Summary
Background
Autophagy is a fundamental cell biological process whereby eukaryotic cells form membranes in the cytoplasm to sequester diverse intracellular targets. Although significant progress has been made in understanding the origins of autophagosomal organelles, the source of lipids that support autophagic membrane formation remain an important open question.
Results
Here we show that lipid droplets as cellular stores of neutral lipids including triglycerides contribute to autophagic initiation. Lipid droplets, as previously shown, were consumed upon induction of autophagy by starvation. However, inhibition of autophagic maturation by blocking acidification or using dominant negative Atg4C74A that prohibits autophagosomal closure, did not prevent disappearance of lipid droplets. Thus, lipid droplets continued to be utilized upon induction of autophagy but not as autophagic substrates in a process referred to as lipophagy. We considered an alternative model whereby lipid droplets were consumed not as a part of lipophagy but as a potential contributing source to the biogenesis of lipid precursors for nascent autophagosomes. We carried out a screen for a potential link between triglyceride mobilization and autophagy, and identified a neutral lipase, PNPLA5, as being required for efficient autophagy. PNPLA5, which localized to lipid droplets, was needed for optimal initiation of autophagy. PNPLA5 was required for autophagy of diverse substrates including degradation of autophagic adaptors, bulk proteolysis, mitochondrial quantity control, and microbial clearance.
Conclusions
Lipid droplets contribute to autophagic capacity by enhancing it in a process dependent on PNPLA5. Thus, neutral lipid stores are mobilized during autophagy to support autophagic membrane formation.
Introduction
The sensu stricto autophagy (macroautophagy) is a fundamental biological process [1]. Dysfunctional autophagy has been linked to human pathologies in aging, cancer, neurodegeneration, and inflammatory diseases [2]. Autophagy degrades bulk cytosol during starvation, and removes toxic protein aggregates, intracellular pathogens, and surplus or damaged organelles such as depolarized mitochondria or lipid droplets (LD) [1–3].
The committed step in mammalian autophagy is the induction and nucleation of a membranous precursor called phagophore that expands and closes into an emblematic structure, the double membrane autophagosome [1]. Autophagosomes sequester portions of the cytoplasm or specific targets and fuse with lysosomes to digest the captured cargo [1]. Each of these steps involves the hierarchical activity of Atg (Autophagy-related) proteins [1]. The formation of the phagophore requires mammalian orthologs of Atg1 (Ulk1/2) [1] and the class III phosphoinositide 3-kinase (PI3K) Vps34 complexed with the mammalian ortholog of Atg6 (Beclin-1) to generate phosphatidylinositol 3-phosphate (PI3P) [4]. A PI3P-binding protein, DFCP1, marks omegasome structures derived from the ER that serve as precursors [5] to the ER-derived autophagosomes [6, 7]. PI3P also recruits mammalian orthologs of Atg18 (WIPI1 and WIPI2) contributing to the subsequent steps in phagophore formation [8, 9].
The source of the autophagosome membrane remains an important question in the field of autophagy [10–12]. Several compartments, including the endoplasmic reticulum (ER) [6, 7], mitochondria-ER contact sites [11], Golgi apparatus [13], and the plasma membrane via recycling endosome [12], have been implicated as contributing sources to autophagosomal membranes. Given that autophagy is a process requiring intense membrane remodeling and consumption, and thus could impinge on the functionality of the organelles proposed to be the membrane sources, we wondered whether the cell may utilize relatively inactive lipid reserves such as its neutral lipid stores to supplement autophagosomal membrane formation.
LDs are dynamic organelles that represent a major depot of cellular neutral lipids such as triglycerides (TG) [14–16]. In addition to their role as substrates for lipophagy [3], the process known as autophagic degradation of LDs, we considered an alternative possibility that LDs might serve as organelles whereby TG stores could be mobilized into phospholipids necessary for autophagosomal membrane formation and growth. We screened TG mobilizing enzymes for their capacity to affect autophagic pathway and found that PNPLA5, a TG lipase [17], was needed for optimal autophagy initiation. We show that LDs and TGs via PNPLA5 contribute lipid intermediates facilitating autophagosome membrane biogenesis.
Results
Increase in cellular lipid droplet content increases autophagic capacity
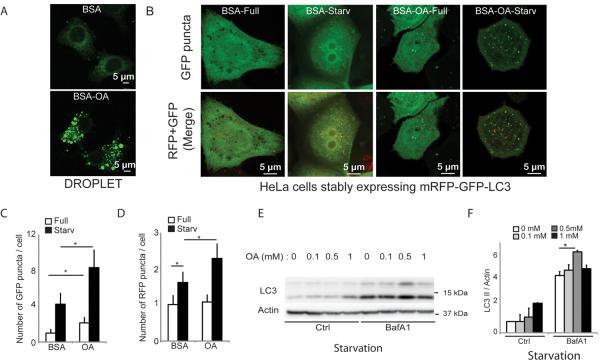
Addition of oleic acid (OA) increases cellular LD content [14–16, 18]. Increase in LDs, following OA pretreatment (Figure 1A), enhanced basal and induced autophagy, as determined by high content imaging analysis of autophagic organelles in HeLa cells stably expressing mRFP-GFP-LC3 (Figure 1B–D and Figure S1A). The enhancement affected both autophagy initiation stage (Figure 1C; GFP-LC3) and maturation (Figure 1D; mRFP-GFP), as determined by the acidification-sensitive GFP and acidification-insenstive mRFP probes [19]. This was confirmed by LC3-II conversion immunoblots (Figure 1E,F and Figure S1B,C). Titration experiments indicated that 500 μM OA was optimal for enhancing starvation-induced autophagy. Further increase (1 mM OA) caused a plateau or inhibition (Figure 1E,F), in keeping with reports that higher concentrations of OA can inhibit autophagic progression [20]. In subsequent experiments, 500 μM OA was used. The standard bafilomycin A1 inhibition assay [19] also showed (Figure 1F) that pretreatment with OA enhanced cellular capacity for initiation of starvation-induced autophagy.
Figure 1. Preformed lipid droplets enhance starvation-induced autophagy.
(A) and (B) HeLa cells stably expressing mRFP-GFP-LC3 were treated for 20 h with BSA alone (BSA) or with BSA-oleic acid (OA; 500 μM OA) and starved (Starv) in EBSS for 90 min or incubated in full medium (Full). (A) Visualization of lipid droplet accumulation. Lipid droplets were stained with Bodipy 493/503. (B) Confocal images of samples (for images in high content acquisition mode see Figure S1). (C) and (D) Number of GFP+ puncta (C) and RFP+ puncta (D) per cell were quantified by high content image acquisition and analysis. Data: means ± s.e. (n=3, where n represents separate experiments; each experimental point in separate experiments contained >500 cells identified by the program as valid primary objects); *, p<0.05 (t-test). (E,F) HeLa cells treated with OA as in A–D with or without bafilomycin A1 (BafA1); LC3-II/actin ratios determined by immunoblotting (E) and densitometry (F). Immunobloting data: means ± s.e., *, p<0.05 (t-test).
