Abstract
Background
Stool DNA testing is a new approach to colorectal cancer detection. Few data are available from the screening setting.
Objective
To compare stool DNA and fecal blood testing for detection of screen-relevant neoplasia (curable-stage cancer, high-grade dysplasia, or adenomas >1 cm).
Design
Blinded, multicenter, cross-sectional study.
Setting
Communities surrounding 22 participating academic and regional health care systems in the United States.
Participants
4482 average-risk adults.
Measurements
Fecal blood and DNA markers. Participants collected 3 stools, smeared fecal blood test cards and used same-day shipment to a central facility. Fecal blood cards (Hemoccult and HemoccultSensa, Beckman Coulter, Fullerton, California) were tested on 3 stools and DNA assays on 1 stool per patient. Stool DNA test 1 (SDT-1) was a precommercial 23-marker assay, and a novel test (SDT-2) targeted 3 broadly informative markers. The criterion standard was colonoscopy.
Results
Sensitivity for screen-relevant neoplasms was 20% by SDT-1, 11% by Hemoccult (P = 0.020), 21% by HemoccultSensa (P = 0.80); sensitivity for cancer plus high-grade dysplasia did not differ among tests. Specificity was 96% by SDT-1, compared with 98% by Hemoccult (P < 0.001) and 97% by HemoccultSensa (P = 0.20). Stool DNA test 2 detected 46% of screen-relevant neoplasms, compared with 16% by Hemoccult (P < 0.001) and 24% by HemoccultSensa (P < 0.001). Stool DNA test 2 detected 46% of adenomas 1 cm or larger, compared with 10% by Hemoccult (P < 0.001) and 17% by HemoccultSensa (P < 0.001). Among colonoscopically normal patients, the positivity rate was 16% with SDT-2, compared with 4% with Hemoccult (P = 0.010) and 5% with HemoccultSensa (P = 0.030).
Limitations
Stool DNA test 2 was not performed on all subsets of patients without screen-relevant neoplasms. Stools were collected without preservative, which reduced detection of some DNA markers.
Conclusion
Stool DNA test 1 provides no improvement over HemoccultSensa for detection of screen-relevant neoplasms. Stool DNA test 2 detects significantly more neoplasms than does Hemoccult or HemoccultSensa, but with more positive results in colonoscopically normal patients. Higher sensitivity of SDT-2 was particularly apparent for adenomas.
Colorectal cancer remains the second most common cause of death among the types of cancer (1). Although screening reduces colorectal cancer mortality (2–6), observed reductions have been modest (6, 7) and more than one half of adults in the United States have not received screening (8). More accurate, user-friendly, and widely distributable tools have the potential to improve screening effectiveness, acceptability, and access.
Several molecular approaches to screening stool for colorectal cancer have been studied and reviewed (9, 10), and stool DNA testing has been jointly endorsed by the American Cancer Society, the U.S. Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology (11). The advantages of stool DNA testing include noninvasiveness, absence of bowel preparation or dietary restrictions, and ease of access via mail courier. However, the reported accuracy of stool DNA tests for the detection of colorectal neoplasia varies. In clinical studies that used different assays and selected groups (12–20), sensitivities ranged from 62% to 100% for colorectal cancer and 27% to 82% for advanced adenomas, with specificities ranging from 82% to 100%. In the only reported multi-center study on asymptomatic average-risk patients (21), a precommercial multitarget DNA assay (SDT-1, a prototype of PreGenPlus, EXACT Sciences, Marlborough, Massachusetts) detected 52% of cases of colorectal cancer, compared with 13% by Hemoccult (P = 0.003), at specificities of 94.4% and 95.2%, respectively.
The accuracy of stool DNA testing is influenced by both biological and technical factors. A panel of markers must be used to accommodate the molecular heterogeneity of colorectal neoplasia, and marker selection critically affects discrimination (9). Unlike occult bleeding, which is intermittent (22), DNA markers seem to be shed continuously by exfoliation (23). Thus, the multiple stool sampling practiced with fecal occult blood tests may not be necessary with stool DNA tests. However, recovery of the minute quantities of human DNA and assay of tumor-specific DNA alterations from stool present technical challenges and require exquisite laboratory sensitivity to achieve optimal detection rates.
Our primary aim was to compare the precommercial stool DNA test (SDT-1), which was studied by Imperiale and colleagues (21), with widely used fecal occult blood tests for the detection of screen-relevant neoplasia, defined as curable-stage colorectal cancer (no distant metastases), high-grade dysplasia, or adenomas larger than 1 cm. A secondary aim was to explore neoplasm detection by another stool DNA test 2 (SDT-2), which uses a more broadly informative marker panel.
Methods
Table 1 lists the genes used in our test panels and defines several key terms.
Table 1.
Definitions
| Gene targets in stool DNA test panels: |
| Test 1: point mutations on K-ras, APC, and p53; microsatellite marker BAT-26; long DNA |
| Test 2: point mutations on K-ras, scanned mutator cluster region of APC, vimentin methylation |
| Screen-relevant neoplasia: colorectal cancer, high-grade dysplasia, adenomas ≥1 cm |
| Sensitivity: rate of test positivity for those with screen-relevant neoplasia |
| Specificity: rate of test negativity for those without screen-relevant neoplasia |
| Test positivity: rate of positive stool test results for individual colonoscopic findings or groups of findings |
Design
We conducted this multicenter, prospective, triple-blinded trial, targeting average-risk persons, from 2001 to 2007. A group of national experts on colorectal cancer screening advised on study design, and institutional review boards at each site approved the study. Because we did not know the effect of diet and medications on DNA assays, patients were randomly assigned at entry to group A (restriction of red meat and therapeutic doses of nonsteroidal anti-inflammatory drugs for 3 days before and during stool collections) or group B (no such restrictions). All patients were asked not to ingest vitamin C for the 3 days before and during stool collections. For the companion test, we chose Hemoccult (Beckman Coulter, Fullerton, California), the most widely used fecal occult blood test, which was used in the trials that established the benefit of screening for fecal occult blood (2–4). As a second companion test, we chose the next-generation guaiac test HemoccultSensa (Beckman Coulter). We compared fecal blood results from 3 stools per patient with stool DNA on 1 stool. Experienced technicians performed stool DNA and occult blood testing in separate central laboratories without knowledge of clinical findings or the results of other tests. All patients who completed stool collections also had colonoscopy, which served as the criterion standard. We did not have access to data until after they had been analyzed by statisticians and released by a data monitoring board.
Participants
We recruited asymptomatic persons age 50 to 80 years who were at average risk for colorectal cancer from communities surrounding 22 participating academic and regional health care systems through direct mail and multi-media advertisements. The exclusion criteria were structural colorectal evaluation (endoscopic or radiographic) within 10 years; fecal blood testing within 1 year; overt rectal bleeding within 1 month; previous colorectal resection; aerodigestive cancer within 5 years; inability to stop therapeutic doses of nonsteroidal anti-inflammatory drugs or anticoagulants; coagulopathy; contraindications to colonoscopy; chemotherapy within 3 months; high-risk conditions for colorectal cancer, such as familial adenomatous polyposis, the Lynch syndrome, or other cancer syndromes; previous colorectal cancer or adenoma; inflammatory bowel disease; or more than 2 first-degree relatives with colorectal neoplasia. Study assistants at each site registered participants and randomly assigned them by using a Web-based management system; distributed fecal blood test cards, stool collection containers, and colonoscopy preparation materials; and provided instructions.
