INTRODUCTION
Patients with cystic fibrosis (CF) invariably suffer from serial respiratory tract infections. Staphylococcus aureus is an important pathogen in this setting (1), and is frequently isolated from patients with CF early in life and can chronically infect or colonize the lungs of these patients (2). In the US, S. aureus is the most prevalent respiratory pathogen among children with CF; in 2011, according to the Cystic Fibrosis Foundation Patient Registry, the overall S. aureus prevalence in CF patient airways was 68% (3). In particular, the prevalence of methicillin-resistant S. aureus (MRSA) in children with CF increased dramatically from 2% in 2001 to 26% in 2011 (3). This increase in MRSA has paralleled the emergence in the late 1990’s of community-associated (CA) MRSA strains among the general population in the US and elsewhere (4). CA-MRSA strains are genetically distinct from healthcare-associated (HA)-MRSA, and one distinguishing feature is the staphylococcal cassette chromosome mec (SCCmec); to date, eleven SCCmec types have been recognized (5, 6). SCCmec types IV and V are primarily associated with CA-MRSA isolates, while SCCmec types I, II and III characterize HA-MRSA. Airways of patients with CF were previously dominated by HA-MRSA strains (22). Most recently, reports in children have noted the dominance of CA-MRSA strain types in CF patients with newly acquired MRSA (7).
Studies of the microbiology of patients with CF have largely focused on the epidemiology of Pseudomonas aeruginosa airway infection (8). It has been proposed that once colonized with P. aeruginosa, patients remain chronically colonized with the same strain type (9). However, it has also been demonstrated that novel strain types may emerge subsequent to periods of antibiotic therapy (9, 10). Similarly, once detected in the respiratory tract of CF patients, MRSA will most likely persist in the airways and be repeatedly isolated from sputum samples (11, 12). CF patients infected with MRSA have more rapid decline in their lung function (as determined by FEV1% predicted), fail to recover lung function following an acute pulmonary exacerbation, receive more courses of intravenous (IV) antibiotic therapy, and have poorer growth and worse survival (13–17).
Data are lacking regarding molecular characterization of MRSA isolates recovered from serial respiratory specimens from CF patients, including characterization of strain variation, factors influencing strain variation, and whether strain variation influences clinical outcome. We demonstrated in a previous analysis (18) that the majority of recurrent MRSA skin infections in healthy children are caused by identical strain types. In the present study, we analyzed isolates of MRSA recovered from serial respiratory cultures from CF patients, using repetitive-sequence polymerase chain reaction (repPCR) to interrogate strain relatedness. Our objective was to determine whether MRSA strains repeatedly isolated from individual CF patients were identical over time or represented acquisition of new strains. We also investigated potential clinical and epidemiological factors contributing to strain variability within an individual patient.
MATERIALS AND METHODS
Study Design
We conducted a retrospective review of all children up to 18 years of age with CF from whom MRSA was isolated from ≥2 respiratory cultures between January 1, 2005 and December 31, 2011. At St. Louis Children’s Hospital (SLCH), CF patients present for quarterly routine outpatient visits in addition to medical care for acute pulmonary exacerbations. At these visits, sputum specimens (or deep throat swabs from patients too young to expectorate) and pulmonary function tests are obtained. Cases were identified based on the presence of an MRSA isolate recovered from the respiratory tract. Cultures were defined as “acute” if obtained during presentation with acute pulmonary symptoms or a change in health status (e.g., antibiotics were prescribed by their medical provider). “Surveillance” cultures were those collected at routine visits when patients were not considered acutely ill, and antibiotics were not prescribed following their visit. Specimen sources included sputum, deep throat swabs, tracheal aspirate and broncheoalveolar lavage. Each 2 consecutive cultures from an individual patient comprised a “pair.” Data collection focused on patient demographics, characterization of illness and hospitalization, time between episodes, and antibiotics prescribed. In addition, pulmonary function tests were reviewed in association with each culture and compared to relatedness of strains. The Washington University Human Research Protection Office approved the study procedures.
