CR + M group compared to controls (Figure 3(a)).
Figure 3.
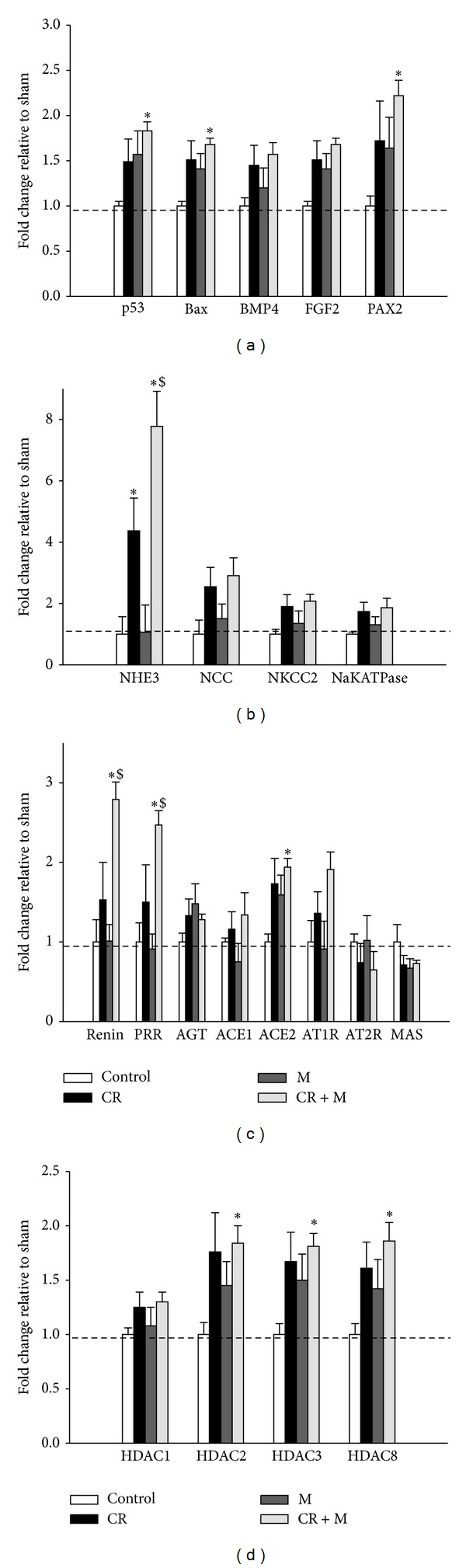
Effect of melatonin and CR on the expression of (a) apoptosis- and nephrogenesis-related genes, (b) sodium transporters, (c) renin-angiotensin system (RAS) components, and (d) class I histone deacetylase (HDAC) in the kidney at 12 weeks of age. *P < 0.05 versus control; # P < 0.05 versus CR; $ P < 0.05 versus M.
3.5. The Effects of Melatonin on Sodium Transporters, RAS, and HDACs
Next, we evaluated two critical pathways involved in hypertension, sodium transporters and RAS components. We found that CR upregulated sodium-hydrogen exchanger 3 (NHE3) expression in the kidney (Figure 3(b)). The increase in renal NHE3 expression was not prevented by melatonin therapy. CR had no effect on the expression of RAS genes in the kidney, including renin, PRR, AGT, ACE, ACE2, AT1R, AT2R, and MAS (Figure 3(c)). Melatonin treatment, on the other hand, upregulated renal expression of renin, PRR, and ACE2 in the CR + M group compared to the control. Because melatonin therapy prevented the elevation of BP in offspring exposed to maternal CR, our data suggested that the antihypertensive effect of melatonin was related to renin, PRR, and ACE2 expression in the CR model. We found that CR did not alter renal expression of class I HDACs in the CR versus control group (Figure 3(d)). However, melatonin therapy increased HDAC-2, -3, and -8 expression in the kidney.
We analyzed the renal protein levels of PRR, ACE2, AT1R, AT2R, and MAS. Melatonin therapy significantly increased renal PRR and ACE2 protein levels in the M and CR + M group compared with the control and CR groups (Figures 4(b) and 4(c)). We observed that renal AT1R, AT2R, and MAS protein levels did not differ among the four groups (Figures 4(d)–4(f)).
Figure 4.
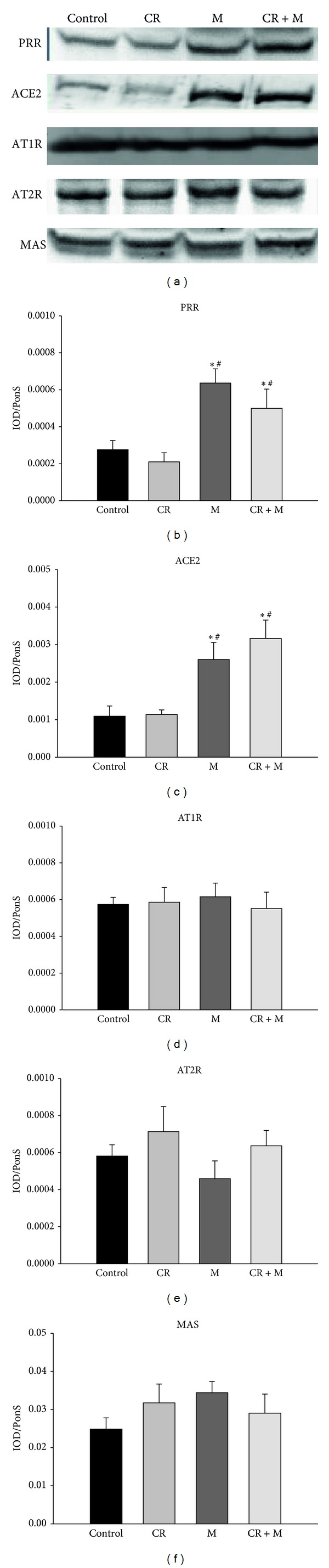
Representative western blots (a) showing prorenin receptor (PRR; 39 kDa), angiotensin-converting enzyme (ACE2; 50 kDa), angiotensin II type 1 (AT1R; 43 kDa) and type 2 (AT2R; 90 kDa), and MAS (37 kDa) proteins in male offspring kidneys at 12 weeks of age. Relative abundance of renal (b) PRR, (c) ACE2, (d) AT1R, (e) AT2R, and (f) MAS is quantified. *P < 0.05 versus control; # P < 0.05 versus CR.
We also determined whether melatonin regulated nephrogenesis-related genes, RAS components, sodium transporters, and HDACs during nephrogenesis. The mRNA levels in rat metanephroi grown in different concentrations of melatonin are shown in Figure 5. We found that low doses of melatonin had no effect on the expression of these genes, whereas high-dose melatonin treatment significantly increased expression of PAX2, renin, PRR, Mas, NHE3, and Na-K-Cl cotransporter 2 in metanephroi.
Figure 5.
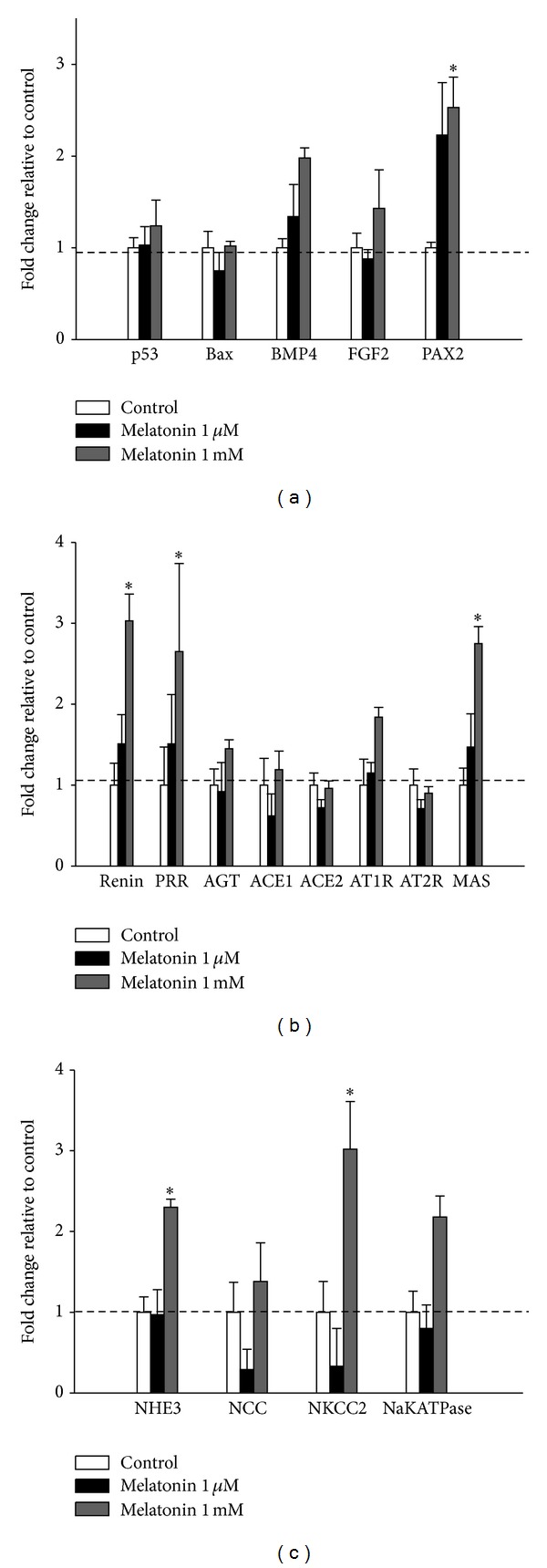
Gene expression of (a) apoptosis- and nephrogenesis-related genes, (b) RAS components, and (c) sodium transporters in the metanephroi of offspring from mothers treated with melatonin (1 μM or 1 mM). *P < 0.05 versus control (n = 5/group).