Stool Collection and Processing
Patients collected 3 stools by using plastic buckets mounted to the toilet seat. Promptly after each individual collection, patients smeared stool onto both windows of their Hemoccult and HemoccultSensa cards and then express-shipped smeared cards and the whole stool (sealed in a bucket in an insulated container cooled with ice packs) to the Mayo Clinic in Rochester, Minnesota. We froze the first stool from each participant whole at −80 °C on receipt and sent it in batches on dry ice to EXACT Sciences (Marlborough, Massachusetts) for DNA assay; each of the subsequent 2 stools were archived in aliquots at −80 °C. If the first stool weighed less than 30 g or was received more than 48 hours after defecation, it was rejected for DNA analysis and the second or third stool (if it met inclusion criteria) was sent for DNA assay.
Stool Assays
DNA Testing
All assays were polymerase chain reaction–based and were run at EXACT Sciences. Stool DNA test 1 was performed as described in Imperiale and colleagues’ study (21). The marker panel for SDT-1 included 21 tumor-specific point mutations (3 on the K-ras gene, 10 on the APC gene, and 8 on the p53 gene); the microsatellite-instability marker BAT-26; and long DNA, a marker for delayed apoptosis, which is characteristic of exfoliated neoplastic colonocytes (12). For SDT-2, sequence-specific DNA markers were detected by acrylamide gel electrophoresis, as described by Whitney and colleagues (24); the panel consisted of 3 tumor-specific markers broadly informative for both colorectal cancer and adenomas (25): K-ras mutations, scanning of APC mutator cluster regions, and methylation of the vimentin gene. We used methods described elsewhere to detect mutant K-ras (12), APC scanning (25), and vimentin gene methylation (20) assays. We defined any positive component marker result according to the manufacturer’s preestablished criteria as a positive test result.
Occult Blood Testing
The manufacturer that developed the Hemoccult and HemoccultSensa cards, without rehydration, trained technicians on-site at the Mayo Clinic. As recommended by the manufacturer, the technicians added the catalyst solution to cards stored at ambient temperature within 48 to 72 hours of collection. We defined a spreading (enlarging) blue color in 60 seconds in any window of the cards as a positive result and any other result as negative.
Colonoscopy
After cathartic preparation, experienced endoscopists performed colonoscopy in all patients. If the examination did not reach the cecum or inspected less than 90% of the mucosa, the patient was disqualified. Endophotographs documented cecal intubation, and the size and location of all lesions were recorded. Costs not covered by third parties were reimbursed by study funding.
Pathologic Examination
Local pathologists examined all endoscopically or surgically sampled lesions. A gastrointestinal pathologist at the coordinating site reexamined all lesions to confirm diagnosis. Classification discrepancies of screen-relevant neoplasms were adjudicated by a second expert pathologist. We categorized patients with multiple neoplasms according to the most advanced lesion. For assay of markers in screen-relevant neoplasms, DNA was extracted from microdissected tissue.
Statistical Analysis
We calculated sample size to ensure adequate power to detect differences in sensitivity comparisons. We powered the study to ensure an adequate number of cases of curable-stage colorectal cancer and high-grade dysplasia and assumed their combined prevalence to be at least 1.5%. A sample size of 2900 would yield an expected 43 curable-stage cancer or high-grade dysplasia cases, which would provide 90% power to detect a 35% improvement in sensitivity of SDT-1 over the Hemoccult test by using a 2-sided McNemar test with α = 0.05 (assuming Hemoccult sensitivity of 25%). The protocol specified interim analyses at one half and three quarters of full enrollment to see whether it was necessary to stop the study early if test sensitivities differed significantly or to revise sample size requirements on the basis of observed prevalence of the target lesion. At the first interim analysis, lesion prevalence was lower than expected, and we readjusted the sample size to 4434 patients. However, before we completed enrollment, the manufacturer altered the SDT-1 assay, which prompted an unplanned interim analysis after 2497 patients. On the basis of these interim results, we stopped SDT-1 testing and began doing the SDT-2 test.
To accomplish a secondary aim of this trial (to see whether restricting diet and medication affects the specificity of the SDT test), we randomly assigned persons to pretest restrictions or no restrictions. The sample size calculated for the sensitivity comparison provided 85% power to detect a 4% difference in specificity between randomization groups. Because SDT specificity was the same in both groups, we pooled the results for all analyses.
We included all patients tested with SDT-1. We compared stool test sensitivities and specificities by using the McNemar test. We used a chi-square test or the Fisher exact test to compare baseline characteristics between cohorts and assay performance in subsets of patients. All P values are 2-sided.
Per agreement with EXACT Sciences, we did the SDT-2 test on all patients with cancer, high-grade dysplasia, and adenomas larger than 2 cm from the full enrollment period as well as on a random sample of 50 patients with 1- to 2-cm adenomas and 75 with normal colonoscopy results. To estimate the population-level sensitivity for the SDT-2 test, we used all case patients tested with SDT-2 and reweighted the calculation to be proportional to the observed prevalence of each screen-relevant neoplasia category in the entire population with screen-relevant neoplasias. Because we did not do the SDT-2 test on all subsets without screen-relevant neoplasia, we could not calculate specificity for screen-relevant neoplasia. To compare test positivity rates in patient subsets, we used the McNemar test.
Role of the Funding Source
The National Cancer Institute funded this study and monitored conduct. EXACT Sciences performed DNA assays at no cost, and Beckman Coulter provided Hemoccult and HemoccultSensa cards at no cost. EXACT Sciences limited SDT-2 coverage to screen-relevant neoplasms and a subset of normal control participants. Neither company influenced study oversight, data analysis, or reporting.
Results
Patients
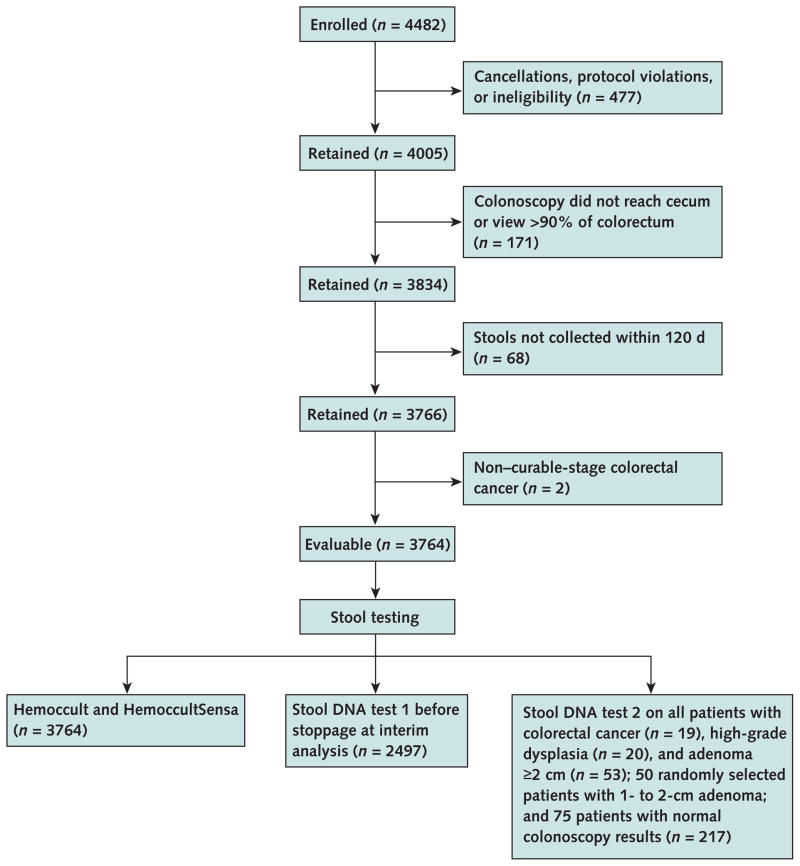
Of the 4482 persons enrolled, 3764 (84%) were evaluable. We excluded 545 patients because of cancellations, protocol violations, or ineligibility; 171 because of incomplete colonoscopies; and 2 because of distant metastases (Figure 1). Table 2 shows demographic and colorectal lesion characteristics. We found screen-relevant neoplasms in 290 (7.7%) patients; 39 had nonmetastatic cancer or high-grade dysplasia and 251 had adenomas that were 1 cm or larger. Major complications from colonoscopy occurred in 4 patients; no procedure-related deaths were reported.