Microbiology and Molecular Characterization of Isolates
The SLCH microbiology laboratory routinely banks MRSA isolates recovered from the airways of patients with CF. These isolates were retrieved for the purposes of this study. Antibiotic susceptibility data were retrieved from the medical record. High-level mupirocin resistance was detected using a 200-µg mupirocin disk (Oxoid Microbiology Products, Hampshire, UK) in accordance with Clinical and Laboratory Standards Institute guidelines (19).
RepPCR was performed on all MRSA isolates to determine relatedness of strains within individual patients using the Diversilab Bacterial Barcodes system (bioMerieux, Durham, NC) as previously described (20, 21). A pair of isolates were considered to be identical if their similarity index (SI) was ≥95%. SCCmec characterization was performed using a multiplex PCR assay that detects and differentiates SCCmec types I–V as previously described (5).
Data Analysis
Variables were compared by Fisher’s exact test or chi-square test (categorical) and Mann-Whitney U test (continuous). P-values ≤0.05 were considered significant. Data analysis was performed using SPSS for Windows version 19.0 (IBM SPSS, Chicago, Illinois). Additionally, we analyzed effects of similar strain types on FEV1% predicted using a mixed random effects model, where similarity was a fixed effect and study subject was random (Mixed Procedure, SAS 9.3, SAS Institute, Cary, NC).
RESULTS
Patient Demographics and Culture Characteristics
We identified 54 CF patients with serial MRSA cultures. More than half (n=29; 54%) were female (Table 1); all were Caucasian and the median age at first positive MRSA culture during the study period was 12.5 years (range 11 days–18 years). The ΔF508 allele was present in 43 of 45 patients (96%) with available genotype; of those, 30 (70%) patients were homozygous and 13 (30%) were heterozygous for the ΔF508 mutation. There were 31 (57%) patients with 2 cultures, 15 (28%) with 3, 4 (7%) with 4, 3 (6%) with 5, and 1 (2%) with 7 cultures, totaling 145 respiratory cultures and 91 pairs of MRSA isolates. Of the 145 cultures, 71 (49%) were surveillance cultures and 74 (51%) were acute cultures. Thirty-eight (70%) patients had at least one acute culture during the study period. The median interval between positive cultures was 310 days (range 8–1573 days). Antibiotic resistance profiles demonstrated that 97.2% of isolates were resistant to erythromycin, 62.1% to clindamycin, 3.5% to trimethoprim-sulfamethoxazole, and 1.4% to mupirocin.
Table 1.
Characteristics of the 54 Cystic Fibrosis patients who maintained identical MRSA strain types over time compared to those who acquired distinct MRSA strains
| Patient Characteristics | Total N=54 (%) |
Distinct N=9 (%) |
Identical N=45 (%) |
p |
|---|---|---|---|---|
| Female | 29 (54) | 3 (33) | 26 (58) | 0.28 |
| Age, years, median [range] | 12.5 [0.03–18] | 12.9 [2–18] | 12.4 [0.03–18] | 0.47 |
| CFTR mutation ΔF508 genotype a (N=45) | 43/45 (93) | 7/7 (100) | 36/38 (92) | 1.00 |
| Number of cultures, median [range] | 2 [2–7] | 2 [2–5] | 2 [2–7] | 0.75 |
Includes both heterozygous (n=13) and homozygous (n=30) ΔF508 mutation CFTR mutations
Strain Typing Data by RepPCR and SCCmec
Overall, 5 strain types were detected by repPCR in our CF population (each strain type representing 53%, 30%, 12%, 4% and 1% of the MRSA isolates, respectively). Forty-five patients maintained the same strain type at all samplings and 9 patients acquired at least one distinct strain type over time as determined by repPCR. A total of 91 pairs were analyzed for strain relatedness. Of these, 81 (89%) pairs were identical strain types and 10 (11%) pairs were distinct strain types. Characteristics of patients who maintained identical strain times over time compared to those who acquired distinct strain types did not differ (Table 1). Table 2 demonstrates characteristics of the culture pairs comprised of identical and distinct strain types. Interval duration between positive cultures ranged from 8 to 1573 days (median 316 days) for identical pairs and 91 to 623 days (median 291 days) for distinct pairs (p=0.61). Prescription of antibiotics effective against MRSA for the first culture in a pair was not associated with acquisition of a distinct strain type (13% vs. 9% not prescribed antibiotics; p=0.74). Considering the clinical scenario at the time cultures were obtained, strain relatedness was similar in pairs of acute cultures (11% distinct), pairs of surveillance cultures (12% distinct) and pairs comprising 1 acute and 1 surveillance culture (10% distinct; p=0.96). Patients with a negative culture between a pair of positive MRSA cultures were more likely to be infected with a distinct strain type at the subsequent episode, although this did not reach statistical significance (20% vs. 8%; p=0.13).