Live imaging and ultrastuctural analyses reveal interactions between autophagosomes and LD organelles
Live imaging of HeLa cells expressing GFP-LC3 (Movie S1, Figure 2A,B) indicated multiple interactions between LDs and LC3-positive structures. The profiles featured in the stills (Figure 2B) show an LC3 structure approaching an LD, contacting it, gyrating around it, separating, and then coming into repeat contacts with the LD. Similar events have been observed by intravital imaging of mouse liver [21] indicated vigorous interactions between LDs and LC3-positive autophagosomes in whole organs in live animal (Movie S2; Figure 2C,D). These LD-autophagosome interactions fell under two categories: short range and long range (Movie S2). The associations were transient in nature and appeared as “kiss-and-run” events between the two organelles.
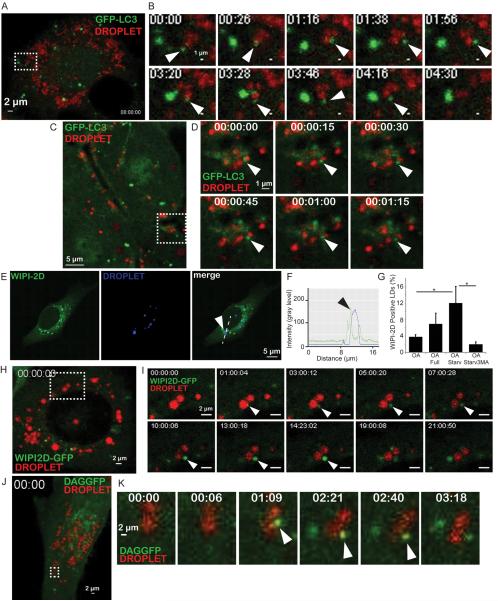
Figure. 2. Imaging analysis of dynamic interactions between lipid droplets and LC3, WIPI2D and a diacylglycerol GFP probes.
(A,B) Still frames from time lapse imaging of GFP-LC3 expressing HeLa cells (Movie S1). HeLa cells expressing GFP-LC3 (green) treated for 20 h with 500 μM OA (BSA-oleic acid), LDs (red) labeled with LipidTox Farred, and live imaging initiated in starvation (EBSS) medium. Time, min:sec. Rectangle in A, area in time-lapse still frames in B. Arrowheads, a GFP-LC3 positive structure (green) interacting with a lipid droplet (red). (C,D) Still frames from intravital imaging of intact liver from GFP-LC3 mice (Movie S2). Time, h:min:sec.. Arrowheads, a GFP-LC3 positive structure (green) interacting with a lipid droplet (red). (E–G) Confocal microscopy analysis of U2OS cells stably expressing GFP WIPI-2D. Lipid droplets were visualized (blue channel) with LipidTox DeepRed. Cells were treated as in A, starved for 2 h and then fixed. Arrowheads, recruitment of WIPI2D to lipid droplets. (F) Two-fluorescence channel line tracings corresponding to dashed lines in images to the left. (G) WIPI-2D positive LDs were quantified by Image J. Cells were pre-treated for 20 h with 500 μM BSA-oleic acid (OA) then starved (OA Starv) or not (OA Full) for 2 h in the absence or presence of 3-MA treatment (OA Starv3MA). Data means ± s.e. (n≥3); *, p<0.05. (H,I) Still frames from live imaging of U2OS cells stably expressing GFP-WIPI2D (Movies S3 and S4). Cells were treated as in A. Time, h:min:sec. Rectangle in H, area in still frames in I. Arrowheads, an emerging GFP-WIPI2D positive (green) structure juxtaposed to an LD, enlarging in size with time. (J,K) Still frames from live imaging of GFP-DAG expressing HeLa cells (Movies S5 and S6). HeLa cells expressing NES-GFP-DAG (green) were treated as in A. Time, min:sec. Rectangle in J, still frames in K. Arrowheads, a GFP-DAG positive (green) structure emerging juxtaposed to LD.
Among several tested early autophagic factors (WIPIs, DFCP1, and ATG16L1 Figure 2 and Figure S2), using fixed cell imaging and biochemical analyses, the mammalian Atg18 orthologs WIPI-1, WIPI-2B (Figure S2A–F) and WIPI-2D (Figure 2E–G)[8, 22] associated with LDs in a manner sensitive to 3-methyl adenine 3MA (Figure S2A–I). Using WIPI-2D-GFP U2OS stable cell line, we observed by live imaging (Movie S3 and S4; Figure 2H,I) the emergence of WIPI2D positive structures on LDs; these WIPI2D profiles grew during autophagic induction by starvation. A time sequence of the growth of a profile of this type is shown in consecutive still frames in Figure 2I. These data suggest that early autophagic structures can grow from LDs.
In keeping with the above findings, we observed in ultrastructural analyses different types of relationships between LDs and autophagic organelles (Figure 3 and Table S1). Examples of phagophores (isolation membranes) in proximity of LDs are shown in Figure 3A–D. A nascent phagophore in contact with an LD via a membrane bridge is shown in Figure 3A,B. In addition, we observed in some images LDs within the lumen of larger autophagosomes with mixed cytoplasmic contents (Figure 3C and D). A quantification of these profiles is shown in Table S1.
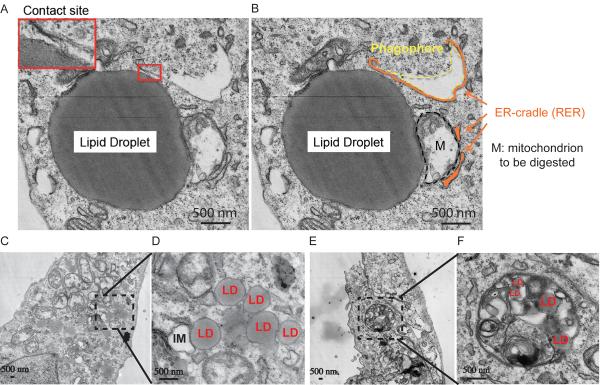
Figure. 3. Ultrastructural analysis of associations between lipid droplets and phagophores.
(A–F) U2OS cells were treated for 24 h with 500 μM BSA-oleic acid and processed for electron microscopy (cumulative analysis in Table S1). (A,D) Examples of autophagic phagophores (isolation membranes) in the proximity of LDs. M, mitochondrion; RER, rough endoplasmic reticulum. Enlarged detail in A, membranous anastomosis between a lipid droplet and a phagophore. (E,F) Examples of LDs, including LDs as substrate for autophagy (lipophagy).
In summary, LDs and early autophagic organelles show a number of interactions that can be intermittent and short-lived, consistent with the possibility that isolation membranes form in the vicinity of autophagosomes. This is further supported by the presence of membrane tethers. In other instances, the ultrastructural images show that LDs can also be a substrate for autophagy consistent with the process termed lipophagy (Figure 3E and F).