Figure 1. Study flow diagram.
Table 2.
Baseline Characteristics and Colorectal Findings
| Characteristic | All Patients (n = 4482) | Evaluable Patients* (n = 3764) | Patients Tested with SDT-1† (n = 2497) | Patients Tested with SDT-2‡ (n = 217) |
|---|---|---|---|---|
| Age, y | ||||
|
| ||||
| Mean (SD) | 63.8 (8.29) | 63.7 (8.25) | 60.4 (7.86) | 66.4 (7.17) |
|
| ||||
| Median (range) | 65 (50–81) | 65 (50–80) | 59 (50–80) | 67 (51–80) |
|
| ||||
| Women, n (%) | 2341 (52.2) | 1964 (52.2) | 1348 (54.0) | 108 (49.8) |
|
| ||||
| White, n (%) | 4184 (93.4) | 3522 (93.6) | 2314 (92.7) | 201 (92.6) |
| Colorectal findings, n (%) | ||||
|
| ||||
| Screen-relevant neoplasia | – | 290 (7.7) | 157 (6.3) | 142 (65.4) |
|
| ||||
| Cancer | ||||
| Stage I | – | 11 (0.3) | 6 (0.2) | 11 (5.1) |
|
| ||||
| Stages II and III | – | 8 (0.2) | 6 (0.2) | 8 (3.7) |
|
| ||||
| Cancer + high-grade dysplasia | – | 39 (1.0) | 22 (0.9) | 39 (18.0) |
|
| ||||
| Adenoma ≥1 cm | – | 251 (6.7) | 135 (5.4) | 103 (47.5) |
|
| ||||
| Adenoma >2 cm | – | 53 (1.4) | 21 (0.8) | 53 (24.4) |
|
| ||||
| Adenoma <1 cm | – | 785 (20.9) | 469 (18.8) | Not tested |
|
| ||||
| Hyperplastic polyps | – | 492 (13.1) | 341 (13.7) | Not tested |
|
| ||||
| Other | – | 86 (2.3) | 57 (2.3) | Not tested |
|
| ||||
| Normal | – | 2111 (56.1) | 1473 (59.0) | 75 (34.6) |
SDT = stool DNA test.
Patients who met all inclusion criteria. Both Hemoccult and HemoccultSensa (Beckman Coulter, Fullerton, California) were performed on all evaluable participants.
On the basis of results from an interim analysis, SDT-1 was terminated.
All participants with cancer, high-grade dysplasia, and adenomas ≥2 cm from the full enrollment period are included, as are random samples from 50 patients with 1- or 2-cm adenomas and 75 with normal colonoscopy results.
Occult Blood Testing: Hemoccult versus HemoccultSensa
Detection sensitivities for the 290 screen-relevant neoplasms found among all 3764 evaluable participants were 10% (95% CI, 7% to 13%) with Hemoccult and 18% (CI, 13% to 22%) with HemoccultSensa (P < 0.001). Based on all 3474 participants without screen-relevant neoplasia, the Hemoccult specificity of 98% (CI, 98% to 99%) was slightly higher than that of HemoccultSensa (97% [CI, 96% to 97%]) (P < 0.001).
Hemoccult and HemoccultSensa positivity rates for the 39 patients with colorectal cancer or high-grade dysplasia were 33% (CI, 19% to 48%) and 44% (CI, 28% to 59%), respectively (P = 0.100). For the 251 patients with adenomas 1 cm or larger, the positivity rates were 6% (CI, 3% to 9%) versus 14% (CI, 9% to 18%) (P = 0.001).
Stool DNA versus Occult Blood Testing
SDT-1 versus Occult Blood Testing
Based on the first 2497 evaluable participants (Table 3), the sensitivity of SDT-1 for screen-relevant neoplasia was higher than that of Hemoccult (20% [CI, 14% to 26%] vs. 11% [CI, 6% to 16%]; P = 0.020) but not that of HemoccultSensa (21% [CI, 15% to 27%]; P = 0.80). For all target lesion groupings, specificities were slightly but significantly lower for SDT-1 than for Hemoccult but not HemoccultSensa, and the positive likelihood ratios for SDT-1 were lower than for either Hemoccult or HemoccultSensa for the more advanced groupings of screen-relevant neoplasms (Table 3).
Table 3.
Summary of Test Performance
| Index Test | Screen-Relevant Neoplasia, n* | Positive Test Result, n | Sensitivity (95% CI) | No Screen-Relevant Neoplasia, n | Negative Test Result, n | Specificity (95% CI) | Positive Likelihood Ratio (95% CI) | Negative Likelihood Ratio (95% CI) |
|---|---|---|---|---|---|---|---|---|
| Hemoccult (n = 2497) | 157 | 17 | 11 (6–16)† | 2340 | 2297 | 98 (98–99)‡ | 5.9 (3–10) | 0.9 (0.9–1.0) |
| HemoccultSensa (n = 2497) | 157 | 33 | 21 (15–27)§ | 2340 | 2258 | 97 (96–97)|| | 6.0 (4–9) | 0.8 (0.8–0.9) |
| SDT-1 (n = 2497) | 157 | 31 | 20 (14–26) | 2340 | 2246 | 96 (95–97) | 4.9 (3–7) | 0.8 (0.8–0.9) |
| SDT-2 (n = 217) | 142 | 66 | 40 (32–49)¶ | 75 | NA** | NA | NA | NA |
NA = not available; SDT = stool DNA test.
Includes curable-stage cancer, high-grade dysplasia, and adenomas ≥1 cm.
P = 0.02 for STD-1 vs. Hemoccult.
P < 0.001 for STD-1 vs. Hemoccult.
P = 0.80 for STD-1 vs. HemoccultSensa.
P = 0.40 for STD-1 vs. HemoccultSensa.
We calculated the weighted sensitvity for SDT-2 with the following equation: . PR = proportion of participants for that category of screen-relevant neoplasia in the entire population with screen-relevant neoplasia. See “Comparison of Stool DNA Tests” for statistical comparisons of SDT-1 and SDT-2 in participants who had both DNA tests performed.
We did not calculate formal specificity because SDT-2 was not performed on all subsets without screen-relevant neoplasia.
Based on test positivity data in subsets of screen-relevant neoplasms (Table 4), SDT-1 had higher detection rates than Hemoccult for 1- to 2-cm adenomas but not for any other subset. Stool DNA test 1 had a lower positivity rate for detecting invasive cancer than did HemoccultSensa (25% [CI, 5% to 57%] vs. 75% [51% to 100%]; P = 0.010).
Table 4.