Table 2.
Characteristics of MRSA culture pairs with identical and distinct MRSA strains
| Pair Characteristics | Total N=91 (%) |
Distinct N=10 (%) |
Identical N=81 (%) |
p |
|---|---|---|---|---|
| Interval between infections, days, median [range] | 310 [8–1573] | 291 [91–623] | 316 [8–1573] | 0.61 |
| Antibiotic effect | ||||
| Antibiotic effective against 1st culture in pair | 48 (53) | 6 (60) | 42 (52) | 0.74 |
| Antibiotic ineffective against 1st culture in pair or no antibiotic prescribed | 43 (47) | 4 (40) | 39 (48) | |
| Clinical presentation | ||||
| 2 acute cultures | 28 (31) | 3 (30) | 25 (31) | 0.96 |
| 2 surveillance cultures | 33 (36) | 4 (40) | 29 (36) | |
| 1 acute and 1 surveillance culture | 30 (33) | 3 (30) | 27 (33) | |
| Negative MRSA culture between pair | ||||
| Yes | 25 (27) | 5 (50) | 20 (25) | 0.13 |
| No | 66 (73) | 5 (50) | 61 (75) | |
| SCCmec type | ||||
| Different | 10 (11) | 5 (50) | 5 (6) | 0.001 |
| Same | 81 (89) | 5 (50) | 76 (94) |
More than half (n = 87; 60%) of patients with MRSA isolated from the respiratory tract had at least one concomitant pathogen isolated. These pathogens included P. aeruginosa (n = 44; 30%), Aspergillus spp. (n = 17; 12%), Haemophilus influenzae (n = 16; 11%), Achromobacter xylosoxidans (n = 7; 5%), Burkholderia cepacia complex (n = 5; 3%), and others (n = 20; 14%: Streptococcus pneumoniae, Group A β-hemolytic Streptococcus, Chrysobacterium sp., Flavobacterium sp., Klebsiella oxytoca, Stenotrophomonas maltophilia, Citrobacter freundii, methicillin-susceptible S. aureus, Pantoea agglomerans, Escherichia coli, Serratia sp., Mycobacterium abscessus, and viruses). Acute MRSA cultures were more likely to be accompanied by P. aeruginosa (39%) than surveillance MRSA cultures (21%; p=0.03).
As acquisition of a unique strain type could correlate with a change in pulmonary status, pulmonary function test results were reviewed for each culture and compared to strain relatedness. There was not a significant difference in the rate of decline in FEV1% predicted between patients having similar strain types (least square mean 0.52, standard error [SE] = 1.9) and those with distinct strain types (least square mean 2.54, SE = 4.1; p=0.66). However, episodes with concomitant pathogens had lower median FEV1% predicted [53 (range 17–138)] than episodes without concomitant pathogens [71 (range 23–144); p=0.006].