3.6. The Effects of Melatonin on Gene Expression during Nephrogenesis
We demonstrated that numerous individual genes were significantly regulated in the kidneys of offspring from melatonin-treated mothers during a critical period of renal development. As shown in Table 4, 439 and 15 genes were upregulated and downregulated, respectively. The most significantly regulated biological theme in the KEGG gene ontology analysis was tryptophan metabolism (Figure 6).
Table 4.
Genes that changed by RPKM > 0.3 in the kidney of melatonin treated offspring versus control at 1 week of age.
| Gene_ID | Gene symbol | Fold changes | Log2 | P value |
|---|---|---|---|---|
| Upregulated: 439 genes | ||||
| ENSRNOG00000038989 | D3ZSD6_RAT | 28.686 | 4.842 | 0.0083 |
| ENSRNOG00000006367 | Slc5a8 | 19.264 | 4.268 | 0.0001 |
| ENSRNOG00000003038 | Sft2d2 | 17.101 | 4.096 | 0.0020 |
| ENSRNOG00000007720 | F1LX97_RAT | 13.841 | 3.791 | 0.0027 |
| ENSRNOG00000019014 | Ndst1 | 12.657 | 3.662 | 0.0003 |
| ENSRNOG00000017434 | Mgat3 | 12.364 | 3.628 | 0.0005 |
| ENSRNOG00000001656 | Kcnj15 | 11.724 | 3.551 | 0.0028 |
| ENSRNOG00000021292 | — | 11.449 | 3.517 | 0.0015 |
| ENSRNOG00000017078 | Sepn1 | 11.107 | 3.473 | 0.0011 |
| ENSRNOG00000030121 | Enpep | 10.970 | 3.456 | 0.0029 |
| ENSRNOG00000005854 | Angpt1 | 10.920 | 3.449 | 0.0165 |
| ENSRNOG00000009944 | LOC314407 | 10.858 | 3.441 | 0.0017 |
| ENSRNOG00000013279 | Scd | 10.775 | 3.430 | 0.0012 |
| ENSRNOG00000001724 | LOC678704 | 10.690 | 3.418 | 0.0011 |
| ENSRNOG00000002463 | LOC682752 | 10.633 | 3.411 | 0.0033 |
| ENSRNOG00000011630 | Ak3l1 | 10.304 | 3.365 | 0.0441 |
| ENSRNOG00000005447 | RGD1311564 | 10.117 | 3.339 | 0.0026 |
| ENSRNOG00000009019 | Slc6a6 | 10.090 | 3.335 | 0.0016 |
| ENSRNOG00000002969 | Itpkb | 9.892 | 3.306 | 0.0020 |
| ENSRNOG00000037307 | Spata22 | 9.876 | 3.304 | 0.0036 |
| ENSRNOG00000039717 | Ipo11 | 9.584 | 3.261 | 0.0069 |
| ENSRNOG00000025372 | Glce | 9.536 | 3.253 | 0.0023 |
| ENSRNOG00000037884 | Oxgr1 | 9.510 | 3.249 | 0.0169 |
| ENSRNOG00000021203 | Atl3 | 9.487 | 3.246 | 0.0056 |
| ENSRNOG00000006787 | Dhcr24 | 9.328 | 3.222 | 0.0023 |
| ENSRNOG00000015038 | Adam10 | 9.279 | 3.214 | 0.0005 |
| ENSRNOG00000002519 | Magt1 | 9.253 | 3.210 | 0.0010 |
| ENSRNOG00000038933 | D3ZF12_RAT | 9.225 | 3.206 | 0.0023 |
| ENSRNOG00000024757 | RGD1310444 | 9.119 | 3.189 | 0.0066 |
| ENSRNOG00000030285 | Epha3 | 9.064 | 3.180 | 0.0032 |
| ENSRNOG00000018338 | Vwa1 | 9.017 | 3.173 | 0.0355 |
| ENSRNOG00000022802 | Tmem184b | 8.982 | 3.167 | 0.0070 |
| ENSRNOG00000013265 | Tgfbr2 | 8.947 | 3.161 | 0.0014 |
| ENSRNOG00000026941 | Tril | 8.934 | 3.159 | 0.0024 |
| ENSRNOG00000020532 | Kcnq1 | 8.904 | 3.154 | 0.0441 |
| ENSRNOG00000018503 | LOC293190 | 8.862 | 3.148 | 0.0172 |
| ENSRNOG00000002198 | LOC685352 | 8.711 | 3.123 | 0.0037 |
| ENSRNOG00000017172 | Fam125b | 8.706 | 3.122 | 0.0291 |
| ENSRNOG00000018554 | — | 8.663 | 3.115 | 0.0067 |
| ENSRNOG00000013963 | IL6RB_RAT | 8.571 | 3.099 | 0.0023 |
| ENSRNOG00000042565 | — | 8.547 | 3.095 | 0.0114 |
| ENSRNOG00000032834 | Hspa13 | 8.544 | 3.095 | 0.0011 |
| ENSRNOG00000002355 | Slc47a1 | 8.474 | 3.083 | 0.0025 |
| ENSRNOG00000011927 | SDC3_RAT | 8.460 | 3.081 | 0.0065 |
| ENSRNOG00000042540 | Mef2a | 8.454 | 3.080 | 0.0368 |
| ENSRNOG00000029216 | Dgcr2 | 8.331 | 3.059 | 0.0175 |
| ENSRNOG00000023725 | LOC689756 | 8.215 | 3.038 | 0.0191 |
| ENSRNOG00000028129 | Fktn | 8.207 | 3.037 | 0.0063 |
| ENSRNOG00000000547 | Tspyl4 | 8.202 | 3.036 | 0.0103 |
| ENSRNOG00000011859 | Eif5a2 | 8.192 | 3.034 | 0.0403 |
| ENSRNOG00000028387 | E9PTK5_RAT | 8.168 | 3.030 | 0.0197 |
| ENSRNOG00000015986 | Rassf8 | 8.134 | 3.024 | 0.0094 |
| ENSRNOG00000029409 | Gstm6l | 8.062 | 3.011 | 0.0292 |
| ENSRNOG00000008895 | Hnf4a | 7.993 | 2.999 | 0.0398 |
| ENSRNOG00000038149 | Defb9 | 7.948 | 2.991 | 0.0437 |
| ENSRNOG00000040287 | Cyp1b1 | 7.890 | 2.980 | 0.0439 |
| ENSRNOG00000010468 | Elovl6 | 7.871 | 2.977 | 0.0394 |
| ENSRNOG00000014524 | F1M9D3_RAT | 7.828 | 2.969 | 0.0077 |
| ENSRNOG00000014209 | Utp6 | 7.793 | 2.962 | 0.0050 |
| ENSRNOG00000013419 | Agphd1 | 7.789 | 2.961 | 0.0031 |
| ENSRNOG00000020653 | S1pr2 | 7.775 | 2.959 | 0.0313 |
| ENSRNOG00000018714 | Arl5b | 7.770 | 2.958 | 0.0078 |
| ENSRNOG00000002408 | Rbm47 | 7.719 | 2.948 | 0.0057 |
| ENSRNOG00000008971 | Hnf4g | 7.715 | 2.948 | 0.0085 |
| ENSRNOG00000011271 | Mcc | 7.688 | 2.943 | 0.0120 |
| ENSRNOG00000002276 | LOC100359714 | 7.641 | 2.934 | 0.0073 |
| ENSRNOG00000009446 | Rxra | 7.607 | 2.927 | 0.0066 |
| ENSRNOG00000019400 | Dag1 | 7.591 | 2.924 | 0.0010 |
| ENSRNOG00000013098 | F1M9J1_RAT | 7.581 | 2.922 | 0.0320 |
| ENSRNOG00000014511 | Alg10 | 7.551 | 2.917 | 0.0076 |
| ENSRNOG00000012490 | Amph | 7.533 | 2.913 | 0.0461 |
| ENSRNOG00000014934 | Fam63b | 7.481 | 2.903 | 0.0193 |
| ENSRNOG00000039630 | LOC290577 | 7.414 | 2.890 | 0.0045 |
| ENSRNOG00000032707 | Egf | 7.368 | 2.881 | 0.0017 |
| ENSRNOG00000015605 | Ptprk | 7.357 | 2.879 | 0.0298 |
| ENSRNOG00000000168 | Gatm | 7.311 | 2.870 | 0.0017 |
| ENSRNOG00000027097 | F1M683_RAT | 7.273 | 2.863 | 0.0087 |
| ENSRNOG00000018109 | Clic4 | 7.251 | 2.858 | 0.0048 |
| ENSRNOG00000008629 | Secisbp2l | 7.236 | 2.855 | 0.0042 |
| ENSRNOG00000019799 | Pcdhgc3 | 7.231 | 2.854 | 0.0246 |
| ENSRNOG00000024089 | Fndc3b | 7.221 | 2.852 | 0.0065 |
| ENSRNOG00000015852 | D4AD82_RAT | 7.192 | 2.846 | 0.0020 |
| ENSRNOG00000006967 | Xiap | 7.151 | 2.838 | 0.0136 |