Test Positivity Rates
| Stool DNA Test and Patient Subset | Positive SDT Result (95% CI), % | Positive Hemoccult Result (95% CI),% | P Value* | Positive HemoccultSensa Result (95% CI), % | P Value† |
|---|---|---|---|---|---|
| SDT-1 | |||||
|
| |||||
| Screen-relevant neoplasms‡ (n = 157) | 20 (14–26) | 11 (6–16) | 0.020 | 21 (15–27) | 0.80 |
|
| |||||
| Site | |||||
| Proximal§ (n = 56) | 20 (9–30) | 7 (2–17) | 0.070 | 11 (3–19) | 0.20 |
|
| |||||
| Distal§ (n = 101) | 20 (12–28) | 13 (6–19) | 0.100 | 27 (18–35) | 0.20 |
|
| |||||
| Cancer (n = 12) | 25 (5–57) | 50 (22–78) | 0.30 | 75 (51–100) | 0.010 |
|
| |||||
| Cancer + high-grade dysplasia (n = 22) | 36 (16–56) | 41 (20–61) | 0.80 | 55 (34–75) | 0.20 |
|
| |||||
| Adenoma ≥1 cm (n = 135) | 17 (11–23) | 6 (2–10) | 0.004 | 16 (9–22) | 0.74 |
|
| |||||
| Adenoma >2 cm (n = 21) | 19 (5–42) | 14 (3–36)|| | 0.60 | 33 (13–54) | 0.30 |
|
| |||||
| Adenoma 1–2 cm (n = 114) | 17 (10–24) | 4 (1–8)|| | 0.004 | 12 (6–18) | 0.40 |
|
| |||||
| Adenoma <1 cm (n = 469) | 4 (2–6) | 3 (1–5) | 0.30 | 5 (3–7) | 0.70 |
|
| |||||
| Hyperplastic polyps (n = 341) | 5 (2–7) | 1 (0.2–3)|| | 0.003 | 4 (2–6) | 0.70 |
|
| |||||
| Other (n = 57) | 5 (1–15) | 2 (0.04–9)|| | 0.30 | 5 (1–15)‡ | 1.00 |
|
| |||||
| Normal (n = 1473) | 4 (3–5) | 2 (1–2) | <0.001 | 3 (2–4) | 0.20 |
|
| |||||
| Age <65 y (n = 1040) | 3 (2–4) | 2 (1–2) | 0.009 | 3 (2–4) | 0.40 |
|
| |||||
| Age >65 y (n = 433) | 5 (3–7) | 2 (1–3) | 0.020 | 3 (2–5) | 0.30 |
| SDT-2 | |||||
|
| |||||
| Screen-relevant neoplasms‡ (n = 142) | 46 (38–55) | 16 (10–22) | <0.001 | 24 (17–31) | <0.001 |
|
| |||||
| Site | |||||
| Proximal§ (n = 56) | 45 (32–58) | 9 (1–16) | <0.001 | 13 (4–21) | <0.001 |
|
| |||||
| Distal§ (n = 86) | 48 (37–58) | 21 (12–30) | <0.001 | 31 (22–41) | 0.020 |
|
| |||||
| Cancer (n = 19) | 58 (36–80) | 47 (25–70) | 0.40 | 63 (41–85) | 0.70 |
|
| |||||
| Cancer + high-grade dysplasia (n = 39) | 49 (33–64) | 33 (19–48) | 0.100 | 44 (28–59) | 0.60 |
|
| |||||
| Adenoma ≥ 1 cm (n = 103) | 46 (35–54) | 10 (4–15) | <0.001 | 17 (9–24) | <0.001 |
|
| |||||
| Adenoma > 2 cm (n = 53) | 57 (43–70) | 13 (4–22) | <0.001 | 25 (13–36) | <0.001 |
|
| |||||
| Adenoma 1–2 cm (n = 50) | 34 (21–47) | 6 (1–17)|| | 0.002 | 8 (2–19)|| | 0.005 |
|
| |||||
| Normal (n = 75) | 16 (8–24) | 4 (1–11)|| | 0.010 | 5 (1–13)|| | 0.030 |
|
| |||||
| Age <65 y (n = 36) | 6 (4–29)|| | 3 (0–15)|| | 0.56 | 6 (1–19)|| | 1.00 |
|
| |||||
| Age >65 y (n = 39) | 26 (12–39) | 5 (1–17)|| | 0.010 | 5 (1–17)|| | 0.010 |
SDT = stool DNA test.
For SDT versus Hemoccult.
For SDT versus HemoccultSensa.
Patients were classified by the most advanced lesion found (in descending order of severity: cancer, high-grade dysplasia, or adenoma). The distribution of screen-relevant neoplasms differs from that in Table 3. See Table 2 for details of patients tested with SDT-2.
Proximal and distal sites are relative to the splenic flexure. “Proximal” includes the cecum and ascending and transverse colon; “distal” includes the splenic flexure, descending colon, sigmoid, and rectum.
The exact CI was used when the numerator was <5.
SDT-2 versus Occult Blood Testing
Table 3 shows the sensitivity of SDT-2, Hemoccult, and HemoccultSensa for screen-related neoplasia. The weighted estimate of SDT-2 sensitivity for screen-relevant neoplasms was 40% (CI, 32% to 49%), compared with the observed Hemoccult and HemoccultSensa sensitivities of 11% (CI, 6% to 16%) and 21% (CI, 15% to 27%), respectively (Table 3). Table 4 shows the test positivity rates for subgroups of colorectal lesions.
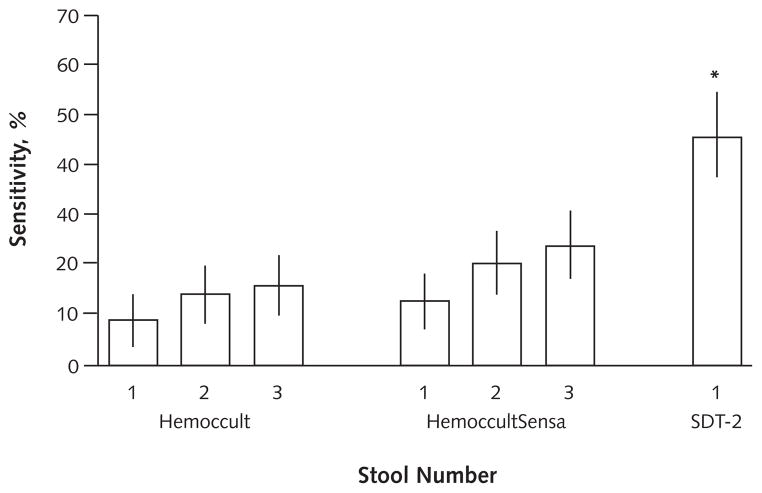
For adenomas 1 cm or larger, the positivity rate was 46% (CI, 35% to 54%) by SDT-2 versus 10% (CI, 4% to 15%) by Hemoccult (P < 0.001) and 17% (CI, 9% to 24%) by HemoccultSensa (P < 0.001). Neoplasm site did not affect detection by SDT-2; however, detection rates were lower for lesions proximal to the splenic flexure than for distal lesions with both Hemoccult (9% [CI, 1% to 16%] vs. 21% [CI, 12% to 30%]; P = 0.060) and HemoccultSensa (13% [CI, 4% to 21%] vs. 31% [CI, 22% to 41%]; P = 0.010). Study comparisons are based on a single stool per patient for SDT-2 and 3 stools per patient for fecal blood tests; differences in test performance are larger if fecal blood test results are analyzed on fewer than 3 stools per patient (Figure 2). For patients with normal colonoscopy results, the positivity rate was 16% (CI, 8% to 24%) by SDT-2, compared with 4% (CI, 1% to 11%) by Hemoccult (P = 0.010) and 5% (CI, 1% to 13%) by Hemoccult-Sensa (P = 0.030).
Figure 2. Stool DNA versus occult blood testing for detection of screen-relevant neoplasia (curable-stage colorectal cancer, high-grade dysplasia, and adenomas ≥1 cm) (n = 142).
Sensitivity is plotted for Hemoccult and HemoccultSensa with 1, 2, and 3 stools per patient and with stool DNA test 2 (SDT-2) on a single stool per patient. Vertical lines represent 95% CIs.
*P < 0.001 versus Hemoccult or HemoccultSensa.
Frequencies of DNA Markers in Neoplastic Tissue
Nearly all of the tissues analyzed from screen-relevant neoplasms contained at least 1 marker from the SDT-2 panel; fewer than two thirds of the tissues contained SDT-1 markers (Table 5).
Table 5.