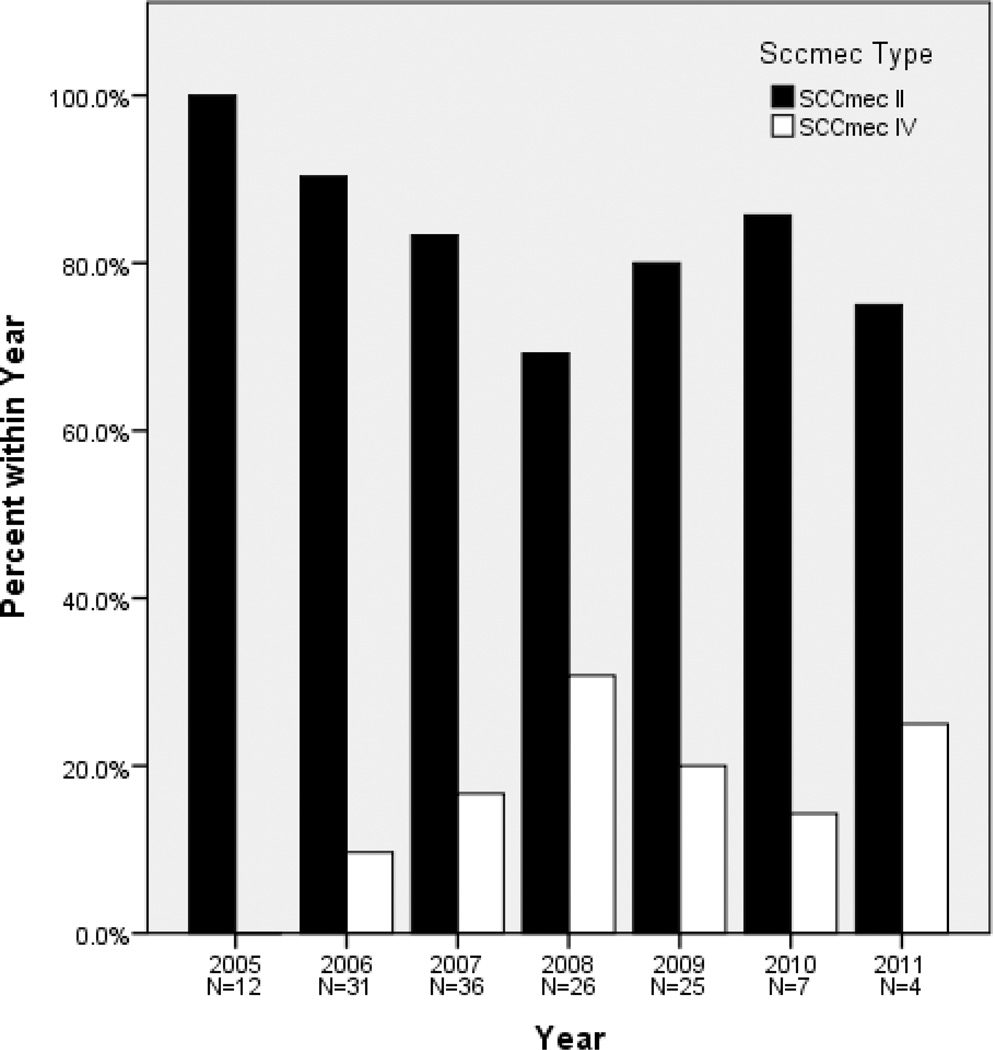
SCCmec typing revealed that 117 (83%) isolates contained SCCmec type II, 24 isolates (17%) possessed SCCmec type IV, and the SCCmec type of the other 4 isolates was not resolvable with our assay. In our cohort, isolates containing SCCmec type IV were first isolated in the year 2006 (Figure). SCCmec type II isolates were resistant to a greater number of antimicrobials compared to SCCmec type IV strains (Table 3). The 2 mupirocin-resistant isolates (recovered from the same patient) carried SCCmec type II (Table 3). Of note, 93% of SCCmec type II strains were clindamycin resistant, compared to 21% of SCCmec type IV strains (p < 0.001). As expected, MRSA culture pairs distinct by repPCR were more likely to have discordant SCCmec types than pairs identical by repPCR (50% vs. 6%; p=0.001) (Table 2). The discordance or concordance in SCCmec types of strains within a pair had no effect on the rate of decline in FEV1% predicted (p=0.57) (Table 4).
Figure.
SCCmec types of 141 MRSA isolates from 54 CF patients from St. Louis Children’s Hospital during a 7-year period. SCCmec type IV strains are emerging in this population.
Table 3.