| ENSRNOG00000031487 | F1LM52_RAT | 7.129 | 2.834 | 0.0477 |
| ENSRNOG00000014866 | Pign | 7.077 | 2.823 | 0.0190 |
| ENSRNOG00000033206 | Entpd5 | 7.060 | 2.820 | 0.0070 |
| ENSRNOG00000037753 | Slc10a2 | 7.002 | 2.808 | 0.0089 |
| ENSRNOG00000040195 | F1LZT0_RAT | 7.001 | 2.808 | 0.0018 |
| ENSRNOG00000042817 | D4A5M8_RAT | 6.925 | 2.792 | 0.0104 |
| ENSRNOG00000005070 | Spopl | 6.920 | 2.791 | 0.0139 |
| ENSRNOG00000006459 | D4AEA4_RAT | 6.871 | 2.781 | 0.0251 |
| ENSRNOG00000012784 | Gtf3c4 | 6.850 | 2.776 | 0.0096 |
| ENSRNOG00000016968 | Gramd4 | 6.838 | 2.774 | 0.0216 |
| ENSRNOG00000004448 | RGD1307051 | 6.819 | 2.770 | 0.0050 |
| ENSRNOG00000021809 | Gpx3 | 6.801 | 2.766 | 0.0008 |
| ENSRNOG00000014183 | Gnaq | 6.801 | 2.766 | 0.0084 |
| ENSRNOG00000012991 | LOC100363275 | 6.798 | 2.765 | 0.0046 |
| ENSRNOG00000013443 | Tm9sf3 | 6.791 | 2.764 | 0.0040 |
| ENSRNOG00000042673 | LOC100359544 | 6.789 | 2.763 | 0.0012 |
| ENSRNOG00000003873 | Cpd | 6.767 | 2.758 | 0.0028 |
| ENSRNOG00000007990 | Adipor2 | 6.762 | 2.758 | 0.0026 |
| ENSRNOG00000007804 | C1galt1 | 6.762 | 2.757 | 0.0109 |
| ENSRNOG00000043256 | D3ZNR8_RAT | 6.720 | 2.749 | 0.0145 |
| ENSRNOG00000015124 | Gpam | 6.720 | 2.748 | 0.0109 |
| ENSRNOG00000004888 | Spred2 | 6.690 | 2.742 | 0.0454 |
| ENSRNOG00000003960 | Tmem27 | 6.682 | 2.740 | 0.0026 |
| ENSRNOG00000015750 | Wnt7b | 6.654 | 2.734 | 0.0218 |
| ENSRNOG00000030763 | Dpp4 | 6.601 | 2.723 | 0.0011 |
| ENSRNOG00000039504 | Q5M885_RAT | 6.562 | 2.714 | 0.0116 |
| ENSRNOG00000032768 | D3Z9G8_RAT | 6.497 | 2.700 | 0.0214 |
| ENSRNOG00000039771 | LOC100361629 | 6.494 | 2.699 | 0.0140 |
| ENSRNOG00000009274 | Fut11 | 6.475 | 2.695 | 0.0354 |
| ENSRNOG00000027938 | RGD1562037 | 6.420 | 2.683 | 0.0117 |
| ENSRNOG00000001335 | Zkscan1 | 6.419 | 2.682 | 0.0077 |
| ENSRNOG00000004978 | Prkacb | 6.379 | 2.673 | 0.0216 |
| ENSRNOG00000005446 | Gna11 | 6.363 | 2.670 | 0.0172 |
| ENSRNOG00000003884 | Acmsd | 6.362 | 2.669 | 0.0262 |
| ENSRNOG00000028190 | D4ACF8_RAT | 6.354 | 2.668 | 0.0432 |
| ENSRNOG00000006338 | Lrp6 | 6.351 | 2.667 | 0.0041 |
| ENSRNOG00000009523 | Rab11fip2 | 6.345 | 2.666 | 0.0470 |
| ENSRNOG00000003759 | Galc | 6.345 | 2.666 | 0.0140 |
| ENSRNOG00000010620 | NDC1_RAT | 6.319 | 2.660 | 0.0275 |
| ENSRNOG00000001821 | Adipoq | 6.306 | 2.657 | 0.0244 |
| ENSRNOG00000038572 | RGD1562646 | 6.293 | 2.654 | 0.0106 |
| ENSRNOG00000026120 | Fam8a1 | 6.282 | 2.651 | 0.0129 |
| ENSRNOG00000025476 | RGD1359349 | 6.243 | 2.642 | 0.0126 |
| ENSRNOG00000019508 | Wars2 | 6.216 | 2.636 | 0.0317 |
| ENSRNOG00000008271 | Fam91a1 | 6.216 | 2.636 | 0.0031 |
| ENSRNOG00000017120 | Abhd2 | 6.208 | 2.634 | 0.0278 |
| ENSRNOG00000010843 | Nhlrc3 | 6.203 | 2.633 | 0.0255 |
| ENSRNOG00000030704 | F1LV74_RAT | 6.139 | 2.618 | 0.0369 |
| ENSRNOG00000002509 | Gnl3l | 6.129 | 2.616 | 0.0124 |
| ENSRNOG00000010841 | Col8a2 | 6.089 | 2.606 | 0.0457 |
| ENSRNOG00000002728 | Btc | 6.088 | 2.606 | 0.0348 |
| ENSRNOG00000027320 | Eif2c1 | 6.082 | 2.605 | 0.0243 |
| ENSRNOG00000009453 | Mobkl2b | 6.072 | 2.602 | 0.0271 |
| ENSRNOG00000007797 | Rbpsuh | 6.069 | 2.602 | 0.0133 |
| ENSRNOG00000017286 | HYES_RAT | 6.064 | 2.600 | 0.0032 |
| ENSRNOG00000002461 | Nid1 | 6.057 | 2.599 | 0.0014 |
| ENSRNOG00000006649 | Thrb | 6.048 | 2.596 | 0.0180 |
| ENSRNOG00000025042 | Pde3a | 6.048 | 2.596 | 0.0189 |
| ENSRNOG00000015916 | Ttc38 | 6.048 | 2.596 | 0.0384 |
| ENSRNOG00000013581 | Extl3 | 6.038 | 2.594 | 0.0093 |
| ENSRNOG00000002332 | MSPD1_RAT | 6.034 | 2.593 | 0.0213 |
| ENSRNOG00000032757 | D3Z903_RAT | 6.032 | 2.593 | 0.0431 |
| ENSRNOG00000029651 | Rdh2 | 6.025 | 2.591 | 0.0258 |
| ENSRNOG00000018588 | Sox4 | 6.019 | 2.590 | 0.0342 |
| ENSRNOG00000012428 | Maf | 6.005 | 2.586 | 0.0483 |
| ENSRNOG00000009506 | Mre11a | 6.005 | 2.586 | 0.0332 |
| ENSRNOG00000028330 | — | 5.987 | 2.582 | 0.0375 |
| ENSRNOG00000034025 | D4A4T5_RAT | 5.979 | 2.580 | 0.0227 |
| ENSRNOG00000007079 | Met | 5.979 | 2.580 | 0.0117 |
| ENSRNOG00000008088 | Btbd3 | 5.979 | 2.580 | 0.0181 |
| ENSRNOG00000017546 | Mylk3 | 5.945 | 2.572 | 0.0224 |
| ENSRNOG00000042333 | Dnal1 | 5.895 | 2.559 | 0.0187 |
| ENSRNOG00000001092 | Kl | 5.873 | 2.554 | 0.0106 |
| ENSRNOG00000016498 | — | 5.836 | 2.545 | 0.0049 |
| ENSRNOG00000037765 | Lims1 | 5.833 | 2.544 | 0.0367 |
| ENSRNOG00000010267 | Klhdc10 | 5.827 | 2.543 | 0.0346 |
| ENSRNOG00000043277 | D3ZIC7_RAT | 5.809 | 2.538 | 0.0206 |
| ENSRNOG00000024799 | D3ZNV9_RAT | 5.803 | 2.537 | 0.0032 |
| ENSRNOG00000004919 | Gns | 5.795 | 2.535 | 0.0282 |
| ENSRNOG00000015080 | Wdfy1 | 5.766 | 2.528 | 0.0292 |
| ENSRNOG00000009565 | Pdk4 | 5.764 | 2.527 | 0.0206 |
| ENSRNOG00000013082 | LCAP_RAT | 5.754 | 2.525 | 0.0322 |
| ENSRNOG00000026501 | Slc6a19 | 5.742 | 2.522 | 0.0406 |
| ENSRNOG00000009597 | Cyp4a1 | 5.740 | 2.521 | 0.0123 |
| ENSRNOG00000011560 | Mtmr9 | 5.738 | 2.521 | 0.0368 |
| ENSRNOG00000022710 | Prrg4 | 5.736 | 2.520 | 0.0269 |
| ENSRNOG00000013469 | LOC100362805 | 5.715 | 2.515 | 0.0059 |
| ENSRNOG00000024640 | RGD1304731 | 5.698 | 2.510 | 0.0080 |
| ENSRNOG00000018952 | Sema3g | 5.692 | 2.509 | 0.0143 |
| ENSRNOG00000020011 | Q66HF5_RAT | 5.677 | 2.505 | 0.0381 |
| ENSRNOG00000012826 | Creb3l2 | 5.665 | 2.502 | 0.0189 |
| ENSRNOG00000032492 | Usp22 | 5.657 | 2.500 | 0.0107 |
| ENSRNOG00000021840 | LOC500046 | 5.644 | 2.497 | 0.0118 |
| ENSRNOG00000034026 | Lclat1 | 5.642 | 2.496 | 0.0223 |
| ENSRNOG00000009153 | Cidec | 5.642 | 2.496 | 0.0432 |