Presence of DNA Markers in Tumor Tissue*
| Marker | SDT-1 Panel
|
SDT-2 Panel
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients, n | K-ras, % | APC†, % | p53, % | BAT-26, % | Full Panel, % | Patients, n | K-ras, % | APC‡, % | Vimentin, % | Full Panel, % | |
| Cancer and high-grade dysplasia | 20 | 45 | 35 | 25 | 0 | 60 | 35 | 51 | 60 | 63 | 94 |
|
| |||||||||||
| Adenoma ≥1 cm | 48 | 42 | 38 | 6 | 2 | 63 | 99 | 39 | 73 | 63 | 98 |
|
| |||||||||||
| All screen-relevant neoplasms§ | 68 | 43 | 37 | 12 | 1 | 62 | 134 | 43 | 69 | 63 | 97 |
SDT = stool DNA test.
Tissues were analyzed from all available samples from patients with cancer and high-grade dysplasia and from a random sample of patients with adenomas ≥1 cm.
The assay targeted 10 high-frequency mutations on the APC gene by using a single-base extension method (see citations in Methods).
The assay targeted any mutation in the mutator cluster region of the APC gene by using the scanning method (see citations in Methods).
Includes curable-stage cancer, high-grade dysplasia, and adenomas ≥1 cm.
Negative Stool DNA Tests in Patients with Screen-Relevant Neoplasia
Incomplete marker recovery from stools, instability of long DNA, and lesion size influenced test results. Recovery of individual markers in stool when they were present in tumor tissue from the same patient was 40% (17 of 42) for SDT-1 and 39% (51 of 130) for SDT-2. The sensitivity of long DNA decreased as the time from defecation to freezing the stool increased; sensitivity for screen-relevant neoplasms decreased from 11% on stools received and frozen 12 to 24 hours after defecation to 1.8% at 24 to 35 hours and to 0% at more than 35 hours after defecation (P = 0.010). The sensitivity of mutation markers did not change over this time range. The median size of screen-relevant neoplasms missed by SDT-2 was 15 mm, compared with 20 mm for detected lesions (P = 0.003). Neither demographic variables nor randomization group affected stool DNA test positivity.
Positive Stool DNA Tests in Patients without Screen-Relevant Neoplasia
Diet and use of nonsteroidal anti-inflammatory drugs affected HemoccultSensa results but not stool DNA test or Hemoccult results. For HemoccultSensa, positivity rates in colonoscopically normal patients randomly assigned to group A (restricted diet and medications) were 2% (CI, 1% to 2%), compared with 4% (CI, 3% to 5%) for those in group B (unrestricted) (P = 0.030). In contrast, positivity rates for the 2 stool DNA tests were unaffected (SDT-1: 4% for group A vs. 3% for group B [P = 0.31]; SDT-2: 19% for group A vs. 13% for group B [P = 0.50]), as were those for Hemoccult (1% for group A vs. 2% for group B; P = 0.170). Age influenced SDT-2 results in patients with normal colonoscopy results, as positivity rates increased from 6% (CI, 4% to 29%) for patients younger than age 65 years to 26% (CI, 12% to 39%) for those 65 years of age or older (P = 0.020) (Table 4). Positivity rates for the component markers of SDT-2 were 6% and 15% for APC scanning, 0% and 8% for methylated vimentin, and 0% and 3% for mutant K-ras in patients younger than age 65 years and those 65 years of age or older, respectively. Other demographic factors did not affect stool DNA test positivity.
Comparison of Stool DNA Tests
We performed the 2 stool DNA tests on 69 patients with screen-relevant neoplasia and 54 with normal colonoscopy results. Positivity rates were higher with SDT-2 than with SDT-1 among the 69 patients with screen-relevant neoplasms (43% [CI, 32% to 55%] vs. 20% [11% to 30%]; P = 0.001), including the subset of 12 patients with colorectal cancer (58% [CI, 28% to 85%] vs. 25% [6% to 57%]; P = 0.050). Positivity rates were higher when comparing results for all 47 patients with adenomas 1 cm or larger (45% [CI, 31% to 59%] vs. 13% [CI, 3% to 22%]; P = 0.003), the 21 patients with adenomas larger than 2 cm (62% [CI, 38% to 82%] vs. 19% [CI, 5% to 42%]; P = 0.003), and the 26 patients with adenomas between 1 and 2 cm (31% [CI, 14% to 52%] vs. 8% [CI, 1% to 25%]; P = 0.030). Test positivity in those with normal colonoscopy results was significantly higher with SDT-2 than with SDT-1 (13% [CI, 5% to 25%] vs. 2% [CI, 1% to 10%]; P = 0.030).
Discussion
In this multicenter study, we found that the precommercial stool DNA test (SDT-1) provides no meaningful improvements over the widely used fecal occult blood tests Hemoccult and HemoccultSensa for detection of screen-relevant colorectal neoplasms. Neoplasm detection rates by SDT-1 were no better than HemoccultSensa for any subset of neoplasms; in fact, HemoccultSensa detected significantly more cases of cancer than did SDT-1. In addition, the estimated positive likelihood ratios for cancer or high-grade dysplasia tended to be larger and the negative likelihood ratios smaller with both fecal blood tests than with SDT-1.
Because of the poor performance of SDT-1, we examined SDT-2 (which targets a potentially more informative marker panel) in a more limited manner. Stool DNA test 2 had significantly better neoplasm detection rates than did the fecal blood tests or SDT-1. We accounted for the higher detection rates of SDT-2 by better detection of advanced adenomas, a finding of particular importance for cancer prevention. Whereas fecal blood testing detected proportionately fewer proximal than distal colorectal neoplasms, corroborating previous findings (26, 27), site did not affect detection by stool DNA testing.
Detection of adenomas is essential for screening to prevent colorectal cancer. In our blinded comparison, using colonoscopy results as the criterion standard, SDT-2 detected 3 times more adenomas that were 1 cm or larger than did SDT-1, 4 times more than did Hemoccult and 3 times more than did HemoccultSensa. Compared with SDT-1, all markers in the SDT-2 panel occur early in the adenoma-to-cancer progression (25, 28, 29) and were collectively more informative for adenomas on our tissue analyses; this accounted for the superior detection rates of SDT-2 over SDT-1. Our finding that guaiac testing is insensitive for detecting adenomas is consistent with previous studies in the screening setting (21, 26, 30, 31). We did not evaluate fecal immunochemical blood tests, and such tests may detect more adenomas than do guaiac tests, because of differences in relative detection rates (32–34). Although most adenomas seem not to bleed, our data suggest that many do shed molecular markers that can be detected in stool.
Technical advances could improve DNA assay sensitivity. Although we found SDT-2 markers in tissue from nearly all screen-relevant neoplasms, the assay detected the markers in stools from the same patient in fewer than one half of cases. Methods to increase stool DNA capture are needed, and some have been reported (20, 24). Preservation of intact DNA sequences in stool could also increase marker recovery. Although long DNA was the most informative panel marker in earlier studies done with minimal processing delays (12, 15, 16), we observed that long DNA degraded during shipment to the study coordinating center and contributed little to neoplasm detection. We (35) and others (36) have shown that adding a preservative to stool at the time of defecation can prevent degradation of long DNA. Finally, because DNA markers are present in such low levels (9), augmented technical sensitivity may further enhance lesion detection (37).
Test positivity among colonoscopically normal patients was significantly more frequent with SDT-2 than with Hemoccult or HemoccultSensa, probably because of the disproportionately elevated positivity rates in older patients; however, the sample size was small and the confidence intervals were relatively wide. False-positive rates for the SDT-2, Hemoccult, and HemoccultSensa tests were similar among participants younger than 65 years of age. Other studies have reported that K-ras mutations (38) and methylated vimentin (20) in stool increase with age. Accumulated DNA alterations may precede endoscopically visible lesions and account for such age-related apparent false-positives. For example, aberrant crypt foci—putative neoplasm precursors not seen by conventional colonoscopy—increase with age (39), and mutant K-ras (39, 40), mutant APC (40), and gene methylation (28, 41) have all been reported in aberrant crypt foci. Whether molecular detection of such preneoplastic field changes predicts future cancer risk is not known.