Consumption of LDs continues when either autophagic maturation or autophagosome closure are blocked
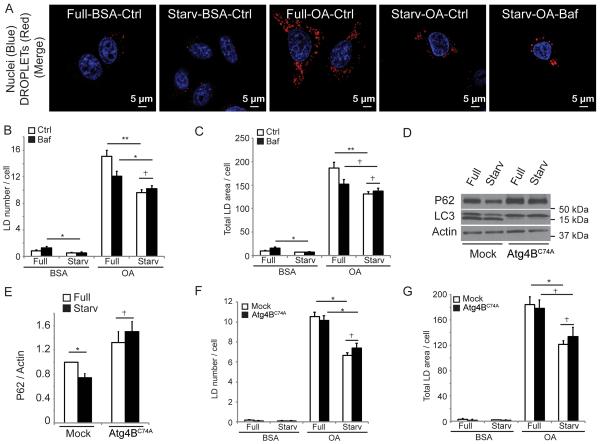
The above observations could be interpreted as LDs being en route for lipophagy or that autophagosomes are being formed in the vicinity or from LDs. To address this, autophagic maturation was blocked in cultured cells with bafilomycin A1. Under these conditions, LDs continued to be consumed during starvation, as quantified by high content automated imaging and analysis of LD numbers and total area (Figures 4A–C and S3A). This was further investigated using 3T3 cells stably expressing Atg4BC74A [23] a dominant negative form of Atg4B which prohibits closure of nascent autophagosomes. We first established that these cells did not support degradation of p62/sequestosome-1, often used as a readout for autophagic degradation (Figure 4D,E). However, Atg4BC74A did permit continuing LD utilization during starvation (Figure 4F,G). Thus, LDs are consumed during starvation-induced autophagy in processes other than autophagic degradation. LD consumption nevertheless depended on early regulators of autophagy initiation, Ulk1 and Ulk2, since LD content was not reduced during starvation in double Ulk1/Ulk2 knockout mouse embryonic fibroblasts (MEFs) (Figure S3B).
Figure. 4. Lipid droplets are consumed during autophagic induction independently of autophagosomal closure and autophagic maturation.
(A) Confocal microscopy image of HeLa cells treated as in Figs 1 and 2 with or without Bafilomycin A1 (Baf) to inhibit autophagic degradation. Cells were stained with Hoechst 33342 (nuclei; blue) and Bodipy 493/503 (lipid droplets; red). Confocal images illustrate high content analysis in (B,C) carried out in epifluoresce mode (Figure S5A). Lipid droplets (LD) number (B) and total LD area (C) were quantified by high content image acquisition and analysis. (D,E) Stable 3T3 cells expressing ATG4B or mStrawberry-ATG4BC74A were treated as in A. p62/actin ratios were determined by immunoblotting (D) and densitometry (E). Immunoblot data, means ± s.e., (n≥3); *, p<0.05. (F,G) Stable 3T3 cells expressing ATG4B or mStrawberry-ATG4BC74A were treated, lipid droplets stained as in A, and analyzed as in B and C. All high content analysis data, means ± s.e (n=3, where n represents separate experiments; each experimental point in separate experiments contained >500 cells identified by the program as valid primary objects); *, p<0.05 †, p≥0.05 (t-test).
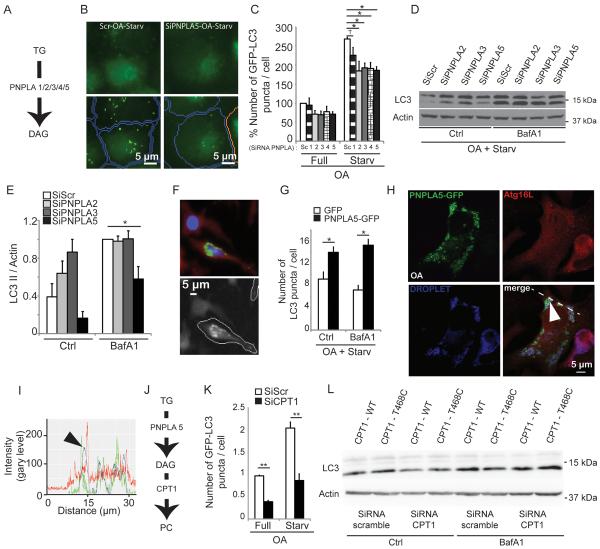
Screen for TG-mobilizing enzymes identifies PNPLA5 as a positive regulator of autophagy
We wondered whether continuing LD consumption was due to a TG turnover in LDs independently of lipophagy. A potential intermediate, diacylglycerol (DAG) formed by lipase action upon TGs, could be used to build phospholipids necessary for autophagosomal membrane formation and growth. We first asked whether TG metabolism enzymes affected autophagy and screened TG-mobilization enzymes, represented by the patatin-like phospholipase domain containing proteins, PNPLA1–5 (Figure S4A and Figure 5A). Autophagy was assessed by high content imaging analysis (Figure 5B). Knockdowns of PNPLAs with the exception of PNPLA1, reduced the numbers and the area of GFP-LC3 puncta under starvation conditions in OA-treated cells (Figure 5C, Figure S4B,C).
Figure 5. Screen for triglyceride metabolism factors identifies PNPLA5 and CPT1 as positive regulators of autophagy.
(A) Triglyceride (TG) catabolic pathway (lipolysis), PNPLAs (1/2/3/4/5), PNPLAs, patatin-like phospholipase domain-containing proteins 1 through 5. (B,C) HeLa cells stably expressing mRFP-GFP-LC3 were transfected twice with scrambled (Scr) control siRNA or siRNAs against PNPLAs. Cells were treated as in Fig. 1. (C) high content image acquisition and analysis. (D,E) Effect of PNPLAs on autophagy induction by measuring LC3-II levels by immunoblotting and densitometry. HeLa cells transfected twice with siRNAs (PNPLA 2,3,5) or scrambled (Scr) were treated as in A with or without Bafilomycin A1 (Baf). Immunoblotting data, means ± s.e. (n≥3); *, p<0.05. (F,G) PNPLA5 overexpression effects on endogenous LC3 puncta. HeLa cells were transfected with GFP or PNPLA5-GFP, treated 20 h with 500 μM BSA-Oleic Acid (OA) and starved for 2 h with or without Bafilomycin A1 (Baf). Endogenous LC3 was stained by immunofluorescence and LC3 dots were quantified within GFP positive cells (as illustrated in fluorescent images in F) by high content image acquisition and analysis in G. (H,I) Confocal microscopy of HeLa cells transfected with PNPLA5-GFP expression plasmid (green cell), ATG16L1 (red) and lipid droplets (LD, LipidTox DeepRed, blue channel). Cells were transfected with PNPLA5-GFP expressing plasmid, treated for 20 h with 500 μM BSA-Oleic Acid (OA), fixed, and lipid droplets stained with LipidTox DeepRed and immunostained for ATG16L1. Arrowhead, colocalization of PNPLA5GFP, ATG16L1 on lipid droplets; dashed line, two-fluorescence channel line tracing shown in panel I. (J) Scheme, enzymes involved in the phospholipid synthesis pathway. (K) HeLa cells stably expressing mRFP-GFP-LC3 were transfected twice with scramble control (Scr) or CPT1 siRNAs. After 24 h, cells were treated for 20 h with 500 μM BSA-Oleic Acid (OA) and starved for 90 min. GFP+ puncta per cell were quantified by high content image acquisition and analysis. (L) Effect of CPT1 on autophagy induction by determininig LC3-II levels. HeLa cells were transfected with siRNAs against CPT1 or scrambled (Scr) control. After 24 h, cells were cotransfected with siRNAs against CPT1 or scrambled (Scr) control and with plasmids expressing wild-type (WT) CPT1-mCherry or mutant CPT1-mCherry T468C (SiRNA resistant construct). Cells were then treated 20 h with 500 μM BSA-Oleic Acid (OA) and starved for 2 h with or without Bafilomycin A1 (Baf) and subjected to immunoblot analysis (see Supplementary Figure S7H for LC3-II/actin ratios determined by densitometry).