Characteristics and Antibiotic Resistance Profiles for SCCmec Types Among MRSA Isolates in Children with Cystic Fibrosis
| Antimicrobial agenta | Prevalence of resistance among MRSA strains | |||
|---|---|---|---|---|
| MRSA Total N = 141 (%) |
SCCmec II N=117 (%) |
SCCmec IV N=24 (%) |
p | |
| Clindamycinb (N=141) | 114 (81) | 109 (93) | 5 (21) | <0.001 |
| Erythromycin (N=141) | 137 (97) | 115 (98) | 22 (92) | 0.134 |
| Trimethoprim-Sulfamethoxazole (N=138) | 5 (4) | 5 (4) | 0 (0) | 1.00 |
| Rifampin (N=113) | 9 (8) | 9 (8) | 0 (0) | 1.00 |
| Tetracycline (N=113) | 6 (5) | 6 (6) | 0 (0) | 1.00 |
| Mupirocin (N=141) | 2 (1) | 2 (2) | 0 (0) | 1.00 |
No linezolid resistance detected
Includes inducible resistance
Table 4.
Change in FEV1% Predicted by Discordance of SCCmec Type of Paired Isolates
| SCCmec Type of 1st Isolate in Pair |
SCCmec Type of 2nd Isolate in Pair |
Number of Pairs | Change in FEV1% Predicted (Least Square Mean, Standard Error) |
|---|---|---|---|
| SCCmec II | SCCmec II | 67 | decline 0.84, 2.0 |
| SCCmec II | SCCmec IV | 8 | decline 0.57, 5.7 |
| SCCmec IV | SCCmec II | 2 | decline 7.5, 10.7 |
| SCCmec IV | SCCmec IV | 10 | increase 2.6, 5.7 |
DISCUSSION
In this study, we describe the epidemiology and genetic background of MRSA in the population of a pediatric tertiary CF center over a 7-year period. The majority of CF patients with recurrent MRSA isolates recovered from the respiratory tract carried identical strain types longitudinally. Despite differences in the clinical status of CF patients harboring MRSA in the airway (acute pulmonary exacerbation vs. routine surveillance), an individual’s strain type generally remained constant over time. When antibiotics were prescribed for the first culture in a pair, no effect was observed on strain relatedness of the subsequent MRSA isolate. These results suggest that even with courses of antibiotic therapy, MRSA strains are not eradicated from the airways of CF patients.
The majority of MRSA isolates recovered from our patient population were characteristic of the predominant HA-MRSA clone (SCCmec type II), although we found that CA-MRSA (SCCmec type IV) has become more prevalent in CF patients in recent years. This is consistent with data reported from other CF centers collected between 2001–2007 in which 20% to 33% of MRSA isolates contained SCCmec type IV (7, 22–24). In healthcare settings worldwide, CA-MRSA strains are emerging as a significant cause of nosocomial infection, overtaking traditional HA-MRSA strains (4), resulting in hospital outbreaks in a variety of adult and pediatric healthcare settings (25–31). This influx of CA-MRSA into the hospitals is especially concerning for the CF population, as this vulnerable population is often exposed to antibiotics. While CA-MRSA strains have traditionally been less resistant to non-β-lactam antibiotics compared with HA-MRSA strains, this selective pressure may result in emergence of, or selection for, multidrug resistance in an already virulent staphylococcal strain (4).
To date, few studies have assessed the molecular epidemiology of repeated MRSA strains isolated from the airways of CF patients over time (22, 24, 32). Consistent with our findings, a study in the US comparing the molecular epidemiology of MRSA isolated from children with CF to healthy individuals over one year (2004–2005) employing multilocus sequence typing (MLST) and SCCmec typing methods found that 65% of CF patients harbored one specific clonal complex (CC), namely CC5, and contained SCCmec type II, while 89% of infections in non-CF children were due to CC8 MRSA isolates that contained SCCmec type IV (22). Cocchi et al., analyzed MRSA strains isolated from CF patients among 9 CF centers in Italy during a 2-year period (2004–2005) using MLST and SCCmec typing. Of 178 MRSA strains identified, 98 isolates belonged to known epidemic lineages. The largest clone was ST8-MRSA-IV which accounted for 29 strains collected from 6 different centers. SCCmec types I, II or III were found in 55% of the MRSA isolates, while 31% carried SCCmec type IV (32).