| ENSRNOG00000028899 | Zbtb33 | 5.633 | 2.494 | 0.0168 |
| ENSRNOG00000001766 | Tfrc | 5.613 | 2.489 | 0.0102 |
| ENSRNOG00000017901 | Acy3 | 5.613 | 2.489 | 0.0044 |
| ENSRNOG00000012095 | Pkia | 5.596 | 2.484 | 0.0339 |
| ENSRNOG00000001796 | Dgkg | 5.573 | 2.479 | 0.0471 |
| ENSRNOG00000004958 | RGD1304605 | 5.563 | 2.476 | 0.0100 |
| ENSRNOG00000025587 | Plagl1 | 5.550 | 2.472 | 0.0289 |
| ENSRNOG00000027540 | Fam102b | 5.536 | 2.469 | 0.0410 |
| ENSRNOG00000001518 | Itga6 | 5.519 | 2.465 | 0.0452 |
| ENSRNOG00000032723 | Eftud1 | 5.515 | 2.463 | 0.0336 |
| ENSRNOG00000002053 | F1M3H3_RAT | 5.491 | 2.457 | 0.0081 |
| ENSRNOG00000003472 | Atp11c-ps1 | 5.473 | 2.452 | 0.0317 |
| ENSRNOG00000003984 | Apln | 5.448 | 2.446 | 0.0337 |
| ENSRNOG00000012453 | RGD1564560 | 5.438 | 2.443 | 0.0046 |
| ENSRNOG00000017846 | Slc44a1 | 5.422 | 2.439 | 0.0293 |
| ENSRNOG00000016921 | Klhl11 | 5.418 | 2.438 | 0.0275 |
| ENSRNOG00000026415 | D4A301_RAT | 5.403 | 2.434 | 0.0280 |
| ENSRNOG00000013798 | Fnbp1l | 5.391 | 2.431 | 0.0098 |
| ENSRNOG00000003620 | Fmo3 | 5.384 | 2.429 | 0.0050 |
| ENSRNOG00000018220 | Pde4dip | 5.377 | 2.427 | 0.0462 |
| ENSRNOG00000000145 | Pik3r3 | 5.352 | 2.420 | 0.0210 |
| ENSRNOG00000008834 | LOC306096 | 5.351 | 2.420 | 0.0356 |
| ENSRNOG00000025882 | Nipal1 | 5.345 | 2.418 | 0.0306 |
| ENSRNOG00000010996 | Mobkl1a | 5.341 | 2.417 | 0.0147 |
| ENSRNOG00000001582 | Bach1 | 5.339 | 2.417 | 0.0199 |
| ENSRNOG00000022309 | D3ZRU8_RAT | 5.313 | 2.410 | 0.0048 |
| ENSRNOG00000015741 | Slc2a13 | 5.298 | 2.406 | 0.0371 |
| ENSRNOG00000014303 | F1M753_RAT | 5.294 | 2.404 | 0.0391 |
| ENSRNOG00000036798 | Dusp3 | 5.284 | 2.402 | 0.0199 |
| ENSRNOG00000012142 | Glyat | 5.283 | 2.401 | 0.0081 |
| ENSRNOG00000024426 | D3ZXW1_RAT | 5.259 | 2.395 | 0.0477 |
| ENSRNOG00000006628 | Dusp16 | 5.256 | 2.394 | 0.0271 |
| ENSRNOG00000026143 | Ckap2l | 5.230 | 2.387 | 0.0271 |
| ENSRNOG00000018867 | Klhdc7a | 5.223 | 2.385 | 0.0489 |
| ENSRNOG00000025296 | Lrrc8a | 5.203 | 2.379 | 0.0176 |
| ENSRNOG00000014508 | Mgll | 5.203 | 2.379 | 0.0137 |
| ENSRNOG00000000589 | RGD1310495 | 5.199 | 2.378 | 0.0372 |
| ENSRNOG00000014234 | Hif1an | 5.192 | 2.376 | 0.0394 |
| ENSRNOG00000008450 | LOC100359539 | 5.178 | 2.372 | 0.0409 |
| ENSRNOG00000010744 | Nrp1 | 5.177 | 2.372 | 0.0072 |
| ENSRNOG00000039837 | RGD1563945 | 5.161 | 2.368 | 0.0466 |
| ENSRNOG00000013177 | Map3k1 | 5.154 | 2.366 | 0.0114 |
| ENSRNOG00000021719 | F1LX81_RAT | 5.153 | 2.365 | 0.0133 |
| ENSRNOG00000024629 | Hadha | 5.116 | 2.355 | 0.0126 |
| ENSRNOG00000014907 | Aldh8a1 | 5.105 | 2.352 | 0.0055 |
| ENSRNOG00000036673 | Sectm1b | 5.098 | 2.350 | 0.0121 |
| ENSRNOG00000024794 | Senp5 | 5.096 | 2.349 | 0.0264 |
| ENSRNOG00000005131 | Lin7c | 5.086 | 2.347 | 0.0289 |
| ENSRNOG00000002225 | Scarb2 | 5.081 | 2.345 | 0.0116 |
| ENSRNOG00000020284 | Prkar2a | 5.077 | 2.344 | 0.0215 |
| ENSRNOG00000014648 | Efnb2 | 5.072 | 2.343 | 0.0303 |
| ENSRNOG00000002488 | Galnt10 | 5.063 | 2.340 | 0.0437 |
| ENSRNOG00000017406 | Atrnl1 | 5.056 | 2.338 | 0.0269 |
| ENSRNOG00000010813 | Tspan14 | 5.048 | 2.336 | 0.0304 |
| ENSRNOG00000000645 | Reep3 | 5.047 | 2.336 | 0.0262 |
| ENSRNOG00000018873 | Fam168a | 5.036 | 2.332 | 0.0160 |
| ENSRNOG00000020253 | RAB1B_RAT | 5.030 | 2.331 | 0.0128 |
| ENSRNOG00000001235 | Gna12 | 5.012 | 2.325 | 0.0149 |
| ENSRNOG00000040215 | F1LZL1_RAT | 5.011 | 2.325 | 0.0302 |
| ENSRNOG00000011619 | Myo9a | 4.988 | 2.319 | 0.0163 |
| ENSRNOG00000039976 | D3ZHG3_RAT | 4.983 | 2.317 | 0.0137 |
| ENSRNOG00000016011 | Plekhg1 | 4.971 | 2.314 | 0.0315 |
| ENSRNOG00000037909 | Ppm1f | 4.964 | 2.312 | 0.0269 |
| ENSRNOG00000016419 | Pdlim5 | 4.962 | 2.311 | 0.0248 |
| ENSRNOG00000023280 | Als2 | 4.952 | 2.308 | 0.0166 |
| ENSRNOG00000005417 | Zhx2 | 4.948 | 2.307 | 0.0430 |
| ENSRNOG00000017671 | Rasa3 | 4.944 | 2.306 | 0.0403 |
| ENSRNOG00000016848 | Fzd4 | 4.942 | 2.305 | 0.0255 |
| ENSRNOG00000003508 | LOC100364400 | 4.942 | 2.305 | 0.0244 |
| ENSRNOG00000012394 | Bcl2l13 | 4.931 | 2.302 | 0.0466 |
| ENSRNOG00000018400 | D4AEL2_RAT | 4.931 | 2.302 | 0.0303 |
| ENSRNOG00000013707 | Spata13 | 4.930 | 2.302 | 0.0445 |
| ENSRNOG00000002039 | LOC100360066 | 4.930 | 2.301 | 0.0436 |
| ENSRNOG00000004563 | Sec24a | 4.917 | 2.298 | 0.0191 |
| ENSRNOG00000020386 | D3ZKH4_RAT | 4.906 | 2.295 | 0.0098 |
| ENSRNOG00000007419 | Pank3 | 4.900 | 2.293 | 0.0128 |
| ENSRNOG00000024533 | Aer61 | 4.889 | 2.290 | 0.0382 |
| ENSRNOG00000027151 | Lrrc58 | 4.886 | 2.289 | 0.0393 |
| ENSRNOG00000030124 | Ptpn11 | 4.869 | 2.284 | 0.0160 |
| ENSRNOG00000006131 | Mettl2 | 4.846 | 2.277 | 0.0271 |
| ENSRNOG00000000407 | Dcbld1 | 4.834 | 2.273 | 0.0412 |
| ENSRNOG00000008061 | Nuak1 | 4.826 | 2.271 | 0.0360 |
| ENSRNOG00000037514 | Qser1 | 4.821 | 2.269 | 0.0136 |
| ENSRNOG00000004959 | Actr2 | 4.807 | 2.265 | 0.0327 |
| ENSRNOG00000028582 | F1M163_RAT | 4.795 | 2.261 | 0.0045 |
| ENSRNOG00000043037 | LOC100366023 | 4.788 | 2.259 | 0.0349 |
| ENSRNOG00000012135 | F1M2H7_RAT | 4.763 | 2.252 | 0.0406 |
| ENSRNOG00000031069 | D4A9A7_RAT | 4.749 | 2.247 | 0.0462 |
| ENSRNOG00000023109 | F1LVL2_RAT | 4.736 | 2.244 | 0.0482 |
| ENSRNOG00000004442 | RGD1311756 | 4.729 | 2.241 | 0.0456 |
| ENSRNOG00000021318 | Epas1 | 4.723 | 2.240 | 0.0138 |
| ENSRNOG00000018099 | Itch | 4.702 | 2.233 | 0.0383 |
| ENSRNOG00000038892 | LOC686123 | 4.691 | 2.230 | 0.0268 |
| ENSRNOG00000000296 | Aqp6 | 4.685 | 2.228 | 0.0310 |