Next, we focused on PNPLAs that yielded an LC3 puncta phenotype (Figure 5C). The PNPLA family members have the following key properties. They contain a conserved lipase catalytic dyad (G-X-S-X-G) in the patatin domain (Figure S4A), [24, 25]. Unlike other PNPLA members, PNPLA1 has no apparent TG-lipase activity and has been suggested to act in phospholipid metabolism instead of TG mobilization [26]. PNPLA2 (ATGL, adipose triglyceride lipase, also known under the name Desnutrin) is the best-studied TG-converting lipase responsible for most of TG hydrolysis in murine white adipose tissue [27], PNPLA3, also known as Adiponutrin can act as an acyltransferase [28] or a lipase [17] but its biosynthetic acyltransferase activity is being the presumed dominant function [28]. We excluded PNPLA4 from further studies since PNPLA4 lacks C-terminal residues found in all other family members and does not localize to LDs [29]. PNPLA5 possess a lipase activity against TGs [17] but PNPLA5 shows some differences in its active site vs. PNPLA2 and 3, suggesting further sub-specialization. Using LC3-II immunoblot assays in presence of Bafilomycin A1 we found that only a PNPLA5 knock-down inhibited LC3-II conversion (Figure 5D,E). We then verified that PNPLA5 affected autophagy initiation by overexpressing PNPLA5-GFP construct in HeLa cells (Figure 5F,G). High content imaging analysis gated on GFP-positive cells (Figure 5F), showed in PNPLA5-expressing cells increased (endogenous) LC3 puncta numbers (Figure 5G) and area (Figure S4D). In keeping with this newly uncovered role of PNPLA5 in autophagy initiation, PNPLA5 colocalized with ATG16L1 on LDs (Figure 5H,I).
CPT1 and LPCAT2 are both positive regulator of autophagy
If the products of PNPLA5 lipase action upon TGs [17] are used to build phospholipids (e.g. following enzymatic conversion of TGs to DAG) for autophagosomal membranes, we reasoned that the de novo synthesis of phosphatidylcholine (PC) or phosphoethanolamine (PE) may be needed to convert DAG to phospholipids during induction of autophagy. We focused on PC. Two major biochemical pathways contribute to the synthesis of PC: the Kennedy pathway for de novo PC synthesis with the participation of cholinephosphotransferase (CPT1, also known as CHPT1; Figure 5J) [30] and the Lands cycle for remodeling of the fatty acid composition of PC species through the concerted actions of phospholipase A2 (PLA2s) and lysophosphatidylcholine acyltransferases (LPCAT1 and 2; Figure S5A) [31]. Thus, we tested whether CPT1 and LPCAT1/2 affected autophagy by high content imaging analysis of LC3 puncta (Figures 5K, S4E–H, and S5B,D). Knockdowns of CPT1 (Figure S4G; a knockdown of CPT1, similarly to knockdown of PNPLA5, did not affect cell viability Figure S4I–M) and LPCAT2 (Figure S5) reduced the numbers and the area of GFP-LC3 puncta under starvation conditions in OA-treated cells (Figure 5K; Figure S4E,F; S5B,C). We furthermore performed LC3 immunoblot analysis and found reduced LC3-II/actin ratio (in the presence of Bafilomycin A1) upon siRNA treatment against endogenous CPT1 (Figure 5L and Figure S4H). In a complementation experiment, an siRNA resistant variant of CPT1 (T468C) reversed this effect (Figure 5L and Figure S4H). Thus, these enzymes of the Kennedy pathway and the Lands cycle were important for LD-based enhancement of autophagic capacity. An efficient knockdown of LPCAT1 could not be obtained (Figure S5D) so we could not rule in or out whether LPCAT1, in addition to LPCAT2, contributes to optimal formation of autophagosomes.
In conclusion, PNPLA5, which generates DAG, and CPT1 that transfers choline to the DAG to form PC, are both required for optimal initiation of autophagy. CPT1 effects may also include direct action on LD formation. Furthermore, the PC remodeling pathway via LPCAT2, known to influence the formation of PC on LDs [32] also affects autophagy.
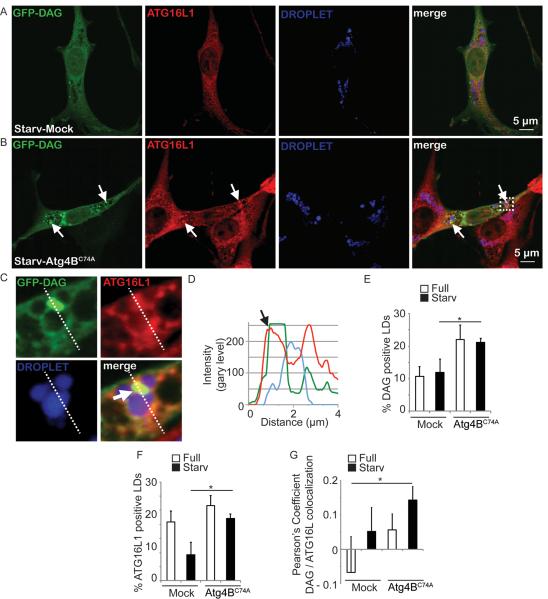
Inhibition of autophagosome closure increases association of DAG and Atg16L1 with LDs
The immediate product of PNPLAs as TG lipases is DAG. Using a DAG-specific GFP probe (NES-GFP-DAG; [33]), we performed time lapse videomicroscopy and found that DAG probe identified newly emerging profiles associated with LDs (Movies S5 and S6; Fig. 2J,K) in addition to the preexisting DAG-positive profiles. We next tested whether DAG appeared on LDs in association with induction of autophagy by starvation (Figure 6). We also tested DAG in relationship to an early autophagic marker, Atg16L1, due to the association of Atg16L1 with LDs (Figure S2M). To trap such putative intermediates, we again employed cells expressing the dominant negative form of Atg4, Atg4BC74A [23], which prevents autophagosome closure and subsequent maturation events. There was an increase of % DAG-positive LDs in Atg4BC74A expressing cells (Figure 6E) relative to mock vector containing cells. This reached statistical significance under autophagy inducing conditions by starvation (Figure 6E). There was a statistically significant increase of % Atg16L1-positive LDs (Figure 6F) under autophagy inducing conditions in Atg4BC74A-expressing cells. A trend in increase of colocalization of Atg16L1 and DAG on LDs was observed when autophagosomal completion was blocked by Atg4BC74A (Figure 6G). An increase in DAG-ATG16L1 colocalization was statistically significant when mock cells in basal (fed) conditions were compared to Atg4BC74A-expressing cells under autophagy inducing conditions (Figure 6G). These observations indicate that DAG, detected by the NES-GFP-DAG probe, was associated with an autophagy initiation marker on LDs upon induction of autophagy by starvation, and that these intermediates were trapped by blocking autophagosomal progression.