Our study differs from the only other multicenter study of stool DNA testing in average-risk patients (21) in several important ways. First, unlike the earlier report (21), we did not find that SDT-1 was better than Hemoccult for detection of invasive cancer. Second, we followed a standardized approach in a central laboratory for guaiac testing, which may account for our much higher Hemoccult sensitivity for cancer. Others have shown that central laboratory testing is more accurate than office testing after digital rectal examination (42, 43), but we do not know whether results from patient-smeared stools are more accurate when processed in a centralized laboratory rather than in a physician’s office. Our study also included the more sensitive HemoccultSensa test. Third, all pathology was reviewed centrally and neoplasm tissue was procured for DNA analyses, allowing us to measure marker recovery rates in the stool. Fourth, our participants were randomly assigned, which demonstrated that neither diet nor nonsteroidal anti-inflammatory drug use affected stool DNA results. Finally, the previous study did not evaluate SDT-2.
Our study provides robust data on the accuracy of guaiac-based occult blood tests in the screening setting. Published results vary widely according to testing algorithms and patient populations, and our Hemoccult results fall in reported ranges from studies done in the screening setting, as reviewed (44). Both Hemoccult and Hemoccult-Sensa achieved high specificity, regardless of diet, on tests performed according to the manufacturer’s instructions and in a centralized laboratory. HemoccultSensa was more sensitive than Hemoccult for all types of screen-relevant neoplasms, and HemoccultSensa detected more than 60% of cases of cancer. However, both tests missed the large majority of the adenomas that were 1 cm or larger. Such low detection rates of premalignant adenomas may account for the reported modest (45) or absent (3, 4, 46) effect of long-term Hemoccult screening on cancer incidence.
Our study has limitations. We did not test all participants or subsets with SDT-2, and those without screen-relevant neoplasm were particularly underrepresented. We used SDT-2 to test samples from all patients with cancer, high-grade dysplasias, or adenomas larger than 2 cm; 50 randomly selected samples from patients who had adenomas measuring between 1 and 2 cm; and 75 randomly selected samples from patients with normal colonoscopy results. We did not test samples from patients with small adenomas or hyperplastic polyps by using SDT-2, and their inclusion may have further increased false-positive rates. In addition, stabilization buffer was not added to stools at time of collection. Use of a stabilization buffer may have improved detection of long DNA and possibly other DNA markers as well.
In summary, stool DNA testing by SDT-1 offered no clear advantages over fecal occult blood testing for screening detection of colorectal neoplasia. Stool DNA test 2, which targets more broadly informative markers, detected significantly more screen-relevant neoplasms than did Hemoccult or HemoccultSensa, and detection was especially improved for adenomas. However, we do not know whether this gain in sensitivity is offset by a loss in specificity. Molecular technology will continue to evolve and provide opportunities for needed assay refinements in stool DNA testing.
Acknowledgments
The authors thank EXACT Sciences for performance of all genetic assays on tissue and stools and Beckman Coulter for provision of Hemoccult and HemoccultSensa cards at no cost; Sara Linker Nord for management at coordination center; Kelli Burger for statistical support; Jacalyn McCormick for secretarial support; H. Samuel Wieand, PhD (University of Pittsburgh), for statistical consulting; and Jane Milburn for assistance on protocol development. The authors also thank the physicians and study coordinators at the participating medical centers for their support (Appendix, available at www.annals.org).
Grant Support: By grants UO1 CA 89389 and UO1 CA 37404 from the National Institutes of Health.
Appendix: Physicians and Study Coordinators
Indiana University Medical Center, Indianapolis, Indiana: Beverly Ann Flamme.
North Central Cancer Treatment Group (NCCTG)-Michigan Cancer Research Consortium, Ann Arbor, Michigan: Naresh T. Gunaratnam, MD; Debra Frick; Marg van der Veen, RN; and Wendy Greer.
Kaiser Permanente Medical Care Program, Sacramento, California: Tashia Orr and Rhonda Gage.
Kaiser Permanente Medical Care Program, Oakland, California: Jo Tucker.
Mayo Clinic Rochester, Rochester, Minnesota: Tammy Neseth and Ann Kolb.
NCCTG-Wichita Community Clinical Oncology Program, Wichita, Kansas: Shaker R. Dakhil, MD, and Marge Good.
Portland Veterans Affairs Medical Center Oregon and Oregon Health & Science University, Portland, Oregon: Chad Sorenson.
Kaiser Permanente Medical Care Program, Walnut Creek, California: Patricia Leighton and Terri Coleman.
University of Arizona, Tucson, Arizona: Kelly Kaltenhauser, RN.
NCCTG-Illinois Oncology Research Association Community Clinical Oncology Program (CCOP), Peoria, Illinois: John W. Kugler, MD, and Jenny Anderson.
NCCTG-Carle Cancer Center CCOP, Urbana, Illinois: Kendrith M. Rowland, Jr., MD; Kena Hahn; Janet Iverson; and Debbie Lessmeister.
NCCTG-Sioux Community Cancer Consortium, Sioux Falls, South Dakota: Loren K. Tschetter, MD, and Gay Heidinger.
NCCTG-Upstate Carolina CCOP, Spartanburg, South Carolina: Kathy Queen, RN, and Pam Bishop.
NCCTG-Meritcare Hospital CCOP, Fargo, North Dakota: Preston D. Steen MD, and Angela Nordstrom.
NCCTG-Siouxland Hematology-Oncology Associates, Sioux City, Iowa: Donald B. Wender, MD, and Sue Legree.
NCCTG-Geisinger Clinic & Medical Center CCOP, Danville, Pennsylvania: Albert M. Bernath, Jr., MD, and Deb Gris-wold.
NCCTG-Toledo Community Hospital Oncology Program CCOP, Toledo, Ohio: Paul L. Schaefer, MD, and Mike Uscio.
NCCTG-Rapid City Regional Oncology Group, Rapid City, South Dakota: Richard C. Tenglin, MD, and Deb Tadlock.
NCCTG-Ochsner CCOP, New Orleans, Louisiana: Carl G. Kardinal, MD, and Kate Rodger.
NCCTG-CentraCare Clinic, St. Cloud, Minnesota: Harold E. Windschitl, MD, and Cheryl Kelley.
NCCTG-Iowa Oncology Research Association CCOP, Des Moines, Iowa: Roscoe F. Morton, MD; Mary Loots, RN; and Janet Mannetter, RN.
Footnotes
Author Contributions: Conception and design: D.A. Ahlquist, D.J. Sargent, C.L. Loprinzi, T.R. Levin, K. Knigge, M.P. Lance, J.E. Allison, M.E. Devens, S.L. Hillman.
Analysis and interpretation of the data: D.A. Ahlquist, D.J. Sargent, D.K. Rex, D.J. Ahnen, K. Knigge, M.P. Lance, L.J. Burgart, S.R. Hamilton, J.E. Allison, J.J. Harrington, S.L. Hillman.
Drafting of the article: D.A. Ahlquist, D.J. Sargent.
Critical revision of the article for important intellectual content: D.A. Ahlquist, D.J. Sargent, C.L. Loprinzi, T.R. Levin, D.K. Rex, D.J. Ahnen, M.P. Lance, J.E. Allison, M.J. Lawson.
Final approval of the article: D.A. Ahlquist, D.J. Sargent, T.R. Levin, D.K. Rex, D.J. Ahnen, M.P. Lance, S.R. Hamilton, J.E. Allison, M.J. Lawson.