| ENSRNOG00000014901 | Uggt1 | 4.684 | 2.228 | 0.0168 |
| ENSRNOG00000019659 | Aspa | 4.680 | 2.227 | 0.0055 |
| ENSRNOG00000010450 | D4ADY9_RAT | 4.662 | 2.221 | 0.0220 |
| ENSRNOG00000011066 | 6-Mar | 4.658 | 2.220 | 0.0264 |
| ENSRNOG00000013121 | Mier3 | 4.647 | 2.216 | 0.0408 |
| ENSRNOG00000030894 | Slco1a6 | 4.640 | 2.214 | 0.0068 |
| ENSRNOG00000004964 | Erbb3 | 4.609 | 2.205 | 0.0351 |
| ENSRNOG00000014135 | Rab11fip4 | 4.607 | 2.204 | 0.0453 |
| ENSRNOG00000005052 | Slc39a9 | 4.594 | 2.200 | 0.0454 |
| ENSRNOG00000005276 | Csnk2a1 | 4.589 | 2.198 | 0.0259 |
| ENSRNOG00000015007 | RGD1565591 | 4.583 | 2.196 | 0.0462 |
| ENSRNOG00000002099 | Wdfy3 | 4.579 | 2.195 | 0.0217 |
| ENSRNOG00000001747 | Pak2 | 4.572 | 2.193 | 0.0178 |
| ENSRNOG00000018226 | Zcchc14 | 4.565 | 2.190 | 0.0441 |
| ENSRNOG00000010702 | Ube3c | 4.564 | 2.190 | 0.0154 |
| ENSRNOG00000010610 | Hpgd | 4.556 | 2.188 | 0.0125 |
| ENSRNOG00000001756 | D3ZDR3_RAT | 4.551 | 2.186 | 0.0486 |
| ENSRNOG00000006335 | Klhl9 | 4.550 | 2.186 | 0.0083 |
| ENSRNOG00000016715 | Kif11 | 4.547 | 2.185 | 0.0159 |
| ENSRNOG00000021916 | Slc16a12 | 4.541 | 2.183 | 0.0224 |
| ENSRNOG00000011250 | Inmt | 4.506 | 2.172 | 0.0125 |
| ENSRNOG00000013140 | Pdzd2 | 4.502 | 2.171 | 0.0305 |
| ENSRNOG00000012440 | Msra | 4.501 | 2.170 | 0.0308 |
| ENSRNOG00000019932 | Ip6k1 | 4.500 | 2.170 | 0.0307 |
| ENSRNOG00000037227 | Yes1 | 4.499 | 2.170 | 0.0412 |
| ENSRNOG00000012054 | Zmpste24 | 4.498 | 2.169 | 0.0179 |
| ENSRNOG00000007370 | Rnf144a | 4.493 | 2.168 | 0.0443 |
| ENSRNOG00000022968 | F1M4Y9_RAT | 4.491 | 2.167 | 0.0400 |
| ENSRNOG00000011340 | D3ZMJ4_RAT | 4.488 | 2.166 | 0.0143 |
| ENSRNOG00000021705 | D3ZXN6_RAT | 4.486 | 2.165 | 0.0229 |
| ENSRNOG00000003865 | Tmigd1 | 4.483 | 2.164 | 0.0072 |
| ENSRNOG00000012105 | F1MAE3_RAT | 4.478 | 2.163 | 0.0346 |
| ENSRNOG00000011312 | F1LQ39_RAT | 4.475 | 2.162 | 0.0366 |
| ENSRNOG00000000127 | F1LT58_RAT | 4.463 | 2.158 | 0.0484 |
| ENSRNOG00000022929 | MTMRC_RAT | 4.438 | 2.150 | 0.0307 |
| ENSRNOG00000033372 | Klhl24 | 4.431 | 2.148 | 0.0197 |
| ENSRNOG00000008332 | Smo | 4.420 | 2.144 | 0.0209 |
| ENSRNOG00000028616 | Pck1 | 4.418 | 2.143 | 0.0219 |
| ENSRNOG00000013281 | Mib1 | 4.415 | 2.142 | 0.0306 |
| ENSRNOG00000011448 | Eri1 | 4.410 | 2.141 | 0.0414 |
| ENSRNOG00000028422 | Rmnd5a | 4.409 | 2.141 | 0.0212 |
| ENSRNOG00000014859 | Rnf152 | 4.404 | 2.139 | 0.0298 |
| ENSRNOG00000001893 | LOC100362453 | 4.397 | 2.137 | 0.0349 |
| ENSRNOG00000018123 | Ccny | 4.396 | 2.136 | 0.0173 |
| ENSRNOG00000016337 | Slc22a1 | 4.394 | 2.135 | 0.0356 |
| ENSRNOG00000003709 | Kmo | 4.389 | 2.134 | 0.0166 |
| ENSRNOG00000019939 | CCND2_RAT | 4.386 | 2.133 | 0.0383 |
| ENSRNOG00000029947 | — | 4.377 | 2.130 | 0.0399 |
| ENSRNOG00000008346 | Itgb6 | 4.372 | 2.128 | 0.0245 |
| ENSRNOG00000008678 | Antxr1 | 4.357 | 2.123 | 0.0237 |
| ENSRNOG00000029924 | Klk1l | 4.344 | 2.119 | 0.0267 |
| ENSRNOG00000043406 | LOC100360800 | 4.341 | 2.118 | 0.0323 |
| ENSRNOG00000012343 | Pdp2 | 4.324 | 2.112 | 0.0419 |
| ENSRNOG00000009899 | D3ZWL1_RAT | 4.306 | 2.106 | 0.0427 |
| ENSRNOG00000003434 | Trove2 | 4.301 | 2.105 | 0.0368 |
| ENSRNOG00000015519 | Ces1d | 4.294 | 2.102 | 0.0253 |
| ENSRNOG00000017439 | Cgnl1 | 4.294 | 2.102 | 0.0236 |
| ENSRNOG00000014700 | Ttc36 | 4.287 | 2.100 | 0.0266 |
| ENSRNOG00000007944 | Edem1 | 4.281 | 2.098 | 0.0367 |
| ENSRNOG00000031263 | Haao | 4.246 | 2.086 | 0.0200 |
| ENSRNOG00000001647 | Ets2 | 4.245 | 2.086 | 0.0357 |
| ENSRNOG00000008652 | RGD1564964 | 4.226 | 2.079 | 0.0153 |
| ENSRNOG00000023202 | Usp15 | 4.217 | 2.076 | 0.0230 |
| ENSRNOG00000016289 | Bmpr1b | 4.212 | 2.075 | 0.0370 |
| ENSRNOG00000015024 | E9PT54_RAT | 4.208 | 2.073 | 0.0252 |
| ENSRNOG00000000555 | Eif4ebp2 | 4.199 | 2.070 | 0.0381 |
| ENSRNOG00000008620 | Smad3 | 4.198 | 2.070 | 0.0440 |
| ENSRNOG00000008619 | Agtrap | 4.198 | 2.070 | 0.0217 |
| ENSRNOG00000009711 | Hepacam2 | 4.196 | 2.069 | 0.0409 |
| ENSRNOG00000015734 | Ube3a | 4.193 | 2.068 | 0.0225 |
| ENSRNOG00000015634 | SMAD4_RAT | 4.189 | 2.067 | 0.0277 |
| ENSRNOG00000042519 | RGD1312026 | 4.182 | 2.064 | 0.0380 |
| ENSRNOG00000007564 | Evc | 4.160 | 2.057 | 0.0289 |
| ENSRNOG00000008372 | Vamp7 | 4.160 | 2.057 | 0.0433 |
| ENSRNOG00000024671 | D4AA13_RAT | 4.157 | 2.056 | 0.0120 |
| ENSRNOG00000004622 | Calcrl | 4.142 | 2.050 | 0.0131 |
| ENSRNOG00000009660 | Enpp6 | 4.140 | 2.050 | 0.0247 |
| ENSRNOG00000014750 | D3ZXU7_RAT | 4.138 | 2.049 | 0.0176 |
| ENSRNOG00000008694 | Miox | 4.134 | 2.048 | 0.0226 |
| ENSRNOG00000004831 | Arid2 | 4.134 | 2.047 | 0.0317 |
| ENSRNOG00000043167 | Itga9 | 4.124 | 2.044 | 0.0349 |
| ENSRNOG00000001770 | Ehhadh | 4.114 | 2.040 | 0.0104 |
| ENSRNOG00000042160 | Tmem167b | 4.112 | 2.040 | 0.0466 |
| ENSRNOG00000018668 | Glg1 | 4.095 | 2.034 | 0.0172 |
| ENSRNOG00000007985 | D4ABH6_RAT | 4.084 | 2.030 | 0.0231 |
| ENSRNOG00000014623 | F1M3F2_RAT | 4.071 | 2.026 | 0.0332 |
| ENSRNOG00000002227 | Kit | 4.056 | 2.020 | 0.0429 |
| ENSRNOG00000016219 | Vnn1 | 4.052 | 2.019 | 0.0115 |
| ENSRNOG00000008322 | E9PTI4_RAT | 4.035 | 2.013 | 0.0418 |
| ENSRNOG00000011358 | Hipk3 | 4.034 | 2.012 | 0.0372 |
| ENSRNOG00000028335 | Fat4 | 4.017 | 2.006 | 0.0190 |
| ENSRNOG00000025554 | Zfp445 | 4.009 | 2.003 | 0.0365 |
| ENSRNOG00000003388 | Cenpf | 3.990 | 1.996 | 0.0146 |
| ENSRNOG00000000614 | Bicc1 | 3.987 | 1.995 | 0.0162 |
| ENSRNOG00000039091 | D3ZRC4_RAT | 3.975 | 1.991 | 0.0145 |
