Abstract
Relapsed pediatric acute lymphoblastic leukemia (ALL) has high rates of treatment failure. Epigenetic regulators have been proposed as modulators of chemoresistance, here we sequence genes encoding epigenetic regulators in matched diagnosis-remission-relapse ALL samples. We find significant enrichment of mutations in epigenetic regulators at relapse with recurrent somatic mutations in SETD2, CREBBP, MSH6, KDM6A and MLL2, mutations in signaling factors are not enriched. Somatic alterations in SETD2, including frameshift and nonsense mutations, are present at 12% in a large de novo ALL patient cohort. We conclude that the enrichment of mutations in epigenetic regulators at relapse is consistent with a role in mediating therapy resistance.
Introduction
Relapse occurs in 15 to 20% of pediatric acute lymphoblastic leukemia (ALL) and 50% of adult ALL patients. Compared to de novo leukemia, relapsed leukemia is chemoresistant and long term survival is poor, however the genetic basis for this is not well understood. Studies of matched diagnosis and relapse ALL samples by SNP arrays demonstrated that 92% of relapsed ALL had DNA copy number variant (CNV) changes from diagnosis. Surprisingly, over half of relapses are missing CNVs present in the predominant clone at diagnosis and appear to have derived from a minor subclone in many cases1. Targeting alterations that are frequently gained at relapse is therefore a rational therapeutic strategy, as these lesions are likely associated with chemotherapy resistance and clonal survival. Unfortunately, most CNV alterations gained at ALL relapse are difficult to target, such as CDKN2A or ETV6 deletion.
Alterations in epigenetic regulators have recently been identified in genome and exome sequencing studies of chemotherapy resistant, very high risk subsets of de novo ALL. For example, 48% of early T-cell precursor (ETP) ALL2 and 60% of hypodiploid ALL3 have somatic mutations in genes encoding epigenetic regulators. In addition, loss of function mutations in the histone acetyltransferase CREBBP have also been described in diagnostic samples from ALL patients who relapse and in samples from relapsed patients4. As the enzymatic activities of CREBBP are directly opposed by histone deacetylases, this provided one possible rationale for why histone deacetylase inhibitors have activity in ALL. These studies suggest a broader paradigm in that mutations in epigenetic regulators may be associated with therapy resistance and can be targeted indirectly through inhibition of the enzymatic activity of opposing regulators. However, thus far, no published study has addressed the mutational frequency of epigenetic regulators at ALL relapse, and whether they are frequently enriched at relapse compared to at diagnosis.
Here we sequence matched sets of diagnosis-remission-relapse samples from pediatric ALL patients, and find a significant enrichment of mutations in epigenetic regulators at relapse, including the H3K36 trimethyltransferase SETD2. We conclude that these mutations are likely involved in chemotherapy resistance and deserve further study to define their role in the prognosis and treatment of ALL.
Results
Somatic mutations in epigenetic regulators in de novo ALL
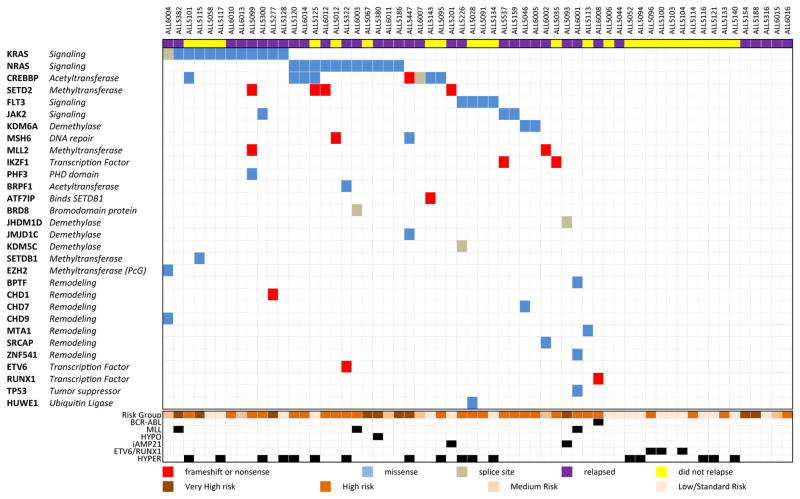
In order to study this, we performed custom exon hybrid capture and Illumina sequencing of 472 known and putative epigenetic regulators and frequently mutated genes in leukemia (Supplementary Table 1) on matched diagnostic-remission samples from 60 pediatric B-ALL patients, and matched relapse samples from 30 of those patients that relapsed. Patients in this cohort were those with pediatric ALL (primarily B-cell), with a range of cytogenetics and risk groups (Supplementary Table 2). In this cohort, relapsed patients had the bone marrow as a site of relapse, with over 60% marrow involvement. 70% of patients had an identified somatic mutation at diagnosis and/or relapse (Fig 1).