Provision of study materials or patients: D.A. Ahlquist, D.J. Sargent, T.R. Levin, D.K. Rex, D.J. Ahnen, M.P. Lance, M.E. Devens.
Statistical expertise: D.J. Sargent, S.L. Hillman.
Obtaining of funding: D.A. Ahlquist, D.J. Sargent, C.L. Loprinzi, M.P. Lance, M.E. Devens.
Administrative, technical, or logistic support: D.A. Ahlquist, D.J. Sargent, C.L. Loprinzi, D.K. Rex, D.J. Ahnen, K. Knigge, S.R. Hamilton, M.E. Devens, J.J. Harrington, S.L. Hillman.
Collection and assembly of data: D.J. Sargent, T.R. Levin, D.K. Rex, D.J. Ahnen, K. Knigge, M.P. Lance, L.J. Burgart, M.J. Lawson, M.E. Devens, J.J. Harrington, S.L. Hillman.
Potential Financial Conflicts of Interest: Consultancies: D.J. Ahnen (EXACT Sciences), J.E. Allison (Fujirebio, Quidel). Stock ownership or options (other than mutual funds): J.E. Allison (IntelligeneScan). Grants received: T.R. Levin (EXACT Sciences), M.P. Lance (National Institutes of Health), M.J. Lawson (EXACT Sciences). The Mayo Clinic is a minor equity investor in EXACT Sciences.
Reproducible Research Statement: Study protocol, statistical code, and data set: Available from Ms. Devens (devens@mayo.edu).
Current author addresses and author contributions are available at www.annals.org.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Mandel JS, Bond JH, Church TR, Snover DC, Bradley GM, Schuman LM, et al. Reducing mortality from colorectal cancer by screening for fecal occult blood. Minnesota Colon Cancer Control Study. N Engl J Med. 1993;328:1365–71. doi: 10.1056/NEJM199305133281901. [DOI] [PubMed] [Google Scholar]
- 3.Hardcastle JD, Chamberlain JO, Robinson MH, Moss SM, Amar SS, Balfour TW, et al. Randomised controlled trial of faecal-occult-blood screening for colorectal cancer. Lancet. 1996;348:1472–7. doi: 10.1016/S0140-6736(96)03386-7. [DOI] [PubMed] [Google Scholar]
- 4.Kronborg O, Jørgensen OD, Fenger C, Rasmussen M. Randomized study of biennial screening with a faecal occult blood test: results after nine screening rounds. Scand J Gastroenterol. 2004;39:846–51. doi: 10.1080/00365520410003182. [DOI] [PubMed] [Google Scholar]
- 5.Winawer SJ, Flehinger BJ, Schottenfeld D, Miller DG. Screening for colorectal cancer with fecal occult blood testing and sigmoidoscopy. J Natl Cancer Inst. 1993;85:1311–8. doi: 10.1093/jnci/85.16.1311. [DOI] [PubMed] [Google Scholar]
- 6.Singh H, Turner D, Xue L, Targownik LE, Bernstein CN. Risk of developing colorectal cancer following a negative colonoscopy examination: evidence for a 10-year interval between colonoscopies. JAMA. 2006;295:2366–73. doi: 10.1001/jama.295.20.2366. [DOI] [PubMed] [Google Scholar]
- 7.Heresbach D, Manfredi S, D’halluin PN, Bretagne JF, Branger B. Review in depth and meta-analysis of controlled trials on colorectal cancer screening by faecal occult blood test. Eur J Gastroenterol Hepatol. 2006;18:427–33. doi: 10.1097/00042737-200604000-00018. [DOI] [PubMed] [Google Scholar]
- 8.Meissner HI, Breen N, Klabunde CN, Vernon SW. Patterns of colorectal cancer screening uptake among men and women in the United States. Cancer Epidemiol Biomarkers Prev. 2006;15:389–94. doi: 10.1158/1055-9965.EPI-05-0678. [DOI] [PubMed] [Google Scholar]
- 9.Osborn NK, Ahlquist DA. Stool screening for colorectal cancer: molecular approaches. Gastroenterology. 2005;128:192–206. doi: 10.1053/j.gastro.2004.10.041. [DOI] [PubMed] [Google Scholar]
- 10.Davies RJ, Miller R, Coleman N. Colorectal cancer screening: prospects for molecular stool analysis. Nat Rev Cancer. 2005;5:199–209. doi: 10.1038/nrc1569. [DOI] [PubMed] [Google Scholar]
- 11.Levin B, Lieberman DA, McFarland B, Smith RA, Brooks D, Andrews KS, et al. American Cancer Society Colorectal Cancer Advisory Group. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA Cancer J Clin. 2008;58:130–60. doi: 10.3322/CA.2007.0018. [DOI] [PubMed] [Google Scholar]
- 12.Ahlquist DA, Skoletsky JE, Boynton KA, Harrington JJ, Mahoney DW, Pierceall WE, et al. Colorectal cancer screening by detection of altered human DNA in stool: feasibility of a multitarget assay panel. Gastroenterology. 2000;119:1219–27. doi: 10.1053/gast.2000.19580. [DOI] [PubMed] [Google Scholar]
- 13.Dong SM, Traverso G, Johnson C, Geng L, Favis R, Boynton K, et al. Detecting colorectal cancer in stool with the use of multiple genetic targets. J Natl Cancer Inst. 2001;93:858–65. doi: 10.1093/jnci/93.11.858. [DOI] [PubMed] [Google Scholar]
- 14.Traverso G, Shuber A, Levin B, Johnson C, Olsson L, Schoetz DJ, Jr, et al. Detection of APC mutations in fecal DNA from patients with colorectal tumors. N Engl J Med. 2002;346:311–20. doi: 10.1056/NEJMoa012294. [DOI] [PubMed] [Google Scholar]
- 15.Tagore KS, Lawson MJ, Yucaitis JA, Gage R, Orr T, Shuber AP, et al. Sensitivity and specificity of a stool DNA multitarget assay panel for the detection of advanced colorectal neoplasia. Clin Colorectal Cancer. 2003;3:47–53. doi: 10.3816/CCC.2003.n.011. [DOI] [PubMed] [Google Scholar]
- 16.Calistri D, Rengucci C, Bocchini R, Saragoni L, Zoli W, Amadori D. Fecal multiple molecular tests to detect colorectal cancer in stool. Clin Gastroenterol Hepatol. 2003;1:377–83. doi: 10.1053/s1542-3565(03)00186-1. [DOI] [PubMed] [Google Scholar]
- 17.Rengucci C, Maiolo P, Saragoni L, Zoli W, Amadori D, Calistri D. Multiple detection of genetic alterations in tumors and stool. Clin Cancer Res. 2001;7:590–3. [PubMed] [Google Scholar]
- 18.Koshiji M, Yonekura Y, Saito T, Yoshioka K. Microsatellite analysis of fecal DNA for colorectal cancer detection. J Surg Oncol. 2002;80:34–40. doi: 10.1002/jso.10096. [DOI] [PubMed] [Google Scholar]
- 19.Leung WK, To KF, Man EP, Chan MW, Hui AJ, Ng SS, et al. Detection of hypermethylated DNA or cyclooxygenase-2 messenger RNA in fecal samples of patients with colorectal cancer or polyps. Am J Gastroenterol. 2007;102:1070–6. doi: 10.1111/j.1572-0241.2007.01108.x. [DOI] [PubMed] [Google Scholar]
- 20.Itzkowitz SH, Jandorf L, Brand R, Rabeneck L, Schroy PC, 3rd, Sontag S, et al. Improved fecal DNA test for colorectal cancer screening. Clin Gastroenterol Hepatol. 2007;5:111–7. doi: 10.1016/j.cgh.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME Colorectal Cancer Study Group. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704–14. doi: 10.1056/NEJMoa033403. [DOI] [PubMed] [Google Scholar]