| ENSRNOG00000030154 | Cyp4a2 | 3.962 | 1.986 | 0.0422 |
| ENSRNOG00000033172 | — | 3.952 | 1.983 | 0.0176 |
| ENSRNOG00000017466 | Kif5b | 3.949 | 1.981 | 0.0128 |
| ENSRNOG00000042879 | D4A3X0_RAT | 3.943 | 1.979 | 0.0454 |
| ENSRNOG00000002146 | Pkd2 | 3.942 | 1.979 | 0.0358 |
| ENSRNOG00000012940 | Vps41 | 3.937 | 1.977 | 0.0280 |
| ENSRNOG00000017291 | Sord | 3.928 | 1.974 | 0.0133 |
| ENSRNOG00000001606 | Adamts5 | 3.923 | 1.972 | 0.0420 |
| ENSRNOG00000016534 | D3ZKX0_RAT | 3.904 | 1.965 | 0.0295 |
| ENSRNOG00000007202 | Sema3d | 3.898 | 1.963 | 0.0254 |
| ENSRNOG00000012436 | Adh6 | 3.897 | 1.962 | 0.0137 |
| ENSRNOG00000016334 | Rod1 | 3.867 | 1.951 | 0.0167 |
| ENSRNOG00000018011 | RGD1564456 | 3.867 | 1.951 | 0.0417 |
| ENSRNOG00000039494 | D4A608_RAT | 3.853 | 1.946 | 0.0328 |
| ENSRNOG00000014976 | Acsm2 | 3.850 | 1.945 | 0.0330 |
| ENSRNOG00000006636 | Otud6b | 3.817 | 1.932 | 0.0439 |
| ENSRNOG00000015849 | Sepp1 | 3.812 | 1.930 | 0.0292 |
| ENSRNOG00000004689 | Ptdss1 | 3.811 | 1.930 | 0.0342 |
| ENSRNOG00000013808 | Ces2g | 3.803 | 1.927 | 0.0345 |
| ENSRNOG00000014673 | Eri2 | 3.791 | 1.922 | 0.0429 |
| ENSRNOG00000009819 | Vezf1 | 3.784 | 1.920 | 0.0450 |
| ENSRNOG00000016758 | Loxl2 | 3.783 | 1.919 | 0.0310 |
| ENSRNOG00000010061 | Gmfb | 3.763 | 1.912 | 0.0466 |
| ENSRNOG00000023021 | Msl2 | 3.746 | 1.906 | 0.0486 |
| ENSRNOG00000039571 | Glod5 | 3.742 | 1.904 | 0.0370 |
| ENSRNOG00000017600 | Ptpn9 | 3.739 | 1.903 | 0.0329 |
| ENSRNOG00000000590 | Naglt1 | 3.718 | 1.895 | 0.0365 |
| ENSRNOG00000011511 | Stk24 | 3.716 | 1.894 | 0.0398 |
| ENSRNOG00000018279 | Sfxn1 | 3.712 | 1.892 | 0.0213 |
| ENSRNOG00000003953 | RB3GP_RAT | 3.709 | 1.891 | 0.0486 |
| ENSRNOG00000024632 | Atf6 | 3.699 | 1.887 | 0.0421 |
| ENSRNOG00000016779 | Fam120a | 3.679 | 1.879 | 0.0257 |
| ENSRNOG00000010379 | Cugbp1 | 3.670 | 1.876 | 0.0295 |
| ENSRNOG00000010780 | Dlc1 | 3.664 | 1.874 | 0.0495 |
| ENSRNOG00000003948 | Llgl1 | 3.659 | 1.871 | 0.0486 |
| ENSRNOG00000016183 | Ipp | 3.647 | 1.867 | 0.0466 |
| ENSRNOG00000017964 | Slc22a25 | 3.567 | 1.835 | 0.0201 |
| ENSRNOG00000039745 | Pm20d1 | 3.557 | 1.831 | 0.0300 |
| ENSRNOG00000010107 | PALLD_RAT | 3.528 | 1.819 | 0.0467 |
| ENSRNOG00000019444 | D4ADJ6_RAT | 3.515 | 1.814 | 0.0265 |
| ENSRNOG00000011260 | Cmbl | 3.513 | 1.813 | 0.0221 |
| ENSRNOG00000013322 | DPOLA_RAT | 3.505 | 1.810 | 0.0498 |
| ENSRNOG00000039278 | Mcart1 | 3.477 | 1.798 | 0.0345 |
| ENSRNOG00000021108 | Slc22a12 | 3.449 | 1.786 | 0.0263 |
| ENSRNOG00000010887 | RGD1309534 | 3.435 | 1.781 | 0.0382 |
| ENSRNOG00000008331 | RGD1309995 | 3.394 | 1.763 | 0.0425 |
| ENSRNOG00000007949 | Rgn | 3.355 | 1.746 | 0.0276 |
| ENSRNOG00000011987 | Cd2ap | 3.343 | 1.741 | 0.0306 |
| ENSRNOG00000042175 | B6VQA7_RAT | 3.331 | 1.736 | 0.0384 |
| ENSRNOG00000012190 | Cldn2 | 3.324 | 1.733 | 0.0347 |
| ENSRNOG00000023972 | F1M6Q3_RAT | 3.323 | 1.733 | 0.0322 |
| ENSRNOG00000011763 | Serp1 | 3.319 | 1.731 | 0.0316 |
| ENSRNOG00000004496 | Rock2 | 3.318 | 1.730 | 0.0337 |
| ENSRNOG00000004677 | Zeb2 | 3.306 | 1.725 | 0.0306 |
| ENSRNOG00000013409 | Gclm | 3.301 | 1.723 | 0.0338 |
| ENSRNOG00000004302 | Pah | 3.270 | 1.709 | 0.0368 |
| ENSRNOG00000010947 | MMP14_RAT | 3.253 | 1.702 | 0.0337 |
| ENSRNOG00000011058 | Utrn | 3.247 | 1.699 | 0.0378 |
| ENSRNOG00000018215 | Slc22a6 | 3.246 | 1.698 | 0.0377 |
| ENSRNOG00000016456 | Il33 | 3.234 | 1.693 | 0.0319 |
| ENSRNOG00000002541 | Pds5a | 3.164 | 1.662 | 0.0449 |
| ENSRNOG00000002680 | Lamc1 | 3.139 | 1.650 | 0.0366 |
| ENSRNOG00000011124 | Eif4g2-ps1 | 3.125 | 1.644 | 0.0380 |
| ENSRNOG00000004009 | Xpnpep2 | 3.118 | 1.641 | 0.0414 |
| ENSRNOG00000010768 | Kpna4 | 3.114 | 1.639 | 0.0499 |
| ENSRNOG00000042249 | F1LTA7_RAT | 3.101 | 1.633 | 0.0422 |
| ENSRNOG00000014166 | Smoc2 | 3.078 | 1.622 | 0.0413 |
| ENSRNOG00000002305 | Slc15a2 | 3.061 | 1.614 | 0.0400 |
| ENSRNOG00000005130 | Ogdh | 3.049 | 1.608 | 0.0465 |
| ENSRNOG00000018086 | Slc22a8 | 3.048 | 1.608 | 0.0418 |
| ENSRNOG00000010814 | Bmpr1a | 3.006 | 1.588 | 0.0432 |
| ENSRNOG00000032885 | CYC_RAT | 2.936 | 1.554 | 0.0478 |
|
| ||||
| Down-regulated Genes: 15 | ||||
| ENSRNOG00000032087 | F1LWC2_RAT | 0.301 | −1.734 | 0.0328 |
| ENSRNOG00000032609 | — | 0.300 | −1.738 | 0.0394 |
| ENSRNOG00000033748 | F1LWC2_RAT | 0.299 | −1.741 | 0.0381 |
| ENSRNOG00000025670 | Shisa3 | 0.295 | −1.759 | 0.0282 |
| ENSRNOG00000029115 | — | 0.284 | −1.815 | 0.0315 |
| ENSRNOG00000011821 | S100a4 | 0.228 | −2.134 | 0.0086 |
| ENSRNOG00000007632 | Zmynd17 | 0.211 | −2.243 | 0.0365 |
| ENSRNOG00000025408 | D3ZTT0_RAT | 0.189 | −2.403 | 0.0422 |
| ENSRNOG00000028844 | Slc9a5 | 0.181 | −2.466 | 0.0282 |
| ENSRNOG00000006889 | Ambp | 0.150 | −2.741 | 0.0177 |
| ENSRNOG00000028730 | D3ZI71_RAT | 0.148 | −2.757 | 0.0450 |
| ENSRNOG00000026067 | Wfdc10 | 0.144 | −2.791 | 0.0123 |
| ENSRNOG00000037374 | D3ZPQ1_RAT | 0.140 | −2.840 | 0.0087 |
| ENSRNOG00000033517 | LOC100360791 | 0.129 | −2.950 | 0.0009 |
| ENSRNOG00000042909 | F1LZX4_RAT | 0.107 | −3.229 | 0.0466 |
| ENSRNOG00000014578 | Fxyd4 | 0.096 | −3.374 | 0.0001 |
Figure 6.

Enzymes of the tryptophan metabolism pathway that are regulated by melatonin therapy in the kidney (red stars). Data were analyzed using the KEGG Pathway feature of the DAVID software.
4. Discussion