Figure 1. Targeted hybrid capture sequencing identified somatic mutations in 70% of BALL patients.
Matched tumor-remission samples from 60 pediatric B-ALL were sequenced with targeted hybrid capture and somatic mutations in any tumor sample were identified. Genes on the left are organized by mutational frequency. The type of alteration is identified for each mutation as frameshift/stop, missense or splice site mutation. MLL: MLL rearranged, HYPO: Hypodiploidy, iAMP21: intrachromosomal amplification of chromosome 21, HYPER: Hyperdiploidy.
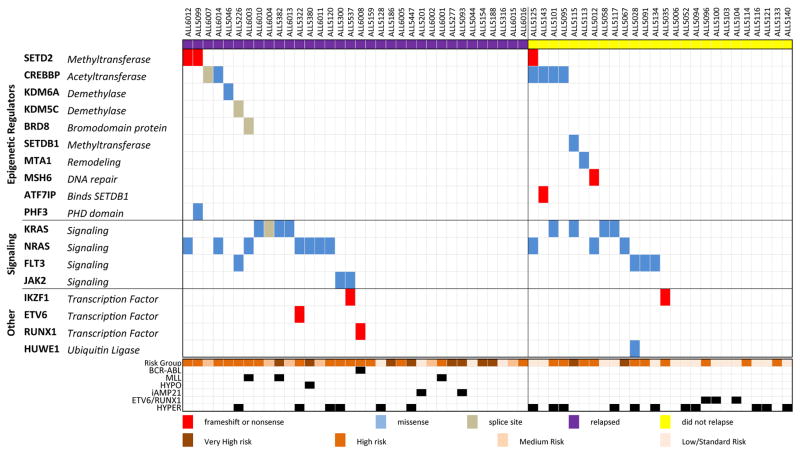
Considering only samples from the time of diagnosis, somatic variants in epigenetic regulators were found in 25% of 60 B-ALL samples (Fig 2), which was similar to de novo T-ALL patients5. In contrast, 50% of B-ALL samples harbored a mutation in a signaling factor at diagnosis with recurrent mutations in the Ras pathway, including activating point mutations in KRAS and NRAS, which were often subclonal (Supplementary Fig. 1A). At diagnosis, recurrent mutations in epigenetic regulator genes were identified in CREBBP (10%) and SETD2 (5%).
Figure 2. Mutations in epigenetic regulators occur in 25% of B-ALL patients at diagnosis, including SETD2.
Matched diagnosis and remission sample pairs from 60 pediatric B-ALL patients were baited with target hybrid capture, Illumina sequenced and somatic mutations were identified. Patients are organized by those that eventually relapsed (on the left), and those that did not (on the right). Final risk category and molecular subtypes listed below. MLL: MLL rearranged, HYPO: Hypodiploidy, iAMP21: intrachromosomal amplification of chromosome 21, HYPER: Hyperdiploidy.