- 22.Ahlquist DA, McGill DB, Fleming JL, Schwartz S, Wieand HS, Rubin J, et al. Patterns of occult bleeding in asymptomatic colorectal cancer. Cancer. 1989;63:1826–30. doi: 10.1002/1097-0142(19900501)63:9<1826::aid-cncr2820630928>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 23.Ahlquist DA. Molecular stool screening for colorectal cancer. Using DNA markers may be beneficial, but large scale evaluation is needed [Editorial] BMJ. 2000;321:254–5. doi: 10.1136/bmj.321.7256.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitney D, Skoletsky J, Moore K, Boynton K, Kann L, Brand R, et al. Enhanced retrieval of DNA from human fecal samples results in improved performance of colorectal cancer screening test. J Mol Diagn. 2004;6:386–95. doi: 10.1016/S1525-1578(10)60536-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kann L, Han J, Ahlquist D, Levin T, Rex D, Whitney D, et al. Improved marker combination for detection of de novo genetic variation and aberrant DNA in colorectal neoplasia. Clin Chem. 2006;52:2299–302. doi: 10.1373/clinchem.2007.070896. [DOI] [PubMed] [Google Scholar]
- 26.Ahlquist DA, Wieand HS, Moertel CG, McGill DB, Loprinzi CL, O’Connell MJ, et al. Accuracy of fecal occult blood screening for colorectal neoplasia. A prospective study using Hemoccult and HemoQuant tests. JAMA. 1993;269:1262–7. [PubMed] [Google Scholar]
- 27.Herzog P, Holtermüller KH, Preiss J, Fischer J, Ewe K, Schreiber HJ, et al. Fecal blood loss in patients with colonic polyps: a comparison of measurements with 51chromium-labeled erythrocytes and with the Haemoccult test. Gastroenterology. 1982;83:957–62. [PubMed] [Google Scholar]
- 28.Chen WD, Han ZJ, Skoletsky J, Olson J, Sah J, Myeroff L, et al. Detection in fecal DNA of colon cancer-specific methylation of the nonexpressed vimentin gene. J Natl Cancer Inst. 2005;97:1124–32. doi: 10.1093/jnci/dji204. [DOI] [PubMed] [Google Scholar]
- 29.Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, et al. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359:235–7. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 30.Lieberman DA, Weiss DG. Veterans Affairs Cooperative Study Group 380. One-time screening for colorectal cancer with combined fecal occult-blood testing and examination of the distal colon. N Engl J Med. 2001;345:555–60. doi: 10.1056/NEJMoa010328. [DOI] [PubMed] [Google Scholar]
- 31.Robinson MH, Kronborg O, Williams CB, Bostock K, Rooney PS, Hunt LM, et al. Faecal occult blood testing and colonoscopy in the surveillance of subjects at high risk of colorectal neoplasia. Br J Surg. 1995;82:318–20. doi: 10.1002/bjs.1800820310. [DOI] [PubMed] [Google Scholar]
- 32.Smith A, Young GP, Cole SR, Bampton P. Comparison of a brush-sampling fecal immunochemical test for hemoglobin with a sensitive guaiac-based fecal occult blood test in detection of colorectal neoplasia. Cancer. 2006;107:2152–9. doi: 10.1002/cncr.22230. [DOI] [PubMed] [Google Scholar]
- 33.Morikawa T, Kato J, Yamaji Y, Wada R, Mitsushima T, Shiratori Y. A comparison of the immunochemical fecal occult blood test and total colonoscopy in the asymptomatic population. Gastroenterology. 2005;129:422–8. doi: 10.1016/j.gastro.2005.05.056. [DOI] [PubMed] [Google Scholar]
- 34.Allison JE, Sakoda LC, Levin TR, Tucker JP, Tekawa IS, Cuff T, et al. Screening for colorectal neoplasms with new fecal occult blood tests: update on performance characteristics. J Natl Cancer Inst. 2007;99:1462–70. doi: 10.1093/jnci/djm150. [DOI] [PubMed] [Google Scholar]
- 35.Zou H, Harrington JJ, Klatt KK, Ahlquist DA. A sensitive method to quantify human long DNA in stool: relevance to colorectal cancer screening. Cancer Epidemiol Biomarkers Prev. 2006;15:1115–9. doi: 10.1158/1055-9965.EPI-05-0992. [DOI] [PubMed] [Google Scholar]
- 36.Olson J, Whitney DH, Durkee K, Shuber AP. DNA stabilization is critical for maximizing performance of fecal DNA-based colorectal cancer tests. Diagn Mol Pathol. 2005;14:183–91. doi: 10.1097/01.pas.0000176768.18423.7e. [DOI] [PubMed] [Google Scholar]
- 37.Zou H, Harrington J, Jiang X, Loprinzi CL, Levin TR, Rex D, et al. Stool DNA screening of colorectal neoplasia: improved adenoma detection by novel digital melt curve assay [Abstract] Gastroenterology. 2008;134:A-109. [Google Scholar]
- 38.Chien CC, Chen SH, Liu CC, Lee CL, Yang RN, Yang SH, et al. Correlation of K-ras codon 12 mutations in human feces and ages of patients with colorectal cancer (CRC) Transl Res. 2007;149:96–102. doi: 10.1016/j.trsl.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 39.Takayama T, Katsuki S, Takahashi Y, Ohi M, Nojiri S, Sakamaki S, et al. Aberrant crypt foci of the colon as precursors of adenoma and cancer. N Engl J Med. 1998;339:1277–84. doi: 10.1056/NEJM199810293391803. [DOI] [PubMed] [Google Scholar]
- 40.Smith AJ, Stern HS, Penner M, Hay K, Mitri A, Bapat BV, et al. Somatic APC and K-ras codon 12 mutations in aberrant crypt foci from human colons. Cancer Res. 1994;54:5527–30. [PubMed] [Google Scholar]
- 41.Derks S, Postma C, Moerkerk PT, van den Bosch SM, Carvalho B, Hermsen MA, et al. Promoter methylation precedes chromosomal alterations in colorectal cancer development. Cell Oncol. 2006;28:247–57. doi: 10.1155/2006/846251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collins JF, Lieberman DA, Durbin TE, Weiss DG Veterans Affairs Cooperative Study 380 Group. Accuracy of screening for fecal occult blood on a single stool sample obtained by digital rectal examination: a comparison with recommended sampling practice. Ann Intern Med. 2005;142:81–5. doi: 10.7326/0003-4819-142-2-200501180-00006. [DOI] [PubMed] [Google Scholar]
- 43.Nakama H, Zhang B, Abdul Fattah AS, Kamijo N. Does stool collection method affect outcomes in immunochemical fecal occult blood testing? Dis Colon Rectum. 2001;44:871–5. doi: 10.1007/BF02234712. [DOI] [PubMed] [Google Scholar]
- 44.Ahlquist D. Approach to the patient with occult gastrointestinal bleeding. In: Yamada T, editor. Textbook of Gastroenterology. Philadelphia: Lippincott Williams & Wilkins; 2003. pp. 724–38. [Google Scholar]
- 45.Mandel JS, Church TR, Bond JH, Ederer F, Geisser MS, Mongin SJ, et al. The effect of fecal occult-blood screening on the incidence of colorectal cancer. N Engl J Med. 2000;343:1603–7. doi: 10.1056/NEJM200011303432203. [DOI] [PubMed] [Google Scholar]
- 46.Kronborg O, Fenger C, Olsen J, Jørgensen OD, Søndergaard O. Randomised study of screening for colorectal cancer with faecal-occult-blood test. Lancet. 1996;348:1467–71. doi: 10.1016/S0140-6736(96)03430-7. [DOI] [PubMed] [Google Scholar]