Figure. 6. Localization analysis of DAG and ATG16L1 juxtaposed to lipid droplets upon overexpression of mutant ATG4BC74A.
(A–G) Diacylglycerol (DAG, green; revealed by the NES-GFP-DAG probe) and ATG16L1 (red) relative to lipid droplets (LD, blue). 3T3 cells expressing ATG4B (A; Mock) or ATG4BC74A (B) proteins were transfected with a plasmid expressing NES-GFP-DAG, treated for 20 h with 500 μM BSA-Oleic Acid, and starved in EBSS for 150 minutes. ATG16L1, immunostatining. Arrows, overlap between ATG16L1 and DAG signals juxtaposed to LD. (C,D) Enlarged area (dashed box in B) and fluorescence line tracings. (E) Quantification of the recruitment of GFP-DAG probe to LDs. DAG positive LDs were quantified by using SlideBook morphometric analysis software (details in Materials and Methods). 3T3 cells expressing ATG4B (Mock) or ATG4BC74A proteins were processed as in A. (F). ATG16L1 positive LDs were quantified using SlideBook morphometric analysis software in 3T3 cells expressing ATG4B (Mock) or ATG4BC74A processed as in A and B. Data, means ± s.e. (n≥3); *, p<0.05. (G) Pearson's coefficients for DAG and Atg16L1 surrounding the droplets (SlideBook morphometry; 3 independent experiments with 5 fields per experiment). Data, means ± s.e. (n≥3); *, p<0.05
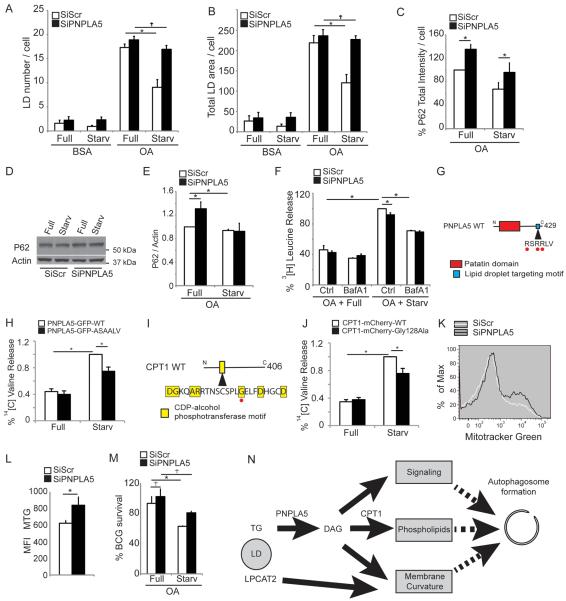
PNPLA5 affects autophagy of diverse cargoes
The above observations collectively indicate that autophagy initiation is associated with LDs, and that LDs support generation of autophagosomes. Autophagosomes derived in part from LDs could be specializing in lipophagy. Alternatively, autophagosomes originating at or from LDs might target other autophagic substrates. We used PNPLA5 knock-downs to differentiate between these two possibilities. PNPLA5 knock-down inhibited LDs consumption (Figure 7A,B, Figure S6). When we tested other autophagy substrates, it turned out that PNPLA5 affected magnitude of these processes as well. PNPLA5 knock-down inhibited degradation of an autophagic adaptor, Sequestosome-1/p62, as one of conventional reporters of selective autophagy (Figure 7C–E).
Figure. 7. PNPLA5 is required for efficient autophagy of diverse substrates.
(A,B) HeLa cells were transfected twice with PNPLA5 or scramble (Scr) siRNA control, treated for 20 h with BSA or with 500 μM BSA-Oleic Acid (OA) and starved (Starv) or not (Full) for 2 h, lipid droplets stained with Bodipy 493/503 (illustrated in Figure S6) and quontified by high content imaging acquisition and analysis. (C–E) HeLa cells transfected and treated as in A; total immunofluorescence intensity of endogenous p62 was quantified in GFP-positive cells by high content image acquisition and analysis (C). (D,E) p62/actin ratios determined by immunoblotting and densitometry. (F) Proteolysis of proteins in HeLa cells. HeLa transfected and treated as in A, in media containing [3H] leucine, were starved or not with or without Bafilomycin A1 (Baf) for 90 min. Leucine release was calculated from radioactivity in the tricarboxylic acid-soluble form relative to total cell radioactivity. (G) Lipid droplet targeting motif of PNPLA5, Red dots, residues that have been mutated to Ala (RSRRLV changed to ASAALV). (H) Proteolysis of long-lived proteins in HeLa cels were transfected with plasmids expressing wild-type (WT) GFP-tagged or ASAALV PNPLA5 proteins and treated 20 h with 500 μM BSA-Oleic Acid (OA) in media containing [14C] valine. Y-axis, valine release from stable proteins. (I) CDP-alcohol phosphotransferase catalytic motif in CPT1. Red dot, Cys-to-Ala mutation. (J) HeLa cells were transfected with plasmids expressing wild-type (WT) mCherry-tagged or CPT1Gly128Ala mutant CPT1 proteins and processed for proteolysis of long-lived proteins as in (H). (K,L) Flow cytometry analysis of cellular mitochondrial content. HeLa were treated as in A (full medium) and stained with MitoTracker Green. (K) histograms; (L) average mean fluorescence intensity (MFI) of MitoTracker Green per cell. Data, means ± s.e. (n≥3); *, p<0.05. (M) Analysis of the role of PNPLA5 in autophagic killing of BCG. RAW 264.7 macrophages were transfected twice with PNPLA5 siRNAs or scramble (Scr) control. Cells were then treated 20 h with 250 μM BSA-Oleic Acid (OA) and infected the day after with BCG. Autophagy was induced 4 h by starvation (Starv). BCG survival, % of control BCG CFU. Data, means ± s.e.m. *, p<0.05. (N) Model for how lipid droplets contribute to autophagosome biogenesis (see discussion).
PNPLA5 was required for optimal proteolysis, since PNPLA5 knockdown reduced autophagy-dependent (i.e. bafilomycin A1-inhibitable component) of proteolysis (Figure 7F). Furthermore, overexpression of a mutant PNPLA5 with altered PNLAP5 LD-targeting motif (RSRRLV) [29] decreased proteolysis of long-lived proteins in response to starvation (Figure 7G,H). This effect on proteolysis was mirrored in a similar experiment carried out with overexpression of a CPT1 catalytic site mutant (CPT1Gly128Ala) [34] (Figure 7I,J).