SETD2 mutations in de novo ALL
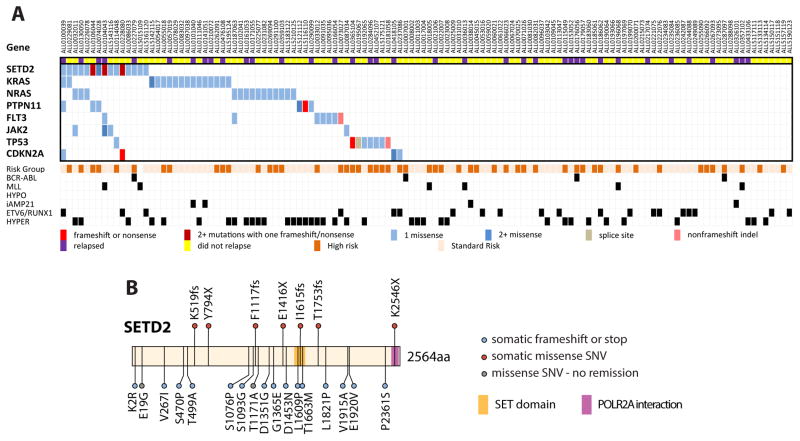
To more accurately determine the frequency of SETD2 mutations, which have not been previously described in B-ALL, the coding regions of SETD2 were PCR amplified and Illumina sequenced in a more representative independent validation cohort of 125 de novo pediatric ALL patients. SETD2 mutations were found in 12% of the cohort and these leukemias also frequently harbored mutations in the Ras pathway (Fig 3A). Four patients harbored multiple SETD2 mutations. In total, we identified 24 SETD2 mutations in 19 out of 185 ALL patients, including seven loss of function frameshift or nonsense mutations, with no obvious hotspot mutations (Fig 3B). Altogether, SETD2 mutations were most prevalent in the MLL rearranged (22%) and ETV6-RUNX1 rearranged (13%) subtypes of ALL (Supplementary table 3). Of note, these two subtypes were underrepresented in the initial cohort, explaining the lower prevalence of SETD2 mutations in that cohort.
Figure 3. SETD2 Mutations in de novo ALL.
A) Amplification and Illumina sequencing of coding exons of SETD2 and other frequently mutated genes in an validation cohort of 125 de novo ALL patients identified 20 additional SETD2 mutations in 15 patients (12%). Relapse status is noted above. Final risk category and molecular subtypes listed below. MLL: MLL rearranged, HYPO: Hypodiploidy, iAMP21: intrachromosomal amplification of chromosome 21, HYPER: Hyperdiploidy. B) Schematic of somatic SETD2 mutations found in ALL patients.
Enrichment of mutations in epigenetic regulators at relapse
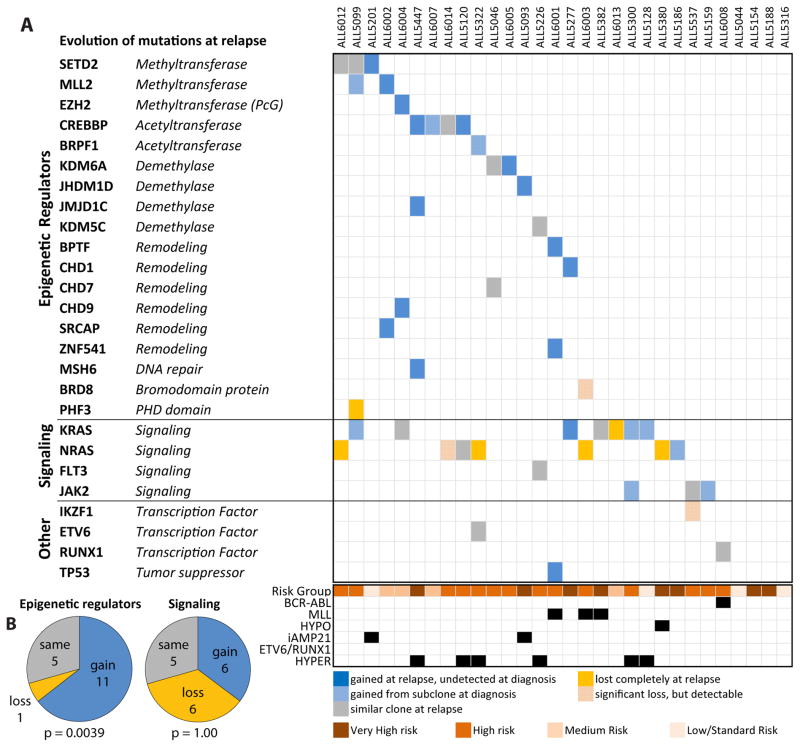
Sequencing of the 30 matched relapsed samples showed a striking gain of mutations in epigenetic regulators, with 17 (57%) harboring a mutation. Of the 11 patients that gained a mutation in an epigenetic regulator at the time of relapse, only three were detectable in a rare subclone at diagnosis. (Fig 4A). As noted above, this frequency is comparable to other poor prognosis de novo ALL subgroups such as hypodiploid and ETP ALL. In addition to the 11 gains, five patients retained the same mutations from diagnosis, and one patient had their clonal mutation reduced to a subclonal allele frequency at relapse (Supplementary Fig. 1B). Details of each identified somatic mutation in each patient and a summary of mutations by molecular subtype are available in the supplemental materials (Supplementary Tables 3 and 4).