The major findings of our study can be summarized as follows: (1) CR offspring developed hypertension at 12 weeks of age and this was prevented by maternal melatonin therapy; (2) melatonin restored the CR-induced increase of plasma ADMA level, decreased l-arginine level, and decreased l-arginine-to-ADMA ratio; (3) CR reduced renal NO level and this was prevented by melatonin; (4) melatonin therapy increased PAX2 mRNA expression in the CR + M group; (5) CR upregulated renin and PRR expression and melatonin suppressed this increase; (6) melatonin therapy significantly increased renal ACE2 protein levels in the M and CR + M group; and (7) the expression of numerous genes was regulated in melatonin-treated offspring kidneys during nephrogenesis.
Our recent work indicates that ADMA-induced NO/reactive oxygen species (ROS) imbalance is involved in the development of hypertension in two different developmental models, maternal caloric restriction, and maternal diabetes [10, 11]. Several lines of evidence in this study indicated that ADMA-induced NO/ROS imbalance is involved in the developmental programming of hypertension in offspring exposed to maternal caloric restriction. First, plasma levels of the endogenous NOS inhibitor ADMA were increased in the CR group. Second, ADMA and l-arginine both compete for NOS and are present in a ratio that maintains NO homeostasis; this ratio was decreased in the plasma in the CR group. Third, maternal CR decreased renal NO levels in the offspring. Thus, alterations in the ADMA-NO pathway might be a major factor involved in programmed adult hypertension in response to maternal CR.
Melatonin is rapidly transferred from maternal to fetal circulation [20]. Administration of melatonin to pregnant rats prevents oxidative stress damage in the brains of offspring [21]. Previously, we showed that melatonin increases NO, restoring NO/ROS balance at the prehypertension stage and leading to lower blood pressure in young SHR [15]. Consistent with these findings, we found that early melatonin therapy in the mother could prevent programmed hypertension in their adult offspring. Thus, we suggest that melatonin has a novel protective effect on programmed hypertension through acting on the ADMA-NO pathway.
In addition to oxidative stress, the RAS plays a fundamental role in the development of hypertension and kidney development [5]. Epigenetic regulation of several RAS components has been reported in different programmed hypertension models [22, 23]. We demonstrated for the first time that melatonin therapy during nephrogenesis increased renin, PRR, and ACE2 expression in the kidney of the adult offspring. Consistent with these data, renal protein levels of PRR and ACE2 were increased in melatonin-treated offspring. Renin-PRR signaling is essential for proper kidney development and is causally linked to hypertension [13]. ACE2 appears to antagonize the effects of ACE through the production of angiotensin (1–7) in a manner that opposes the development of hypertension [24]. Surprisingly, melatonin therapy increased ACE2 expression in the kidney and prevented CR-induced programmed hypertension, despite the presence of increased renin and PRR expression. Notably, melatonin upregulated several RAS components and had reciprocal effects on vasodilation and vasoconstriction in rats at 3 months of age. Future studies are required to clarify the underlying mechanisms involved in the differential regulation of RAS components by melatonin.
Long-term amelioration of hypertension by melatonin therapy during gestation and lactation may be due to epigenetic changes in the kidney during a critical period of nephrogenesis. We found that melatonin upregulated HDAC-2, -3, and -8 expression in the kidney in CR + M group. This finding is consistent with that of our previous study showing that melatonin increased the expression of both class I and class II HDACs in vitro [25]. Given that melatonin increased class I HDACs expression and that HDACs are primarily thought to repress gene transcription, melatonin likely upregulates gene expression. Conversely, melatonin is known as a class III HDAC inhibitor [17]. Thus, melatonin might have dual effects on HDACs to epigenetically regulate gene expression. To the best of our knowledge, our study is the first to document altered expression of more than 400 genes in the kidney in response to melatonin and implicates melatonin in the protection from programmed hypertension in adult life. Notably, our data imply that melatonin is liable to induce, but not suppress, gene expression in the developing kidney. Using the KEGG database, several biological pathways were proposed to be regulated by melatonin including focal adhesion signaling, the peroxisome proliferator-activated receptors signaling pathway, fatty acid metabolism, the transforming growth factor β signaling pathway, and the Wnt signaling pathway. These findings suggest that melatonin might have a global epigenetic effect during nephrogenesis. Interestingly, the most significantly regulated biological theme was tryptophan metabolism, indicating that melatonin might have a negative feedback effect on its precursor tryptophan. Notably, maternal melatonin therapy has adverse effects on survival and renal growth in Wistar-Kyoto rats [26]. Because our data showed that maternal melatonin therapy had strong epigenetic effects, further evaluation is warranted to determine whether early melatonin therapy causes long-term epigenetic changes that lead to adverse effects in adulthood.