Mitophagy decreased in cells subjected to PNPLA5 knockdown, as shown by increase in mitochondrial content per cell measured by MitoTracker Green (Figure 7K,L). Finally, elimination of an intracellular microbe (Mycobacterium bovis BCG) by xenophagy was reduced upon PNPLA5 knockdown (Figure 7M). These findings demonstrate that PNPLA5 plays a role in autophagy of diverse substrates including an autophagic adaptor-mediated processes, organelles (mitophagy), bacteria (xenophagy) and bulk autophagy of the cytosol, and suggests a model in which autophagy initiation at sites controlled by PNPLA5 (e.g. LDs as shown here) affects autophagy in general and not just the autophagy engaged in lipophagy.
Discussion
This work shows that lipid droplets enhance autophagic capacity of a mammalian cell. In the absence of an LD build-up, autophagy still occurs but at a lower level. The enhancement of autophagic capacity depends on PNPLA5, a member of the patatin-like phospholipase domain-containing proteins that mobilize neutral lipids. In addition, CPT1, an enzyme important for de novo phospholipid synthesis, and LPCAT2, an enzyme engaged in PC remodeling, support lipid droplet-dependent enhancement of autophagy. Lipid droplets as stores of neutral lipids [15, 16] represent a contributing source of membrane precursors for formation of autophagosomes (Figure 7N). By mobilizing the precipitated out lipid matter, i.e. the TGs within LDs, cells are able to safely build new autophagosomes without unnecessarily compromising integrity and functionality of the pre-existing organelles.
As an alternative or in addition to the above model, DAG generated by PNPLA5 may affect signaling events or curvature of the autophagic membrane (Figure 7N). In the context of membrane curvature, the driving force for curving of phagophore membranes to eventually close into fully formed ovoid or globular autophagosomes is still not clear. We propose here that the remodeling (with potential participation of Lands cycle enzymes such as LPCAT2) and exchange of phospholipids between lipid droplets that are delimited by a hemi-membrane (phospholipid monolayer) and the growing autophagosome delimited by a bilayer membrane may lead to an asymmetric growth from the outside and thus induce curvature. Such exchange could take place during repeat dynamic kiss-and-run interactions evident in our time lapse imaging of cultured cells and intravital organ imaging studies.
Although our study expands the number of proposed compartments involved in autophagosome biogenesis [6, 7, 10, 11, 13] it is fundamentally different from reporting a yet another source contributing membranes to autophagosmes. The paradigm shift here posits utilization of neutral lipid stores while sparing preformed membranes of another functional compartment. It however does not contradict the current models of autophagosome formation. For example, LDs interact extensively with the ER [16, 35], which is the site where DFCP1-positive transient omegasome precursors cradle the nascent autophagic phagophores [5]. Not surprisingly, LDs are found in the vicinity of autophagosomes while they are still connected with the ER [6, 7]. A recent study furthermore implicates contact sites between the ER and mitochondria in autophagosome formation [11]. Thus, the participation LDs in autophagosome formation and function is likely intertwined with roles of other compartments that contribute to autophagosomal biogenesis [10].
Mizushima and colleagues [36] have shown that an Atg5 knockdown affects only the number of LDs and not the total volume of a cellular LD content [36]. This is in keeping with the relationships uncovered in our study: a surface to volume reduction in Atg5-deficient cells can be explained by an altered phospholipid-to-neutral lipids ratio resulting in a coalescence of neutral lipids into a structure with a smaller in total size phospholipid monolayer enveloping the neutral lipid cores of LDs. In the present study, we have also shown that early autophagy factors WIPIs (mammalian Atg18 homologs) are recruited to LDs upon induction of autophagy by starvation. Another autophagy factor, Atg2, cooperating with WIPIs, localizes to both autophagosomes and lipid droplets [36].
Lipophagy contributes via lysosomal lipases to lipolysis and breakdown of TGs into free fatty acids for energy consumption via β-oxidation in mitochondria in different cell types including hepatocytes [3]. In contrast, we found that consumption of LDs was unaffected by blocking completion of autophagosome closure or phagosomal maturation. This indicates that LDs continued to be utilized upon induction of autophagy but not as an autophagic cargo. However, it is likely that under physiological conditions both processes, autophagosome biogenesis and lipolysis via autophagy, occur in a balanced fashion. It will be of interest to examine how these two functions are coordinated. This is of particular significance in the context of a fine balance between lipid storage and lipolysis that needs to be carefully buffered lest lipotoxicity of free fatty acids occurs [15, 16]. For example, increasing capacity of storing lipids in their neutral form such as TGs can paradoxically prevent diabetes even in the face of obesity [37]. Based on our results, autophagy activation could contribute to protection against lipotoxicity by utilizing TGs to build phospholipid membranes rather than bulk-converting all neutral lipids into potentially toxic fatty acids especially under conditions when mitochondria may not have the functional capacity for excessive β-oxidation.
Lipid droplets are intracellular depots of excess lipids that are sequestered in a neutral form such as TGs and cholesterol esters for storage, avoidance of lipotoxicity, and controlled mobilization [14–16]. Our data together with the prior reports connecting autophagy with LD utilization [3, 38, 39] may have import on a range of diseases associated with reduced tissue capacity for lipid storage vs a need to deal with surplus of toxic unesterified fatty acids and cholesterol [15]. A further example of uncoordinated or excessive lipolysis, where autophagy role needs to be explored, is cancer-associated cachexia [40]. It will be of interest to study in depth the relationships uncovered here as they may pertain to atherosclerosis, obesity, diabetes and insulin resistance with elevated intracellular deposition of neutral lipids in adipocytes, smooth muscle cells and macrophages mirrored by elevated plasma levels of low-density lipoprotein. These conditions may be influenced by the role of LD consumption in biogenesis of autophagic membranes.
Experimental Procedures
Knockdowns with siRNAs, knockdown validation, and mutagenesis
SMARTpool SiGENOME siRNAs procedures, and RT-PCR primers used for knockdown validation are listed in Supplementary Materials. Site-directed mutagenesis was performed using a Quick Change kit (Stratagene).
Media and pharmacological agonists and inhibitors of autophagy
Autophagy was induced by starvation in Earle's balanced salt solution (EBSS). Oleic acid was complexed at room temperature with fatty acid-free bovine serum albumin as previously described [18]. Cells were treated with 100 nM bafilomycin A1 and 10 mM 3-MA.
High content image acquisition and analysis and other imaging techniques
An automated high content analysis system (Cellomics Array Scan, Thermo Scientific) was used to acquire images by computer-driven (operator independent) collection of 49 valid fields per well with cells in 96 well plates, with >500 cellls (identified by the program as valid primary object) per each sample. Objects were morphometrically and statistically analyzed using the iDev software. Computer-driven identification of primary and secondary objects was based on predetermined parameters, and fluorescent objects (cells, puncta, droplets, total cytoplasm) were quantified using a suite of applicable parameter (including number of objects per cell; total area per cell; total intensity). Nuclei and its associated cytoplasm were sequentially segmented using the Hoechst channel, GFP fluorescent puncta or endogenous LC3 and p62 were revealed by fluorescent antibody staining. Bodipy 493/503, LipidTOX™ Red and LipidTOX™ DeepRed were used to stain lipid droplets. Some studies were carried out using Opera WQEHS and Acapella software (Perkin Elmer; see Supplemental Information). Other methods (time lapse imaging of cultured cells and intravital microscopy are described in Supplementary Information.