Figure 4. Relapsed B-ALL is enriched for mutations in epigenetic regulators.
A) Gain or loss of somatic mutations in matched diagnosis-relapse samples from 30 pediatric B-ALL patients. Clonal mutations at relapse which were detected in a minor subclone at diagnosis and clonal mutations at diagnosis which became subclonal at relapse are shaded. MLL: MLL rearranged, HYPO: Hypodiploidy, iAMP21: intrachromosomal amplification of chromosome 21, HYPER: Hyperdiploidy. B) Comparison of the number of patients which gain, lose, or have the same number of mutations in epigenetic regulators and signaling factors at relapse. The McNemar test was used to calculate the P-values.
By McNemar test, there was a statistically significant enrichment of the entire epigenetic group (p=0.0039) (Fig 4B) and SETD2 and CREBBP together (p=0.045). In contrast, there was not a significant enrichment of somatic mutations in signaling factors such as NRAS, KRAS, FLT3 and JAK2, as individual genes, or as a group. This was because there were as many patients gaining signaling mutations or mutational allele frequency as those losing them (six, McNemar test p=1.00) (Fig 4B). The selective enrichment of epigenetic regulators is consistent with the hypothesis that mutations in genes encoding epigenetic regulators mediate chemotherapy resistance. Conversely, oncogenic mutations in signaling factors showed no enrichment, with frequent losses and gains, implying no direct role in modulating drug resistance and non-dependence on any particular mutation. Thus, solely targeting signaling factors by specific inhibition may not circumvent the development of chemoresistance. In total, recurrently mutated epigenetic regulators included MLL2, KDM6A and MSH6, as well as additional mutations in SETD2 and CREBBP. KDM6A encodes for a H3K27 demethylase. We recently reported somatic KDM6A mutations in 4% of pediatric ALL at diagnosis, with mutations clustered in the catalytic jmjC domain6. SETD2 is the sole mammalian H3K36 trimethyltransferase and mutations in it have been described in ETP T-ALL2 and clear cell renal cell carcinoma7, but SETD2 has not been found to be recurrently mutated in large cohorts of non-ETP T-ALL2,5,8 and AML9 patients.
MSH6 is a vital component of the DNA mismatch repair (MMR) machinery that localizes to chromatin by binding to the H3K36 trimethyl mark that SETD2 makes. SETD2 knockdown has been shown to cause mis-localization of MSH6 and microsatellite instability and a mutator phenotype in several cell types10.
Discussion
Here, we report that somatic mutations in epigenetic regulators are enriched at relapse in pediatric B-ALL with CREBBP, KDM6A, MLL2, SETD2 and MSH6 as recurrently mutated genes. This finding suggests that therapy may be applying pressure to acquire or select for rare subclones which possess these mutations. As noted above, SETD2 and MSH6 have been linked together in DNA mismatch repair via the H3K36 trimethyl mark. We speculate that the loss-of-function mutations identified in SETD2 and MSH6 may lead to clonal survival by at least two possible mechanisms. One, a mutator phenotype induced by SETD2 inactivation could increase the mutational diversity and thus adaptability of the leukemia, leading to clonal survival. Two, since intact MMR is important for triggering apoptosis and/or cell cycle arrest in response to many DNA damaging chemotherapies, SETD2 loss may lead to chemotherapy tolerance. For example, leukemia cells lines with defective MMR show chemoresistance to the thiopurines 6-MP and 6-TG11, which are commonly used in ALL therapy. These mechanisms are potentially reversible by inhibiting the epigenetic regulators that oppose SETD2 activity such as the H3K36 demethylases.