Previously, we showed that maternal CR reduces nephron numbers in offspring [10]. Increases in renal apoptosis and impaired expression of nephrogenesis-related genes may contribute to this reduction. In contrast to several earlier reports [27, 28], we found that apoptosis- and nephrogenesis-related genes were not altered in maternal CR-induced programmed hypertension. Of note, we showed for the first time that melatonin treatment upregulated PAX2 mRNA in metanephroi. Because PAX2 plays a crucial role in kidney development and is associated with various congenital renal and ureteral malformations, further studies are warranted to understand the epigenetic regulation of melatonin on PAX2 during nephrogenesis.
We conclude that prenatal melatonin therapy offsets the effects of maternal CR-induced programmed hypertension in adult offspring, primarily through the restoration of the ADMA-NO balance in the kidney. Our data suggested that a critical window exists during nephrogenesis in which the adult BP can be modified. Moreover, we showed that melatonin can modulate type I HDACs and serve as an inducer of gene expression in the developing kidney. The implications of melatonin-induced epigenetic changes on programmed hypertension in later life remain to be explored.
Acknowledgments
This work was supported by Grant NSC 101-2314-B-182A-021-MY3 from the National Science Council (Taiwan) and Grants CMRPG8B0172 and CMRPG8C0041 from Chang Gung Memorial Hospital (Kaohsiung, Taiwan).
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Ojeda NB, Grigore D, Alexander BT. Developmental programming of hypertension: insight from animal models of nutritional manipulation. Hypertension. 2008;52(1):44–50. doi: 10.1161/HYPERTENSIONAHA.107.092890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagby SP. Maternal nutrition, low nephron number, and hypertension in later life: pathways of nutritional programming. Journal of Nutrition. 2007;137(4):1066–1072. doi: 10.1093/jn/137.4.1066. [DOI] [PubMed] [Google Scholar]
- 3.McMillen IC, Robinson JS. Developmental origins of the metabolic syndrome: prediction, plasticity, and programming. Physiological Reviews. 2005;85(2):571–633. doi: 10.1152/physrev.00053.2003. [DOI] [PubMed] [Google Scholar]
- 4.Alwasel SH, Ashton N. Prenatal programming of renal sodium handling in the rat. Clinical Science. 2009;117(2):75–84. doi: 10.1042/CS20080294. [DOI] [PubMed] [Google Scholar]
- 5.Vehaskari VM, Stewart T, Lafont D, Soyez C, Seth D, Manning J. Kidney angiotensin and angiotensin receptor expression in prenatally programmed hypertension. The American Journal of Physiology—Renal Physiology. 2004;287(2):F262–F267. doi: 10.1152/ajprenal.00055.2004. [DOI] [PubMed] [Google Scholar]
- 6.Dennery PA. Oxidative stress in development: nature or nurture? Free Radical Biology and Medicine. 2010;49(7):1147–1151. doi: 10.1016/j.freeradbiomed.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Wilcox CS. Oxidative stress and nitric oxide deficiency in the kidney: a critical link to hypertension? The American Journal of Physiology—Regulatory Integrative and Comparative Physiology. 2005;289(4):R913–R935. doi: 10.1152/ajpregu.00250.2005. [DOI] [PubMed] [Google Scholar]
- 8.Baylis C. Arginine, arginine analogs and nitric oxide production in chronic kidney disease. Nature Clinical Practice Nephrology. 2006;2(4):209–220. doi: 10.1038/ncpneph0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tain Y-L, Huang L-T. Asymmetric dimethylarginine: clinical applications in pediatric medicine. Journal of the Formosan Medical Association. 2011;110(2):70–77. doi: 10.1016/S0929-6646(11)60012-0. [DOI] [PubMed] [Google Scholar]
- 10.Tain Y-L, Hsieh C-S, Lin I-C, Chen C-C, Sheen J-M, Huang L-T. Effects of maternal l-citrulline supplementation on renal function and blood pressure in offspring exposed to maternal caloric restriction: the impact of nitric oxide pathway. Nitric Oxide. 2010;23(1):34–41. doi: 10.1016/j.niox.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 11.Tain YL, Lee WC, Hsu CN, et al. Asymmetric dimethylarginine is associated with developmental programming of adult kidney disease and hypertension in offspring of streptozotocin-treated mothers. PLoS ONE. 2013;8(2) doi: 10.1371/journal.pone.0055420.e55420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S, Bellew C, Yao X, et al. Histone deacetylase (HDAC) activity is critical for embryonic kidney gene expression, growth, and differentiation. The Journal of Biological Chemistry. 2011;286(37):32775–32789. doi: 10.1074/jbc.M111.248278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song R, Van Buren T, Yosypiv IV. Histone deacetylases are critical regulators of the renin-angiotensin system during ureteric bud branching morphogenesis. Pediatric Research. 2010;67(6):573–578. doi: 10.1203/PDR.0b013e3181da477c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galano A, Tan DX, Reiter RJ. On the free radical scavenging activities of melatonin's metabolites, AFMK and AMK. Journal of Pineal Research. 2013;54(3):245–257. doi: 10.1111/jpi.12010. [DOI] [PubMed] [Google Scholar]
- 15.Tain Y-L, Huang L-T, Lin I-C, Lau Y-T, Lin C-Y. Melatonin prevents hypertension and increased asymmetric dimethylarginine in young spontaneous hypertensive rats. Journal of Pineal Research. 2010;49(4):390–398. doi: 10.1111/j.1600-079X.2010.00806.x. [DOI] [PubMed] [Google Scholar]
- 16.Korkmaz A, Rosales-Corral S, Reiter RJ. Gene regulation by melatonin linked to epigenetic phenomena. Gene. 2012;503(1):1–11. doi: 10.1016/j.gene.2012.04.040. [DOI] [PubMed] [Google Scholar]
- 17.Jung-Hynes B, Reiter RJ, Ahmad N. Sirtuins, melatonin and circadian rhythms: building a bridge between aging and cancer. Journal of Pineal Research. 2010;48(1):9–19. doi: 10.1111/j.1600-079X.2009.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tain Y-L, Baylis C. Determination of dimethylarginine dimethylaminohydrolase activity in the kidney. Kidney International. 2007;72(7):886–889. doi: 10.1038/sj.ki.5002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 20.Okatani Y, Wakatsuki A, Kaneda C. Melatonin increases activities of glutathione peroxidase and superoxide dismutase in fetal rat brain. Journal of Pineal Research. 2000;28(2):89–96. doi: 10.1034/j.1600-079x.2001.280204.x. [DOI] [PubMed] [Google Scholar]
- 21.Watanabe K, Wakatsuki A, Shinohara K, Ikenoue N, Yokota K, Fukaya T. Maternally administered melatonin protects against ischemia and reperfusion-induced oxidative mitochondrial damage in premature fetal rat brain. Journal of Pineal Research. 2004;37(4):276–280. doi: 10.1111/j.1600-079X.2004.00167.x. [DOI] [PubMed] [Google Scholar]
- 22.Bogdarina I, Welham S, King PJ, Burns SP, Clark AJL. Epigenetic modification of the renin-angiotensin system in the fetal programming of hypertension. Circulation Research. 2007;100(4):520–526. doi: 10.1161/01.RES.0000258855.60637.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goyal R, Goyal D, Leitzke A, Gheorghe CP, Longo LD. Brain renin-angiotensin system: fetal epigenetic programming by maternal protein restriction during pregnancy. Reproductive Sciences. 2010;17(3):227–238. doi: 10.1177/1933719109351935. [DOI] [PubMed] [Google Scholar]
- 24.Feng Y, Xia H, Santos RA, Speth R, Lazartigues E. Angiotensin-converting enzyme 2: a new target for neurogenic hypertension. Experimental Physiology. 2010;95(5):601–606. doi: 10.1113/expphysiol.2009.047407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma R, Ottenhof T, Rzeczkowska PA, Niles LP. Epigenetic targets for melatonin: induction of histone H3 hyperacetylation and gene expression in C17.2 neural stem cells. Journal of Pineal Research. 2008;45(3):277–284. doi: 10.1111/j.1600-079X.2008.00587.x. [DOI] [PubMed] [Google Scholar]
- 26.Singh HJ, Keah LS, Kumar A, Sirajudeen KNS. Adverse effects of melatonin on rat pups of Wistar-Kyoto dams receiving melatonin supplementation during pregnancy. Experimental and Toxicologic Pathology. 2012;64(7-8):751–752. doi: 10.1016/j.etp.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Dickinson H, Walker DW, Wintour EM, Moritz K. Maternal dexamethasone treatment at midgestation reduces nephron number and alters renal gene expression in the fetal spiny mouse. The American Journal of Physiology—Regulatory Integrative and Comparative Physiology. 2007;292(1):R453–R461. doi: 10.1152/ajpregu.00481.2006. [DOI] [PubMed] [Google Scholar]
- 28.Abdel-Hakeem AK, Henry TQ, Magee TR, et al. Mechanisms of impaired nephrogenesis with fetal growth restriction: altered renal transcription and growth factor expression. The American Journal of Obstetrics and Gynecology. 2008;199(3):252.e1–252.e7. doi: 10.1016/j.ajog.2008.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]