Supplementary Material
Movie S1 Oleic acid pretreated HeLa cells expressing GFP-LC3 were labelled with LipidToxFarred (red) for 5 min. Cells were starved and imaged by confocal microscopy in time-lapse. LC3-GFP vesicles (green) and lipid droplets (red). Time, min:sec. Scale bar, 2 μm.
Movie S2 An GFP-LC3 mouse was anesthetized, the liver exposed, bathed for 30 min with 0.1 mg/ml BODIPY 665 to label lipid droplets, and imaged by confocal microscopy in time-lapse. LC3-GFP vesicles (green) and lipid droplets (red) were imaged in the cytoplasm of hepatocytes located 20 μm from the surface of the organ. Time, h:min:sec. Scale bar, 5 μm.
Movie S3 Oleic acid pretreated U2OS cells expressing GFP-WIPI2D were labelled with LipidToxFarred (red) for 5 min. Cells were starved and imaged by confocal microscopy in time-lapse. LC3-GFP vesicles (green) and lipid droplets (red). Time, h:min:sec. Scale bar, 2 μm.
Movie S4 Oleic acid pretreated U2OS cells expressing GFP-WIPI2D were labelled with LipidToxFarred (red) for 5 min. Cells were starved and imaged by confocal microscopy in time-lapse. LC3-GFP vesicles (green) and lipid droplets (red). Time, h:min:sec. Scale bar, 2 μm.
Movie S5 Oleic acid pretreated HeLa cells expressing GFP-DAG were labelled with LipidToxFarred (red) for 5 min. Cells were starved and imaged by confocal microscopy in time-lapse. LC3-GFP vesicles (green) and lipid droplets (red). Time, min:sec. Scale bar, 2 μm.
Movie S6 Oleic acid pretreated HeLa cells expressing GFP-DAG were labelled with LipidToxFarred (red) for 5 min. Cells were starved and imaged by confocal microscopy in time-lapse. LC3-GFP vesicles (green) and lipid droplets (red). Time, min:sec. Scale bar, 2 μm.
Highlights
Neutral lipid stores increase autophagic capacity of the cell
Optimal autophagy initiation requires triglyceride phospholipase PNPLA5
Lipid droplet phospholipids enhance autophagosomal membrane biogenesis
Lipid droplets enable effective autophagy of diverse substrates
Acknowledgments
We thank Patrice Codogno for his generous support of ND and Chantal Bauvy for experimental help. We acknowledge W. Ornatowski, D. Bhattacharya and M. Mudd for assistance. We are grateful to N. Mizushima for GFP-LC3 transgenic mice, T. Yoshimori for NIH3T3 cells stably expressing Atg4B or mStrawberry-Atg4BC74A, N. Ktistakis for HEK-293 stably expressing GFP-DFCP1, D. Rubinsztein for mRFP-GFP-LC3 HeLa cells, S. Tooze for spontaneously immortalized ULK1/2 DKO MEFs cells and X. M. Yin and T. Balla for plasmids expressing respectively GFP-Atg16L1 and NES-GFP-DAG. N.D. acknowledges facilities support by Conseil de la Region Ile-de-France (program Sesame 2007, project Imagopole, S. Shorte) and from the Fondation Française pour la Recherche Médicale (FRM, Programme Grands Equipements) to N. Aulner, the Necker Institute Imaging Facility, the Fondation Imagine and the Imagopole-PFID France-BioImaging infrastructure supported by the French National Research Agency (ANR-10-INSB-04-01, “Investments for the future” for advice and access to the Opera system. ND is recipient of a Fondation pour la Recherche Médicale (FRM) fellowship. Studies in Patrice Codogno's laboratory are supported by institutional funding from INSERM, University Paris-Descartes, and grants from ANR and INCa. T.P-C. acknowledges the support by DFG SFB773/TP A03. E.F.C. was supported by a minority supplement grant AI042999-13S1. This work was supported by grant R01 AI042999 from National Institutes of Health to V.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Other methods
Additional materials and methods are described in Supplementary Information.
The authors declare no conflict of interest.
References
- 1.Mizushima N, Yoshimori T, Ohsumi Y. The role of atg proteins in autophagosome formation. Annual review of cell and developmental biology. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 2.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nature reviews. Drug discovery. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 5.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nature cell biology. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 7.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 8.Proikas-Cezanne T, Waddell S, Gaugel A, Frickey T, Lupas A, Nordheim A. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene. 2004;23:9314–9325. doi: 10.1038/sj.onc.1208331. [DOI] [PubMed] [Google Scholar]
- 9.Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010;6 doi: 10.4161/auto.6.4.11863. [DOI] [PubMed] [Google Scholar]
- 10.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 11.Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013 doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 12.Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154:1285–1299. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yen WL, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, Hughson FM, Baba M, Klionsky DJ. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol. 2010;188:101–114. doi: 10.1083/jcb.200904075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujimoto T, Parton RG. Not just fat: the structure and function of the lipid droplet. Cold Spring Harbor perspectives in biology. 2011;3 doi: 10.1101/cshperspect.a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruggles KV, Turkish A, Sturley SL. Making, baking, and breaking: the synthesis, storage, and hydrolysis of neutral lipids. Annual review of nutrition. 2013;33:413–451. doi: 10.1146/annurev-nutr-071812-161254. [DOI] [PubMed] [Google Scholar]
- 16.Krahmer N, Farese RV, Jr., Walther TC. Balancing the fat: lipid droplets and human disease. EMBO molecular medicine. 2013;5:905–915. doi: 10.1002/emmm.201100671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lake AC, Sun Y, Li JL, Kim JE, Johnson JW, Li D, Revett T, Shih HH, Liu W, Paulsen JE, et al. Expression, regulation, and triglyceride hydrolase activity of Adiponutrin family members. J Lipid Res. 2005;46:2477–2487. doi: 10.1194/jlr.M500290-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.Brasaemle DL, Wolins NE. Bonifacino Juan S., et al., editors. Isolation of lipid droplets from cells by density gradient centrifugation. Current protocols in cell biology / editorial board. 2006;Chapter 3(Unit 3):15. doi: 10.1002/0471143030.cb0315s29. [DOI] [PubMed] [Google Scholar]
- 19.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koga H, Kaushik S, Cuervo AM. Inhibitory effect of intracellular lipid load on macroautophagy. Autophagy. 2010;6:825–827. doi: 10.1096/fj.09-144519. [DOI] [PubMed] [Google Scholar]