Interestingly, alterations in other factors which modify H3K36 methylation, such as NSD1, WHSC1/NSD2, NSD3, and KDM4A have been associated with ALL, AML, multiple myeloma, breast cancer and myelodysplastic syndrome12. Several recent reports have identified recurrent activating WHSC1/NSD2 E1099K mutations in many ALL cell lines and 20% of ETV6-RUNX1 translocated ALL patients13,14
The genetic basis for chemoresistance and poor outcomes in relapsed ALL is beginning to be defined. This study identifies mutations in genes encoding epigenetic regulators as frequently gained at the time of relapse. Although this study was not powered to determine a prognostic effect of these mutations in this modest cohort of patients, the enrichment of mutations in epigenetic regulators in relapsed B-ALL and other chemoresistant, high risk subtypes of de novo leukemia is quite suggestive of a role in clonal survival. Epigenetic regulators deserve particular attention because they are amenable to pharmacological modulation. Histone deacetylase inhibitors are currently in clinical use15, and histone methyltransferase inhibitors, such as DOT1L inhibitors16, are in clinical development. Although these epigenetic therapies are first being evaluated in the relapsed setting, our work also suggests that incorporating epigenetic therapies in upfront therapy, particularly in patients with identified mutations, could be a strategy to prevent the development of chemoresistance and relapse.
Further studies are warranted to define the prognostic value of mutations in epigenetic regulators in ALL, their effects on chemotherapy resistance and whether therapeutic modulation of H3K36 methylation levels is a viable therapeutic strategy to prevent and treat relapse in patients with leukemia.
Methods
Patient materials
Subjects were pediatric acute lymphoblastic patients enrolled and banked on upfront therapy protocols DFCI 00-001, DFCI 05-001, AIEOP-2000, AIEOP-2006 or Interfant-99 and their families each gave informed consent for future biological research on their banked samples under protocols approved by the Institutional Review Board at each participating institution and Dana Farber Cancer Institute. Bone marrow specimens were primarily used, but peripheral blood was used for remission samples, if no bone marrow sample was available. Diagnostic and relapse marrow samples had at least 60% blasts by morphology. Mononuclear cells from each sample were isolated with Ficoll density gradient centrifugation, and genomic DNA was extracted.
Exon capture
For exon capture, 1–2ug of genomic DNA was sheared with a Covaris E100 to an average size of 200bp. Sheared DNA was end repaired, a-tailed, and ligated to barcoded adapters as per the Illumina Truseq Library Preparation protocol. Libraries were amplified with the Kapa Realtime Library Amplification kit into order to minimize the number of PCR cycles used (range 4–12). Library was then quantified by Kapa Realtime Quantification Kit and pooled up to 6 samples per lane in equimolar amounts totaling 500ng of DNA. Each pool was then hybridized to a custom SureSelect In Solution Hybrid Capture RNA baitset, consisting of 19,952 baits, spanning 2.35 Mb and 472 genes (Supplementary Table 1). Each capture was washed and amplified with the Kapa Realtime Library Amplification Kit and submitted for sequencing on an Illumina HiSeq 2000 100bp single end run.
Amplicon sequencing
192 primer pairs spanning the coding regions of SETD2, NRAS, KRAS, FLT3, JAK2, PTPN11, TP53 and CDKN2A were designed and pooled into 48 primer pools and used for amplification in the Fluidigm All Access Array (Supplementary Table 5). 50ng of genomic DNA samples from 128 patients, 48 samples at a time with 48 primers pools, were amplified by the Fluidigm All Access microfluidics system and each tagged with a unique barcode. All samples were then pooled into one Illumina Hiseq 2000 100bp single end run.
Variant Calling
Fastq files were aligned to hg19 version of the human genome with BWA. SNP, indel and copy number variant calling was performed with samtools mpileup and Varscan 2.2.3 somatic option. Annovar was used to annotate variants. Relapsed and diagnostic variants were linked and variants were filtered for >=15% allele frequency and 20x coverage in either tumor sample, <5% and 10x coverage in the remission sample, bidirectional variant reads, minimal strand bias, and minimal local noise. Variants were visualized in the Integrated Genome Viewer to eliminate variants with clustered read position and variants caused by indel artifacts. Sanger sequencing was used to validate a sample of somatic variants found by next generation sequencing to validate filtering criteria. Every filtered somatic variant position was then forced called on every sample to examine the baseline noisiness of each position. Those with mean baselines >1% allele frequency in remission samples were excluded. Variants were also filtered to only those positions within 50 base pairs of those included in the custom capture baitset. Variants were considered somatic if they were present in less than 5% allele frequency in the paired remission sample (to account for high minimal residual disease).
Supplementary Material
Acknowledgments
We are grateful to C. Lindsley, S. Jaiswal, A. Krivtsov, M. Correll, R. Rubio, F. Abderazzaq and Y. Wang for technical advice and valuable discussions. We would also like to thank S. Hunt, E. Swaim, J. Allen, H. Wang and A. Lack for assistance obtaining clinical specimens. B.G.M. was supported by the National Institutes of Health (5T32CA136432-03), the Lady Tata Memorial Trust, the William Lawrence and Blanche Hughes Foundation, the Abraham Fellowship Fund, and the Friends of Dana Farber. Additional research support was provided by the National Institutes of Health (CA140575 and CA068484), the Charles H. Hood Foundation, and the Leukemia and Lymphoma Society.
Footnotes
Contributions
B.G.M., L.B.B, and S.A.A. conceived and designed the project. K.M.M. and B.G.M. performed the sequencing library preparation and preparing of samples for Sanger sequencing. B.G.M. and P.V.G. performed the informatics analysis of the sequencing data. M.H.H., L.B.S., S.E.S. and L.L.N. provide patient samples for analysis. K.S. and D.S.N. provided clinical annotation and biostatistical analysis. B.G.M. wrote the manuscript and S.A.A., L.B.B. and B.L.E edited the manuscript. S.A.A. and B.L.E jointly directly the work.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Mullighan CG, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322:1377–80. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmfeldt L, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nature genetics. 2013;45:242–52. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mullighan CG, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–9. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Keersmaecker K, et al. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nature genetics. 2013;45:186–190. doi: 10.1038/ng.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mar B, et al. Sequencing histone-modifying enzymes identifies UTX mutations in acute lymphoblastic leukemia. Leukemia. 2012;26:1881–1883. doi: 10.1038/leu.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalgliesh G, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzoneva G, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nature Medicine. 2013;19:368–371. doi: 10.1038/nm.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. The New England journal of medicine. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F, et al. The Histone Mark H3K36me3 Regulates Human DNA Mismatch Repair through Its Interaction with MutSalpha. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diouf B, et al. Somatic deletions of genes regulating MSH2 protein stability cause DNA mismatch repair deficiency and drug resistance in human leukemia cells. Nature Medicine. 2011;17:1298–1303. doi: 10.1038/nm.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner E, Carpenter P. Understanding the language of Lys36 methylation at histone H3. Nature reviews. Molecular cell biology. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaffe JD, et al. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat Genet. 2013;45:1386–91. doi: 10.1038/ng.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oyer J, et al. Point mutation E1099K in MMSET/NSD2 enhances its methyltranferase activity and leads to altered global chromatin methylation in lymphoid malignancies. Leukemia. 2013 doi: 10.1038/leu.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spiegel S, Milstien S, Grant S. Endogenous modulators and pharmacological inhibitors of histone deacetylases in cancer therapy. Oncogene. 2012;31:537–51. doi: 10.1038/onc.2011.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daigle S, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.