- 21.Masedunskas A, Sramkova M, Parente L, Sales KU, Amornphimoltham P, Bugge TH, Weigert R. Role for the actomyosin complex in regulated exocytosis revealed by intravital microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13552–13557. doi: 10.1073/pnas.1016778108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Proikas-Cezanne T, Ruckerbauer S, Stierhof YD, Berg C, Nordheim A. Human WIPI-1 puncta-formation: a novel assay to assess mammalian autophagy. FEBS Lett. 2007;581:3396–3404. doi: 10.1016/j.febslet.2007.06.040. [DOI] [PubMed] [Google Scholar]
- 23.Fujita N, Hayashi-Nishino M, Fukumoto H, Omori H, Yamamoto A, Noda T, Yoshimori T. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008;19:4651–4659. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lass A, Zimmermann R, Oberer M, Zechner R. Lipolysis - a highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog Lipid Res. 2011;50:14–27. doi: 10.1016/j.plipres.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rydel TJ, Williams JM, Krieger E, Moshiri F, Stallings WC, Brown SM, Pershing JC, Purcell JP, Alibhai MF. The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a Ser-Asp catalytic dyad. Biochemistry. 2003;42:6696–6708. doi: 10.1021/bi027156r. [DOI] [PubMed] [Google Scholar]
- 26.Grall A, Guaguere E, Planchais S, Grond S, Bourrat E, Hausser I, Hitte C, Le Gallo M, Derbois C, Kim GJ, et al. PNPLA1 mutations cause autosomal recessive congenital ichthyosis in golden retriever dogs and humans. Nature genetics. 2012;44:140–147. doi: 10.1038/ng.1056. [DOI] [PubMed] [Google Scholar]
- 27.Schweiger M, Schreiber R, Haemmerle G, Lass A, Fledelius C, Jacobsen P, Tornqvist H, Zechner R, Zimmermann R. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J Biol Chem. 2006;281:40236–40241. doi: 10.1074/jbc.M608048200. [DOI] [PubMed] [Google Scholar]
- 28.Kumari M, Schoiswohl G, Chitraju C, Paar M, Cornaciu I, Rangrez AY, Wongsiriroj N, Nagy HM, Ivanova PT, Scott SA, et al. Adiponutrin functions as a nutritionally regulated lysophosphatidic acid acyltransferase. Cell metabolism. 2012;15:691–702. doi: 10.1016/j.cmet.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murugesan S, Goldberg EB, Dou E, Brown WJ. Identification of diverse lipid droplet targeting motifs in the PNPLA family of triglyceride lipases. PloS one. 2013;8:e64950. doi: 10.1371/journal.pone.0064950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibellini F, Smith TK. The Kennedy pathway--De novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life. 2010;62:414–428. doi: 10.1002/iub.337. [DOI] [PubMed] [Google Scholar]
- 31.Shindou H, Hishikawa D, Harayama T, Yuki K, Shimizu T. Recent progress on acyl CoA: lysophospholipid acyltransferase research. J Lipid Res. 2009;50(Suppl):S46–51. doi: 10.1194/jlr.R800035-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moessinger C, Kuerschner L, Spandl J, Shevchenko A, Thiele C. Human lysophosphatidylcholine acyltransferases 1 and 2 are located in lipid droplets where they catalyze the formation of phosphatidylcholine. J Biol Chem. 2011;286:21330–21339. doi: 10.1074/jbc.M110.202424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim YJ, Guzman-Hernandez ML, Balla T. A highly dynamic ER-derived phosphatidylinositol-synthesizing organelle supplies phosphoinositides to cellular membranes. Dev Cell. 2011;21:813–824. doi: 10.1016/j.devcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams JG, McMaster CR. Scanning alanine mutagenesis of the CDP-alcohol phosphotransferase motif of Saccharomyces cerevisiae cholinephosphotransferase. J Biol Chem. 1998;273:13482–13487. doi: 10.1074/jbc.273.22.13482. [DOI] [PubMed] [Google Scholar]
- 35.Robenek H, Hofnagel O, Buers I, Robenek MJ, Troyer D, Severs NJ. Adipophilin-enriched domains in the ER membrane are sites of lipid droplet biogenesis. Journal of cell science. 2006;119:4215–4224. doi: 10.1242/jcs.03191. [DOI] [PubMed] [Google Scholar]
- 36.Velikkakath AK, Nishimura T, Oita E, Ishihara N, Mizushima N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Molecular biology of the cell. 2012;23:896–909. doi: 10.1091/mbc.E11-09-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franckhauser S, Munoz S, Pujol A, Casellas A, Riu E, Otaegui P, Su B, Bosch F. Increased fatty acid re-esterification by PEPCK overexpression in adipose tissue leads to obesity without insulin resistance. Diabetes. 2002;51:624–630. doi: 10.2337/diabetes.51.3.624. [DOI] [PubMed] [Google Scholar]
- 38.Khaldoun SA, Emond-Boisjoly MA, Chateau D, Carriere V, Lacasa M, Rousset M, Demignot S, Morel E. Autophagosomes contribute to intracellular lipid distribution in enterocytes. Molecular biology of the cell. 2013 doi: 10.1091/mbc.E13-06-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Zutphen T, Todde V, de Boer R, Kreim M, Hofbauer H, Wolinski H, Veenhuis M, van der Klei IJ, Kohlwein SD. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Molecular biology of the cell. 2013 doi: 10.1091/mbc.E13-08-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Das SK, Eder S, Schauer S, Diwoky C, Temmel H, Guertl B, Gorkiewicz G, Tamilarasan KP, Kumari P, Trauner M, et al. Adipose triglyceride lipase contributes to cancer-associated cachexia. Science. 2011;333:233–238. doi: 10.1126/science.1198973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1 Oleic acid pretreated HeLa cells expressing GFP-LC3 were labelled with LipidToxFarred (red) for 5 min. Cells were starved and imaged by confocal microscopy in time-lapse. LC3-GFP vesicles (green) and lipid droplets (red). Time, min:sec. Scale bar, 2 μm.
Movie S2 An GFP-LC3 mouse was anesthetized, the liver exposed, bathed for 30 min with 0.1 mg/ml BODIPY 665 to label lipid droplets, and imaged by confocal microscopy in time-lapse. LC3-GFP vesicles (green) and lipid droplets (red) were imaged in the cytoplasm of hepatocytes located 20 μm from the surface of the organ. Time, h:min:sec. Scale bar, 5 μm.
Movie S3 Oleic acid pretreated U2OS cells expressing GFP-WIPI2D were labelled with LipidToxFarred (red) for 5 min. Cells were starved and imaged by confocal microscopy in time-lapse. LC3-GFP vesicles (green) and lipid droplets (red). Time, h:min:sec. Scale bar, 2 μm.
Movie S4 Oleic acid pretreated U2OS cells expressing GFP-WIPI2D were labelled with LipidToxFarred (red) for 5 min. Cells were starved and imaged by confocal microscopy in time-lapse. LC3-GFP vesicles (green) and lipid droplets (red). Time, h:min:sec. Scale bar, 2 μm.
Movie S5 Oleic acid pretreated HeLa cells expressing GFP-DAG were labelled with LipidToxFarred (red) for 5 min. Cells were starved and imaged by confocal microscopy in time-lapse. LC3-GFP vesicles (green) and lipid droplets (red). Time, min:sec. Scale bar, 2 μm.
Movie S6 Oleic acid pretreated HeLa cells expressing GFP-DAG were labelled with LipidToxFarred (red) for 5 min. Cells were starved and imaged by confocal microscopy in time-lapse. LC3-GFP vesicles (green) and lipid droplets (red). Time, min:sec. Scale bar, 2 μm.