Conflicting evidence exists regarding the effect of antibiotics on the epidemiology of bacterial strains harbored by CF patients. While little is known about MRSA in particular, a study investigating P. aeruginosa in CF patients found that the emergence of distinct strains in the same CF patient was often associated with courses of antibiotic therapy (10). In a European study (33), stringent infection control and limited courses of IV antipseudomonal antibiotics were associated with maintaining a consistent strain type over time. Interestingly, at another CF center where less infection control measures were implemented, more courses of IV antipseudomonal chemotherapy were prescribed, and a broader armamentarium of antipseudomonal agents were used, strain replacement by 2 new predominant strain types occurred (33). These findings suggest that implementing infection control measures to prevent transmission between patients, while also limiting the spectrum and frequency of antipseudomonal antibiotics prescribed, modifies the epidemiology of chronic P. aeruginosa infections in CF patients.
In 2003, the US Cystic Fibrosis Foundation (CFF) and the Society for Healthcare Epidemiology of America (SHEA) published comprehensive infection control guidelines for CF patients, aiming to limit transmission of antibiotic-resistant pathogens (34). The guidelines recommend contact precautions for all CF patients infected or colonized with such resistant organisms and encourage improved respiratory and hand hygiene and appropriate cleaning of equipment and environmental surfaces. These guidelines have led to an ongoing clinical trial (clinicaltrials.gov identifier NCT01349192) for CF patients with recent MRSA acquisition, evaluating an eradication protocol including oral antimicrobials, nasal mupirocin, topical chlorhexidine body washes, and environmental decontamination.
Our data demonstrate that once a particular MRSA strain is isolated from the respiratory tract of a patient with CF, this strain type will most likely persist for a long period of time, even in the setting of intermittent antibiotic therapy. It is postulated that S. aureus employs a number of adaptive strategies in order to chronically persist in the biofilm of the CF lung, which may manifest as modulation of expression of genes associated with metabolism and virulence, conversion to small-colony variants, and acquisition or induction of antimicrobial resistance (35–38). Each of these phenotypic and genetic changes are complex biological phenomenon and appropriate for entire review articles on their own. However, specifically, the phenotype of S. aureus known as “small colony variants,” slowly-growing, often antibiotic resistant strains, have been associated with increased severity of lung disease in CF patients (39). These isolates can be difficult to detect in clinical cultures and it is thought that this adaptation facilities the persistence of these organisms in the hostile environment of the CF lung, especially in the context of ongoing antimicrobial therapy. The sustained presence of MRSA strain types despite intermittent antibiotic therapy supports this theory.
Our study is limited by its retrospective study design, such that data regarding clinical response and adherence to antibiotic therapy were not available. In addition, our data were collected from a single center, and thus the generalizability of our results may be limited. However, we were able to analyze a large collection of MRSA isolates from a unique patient population using two molecular methods over an extended period of time. In addition, microbiologic data were linked to patients’ clinical characteristics.
In conclusion, while the majority of MRSA strains carried by CF patients were traditional HA-MRSA strains, contemporary CA-MRSA strains were detected more frequently in recent years, a trend likely to continue. This study demonstrates that MRSA isolated from CF airways largely reflects the persistence of the same strain type over time. As antibiotics do not appear to influence strain persistence, future studies are needed to focus on prevention of acquisition rather than eradication of persistent MRSA.
ACKNOWLEDGMENTS
We wish to thank Drs. Albert Faro, David Warren, and Alexis Elward for guidance with study design; Dr. Michael Wallendorf for assistance with statistical analyses; and Drs. John Dickinson and David Hunstad for critical review of the manuscript. We also thank Rachel Collins for assistance in performing repPCR and the SLCH microbiology technologists for their support in banking MRSA isolates.
Source of Funding: This work was supported by National Institutes of Health grants K23-AI091690 and UL1-TR000448, and the Children’s Discovery Institute of Washington University and Saint Louis Children’s Hospital. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official view of the National Institutes of Health.
Footnotes
Conflicts of Interest: No other financial disclosures for all the authors.
REFERENCES
- 1.Goss CH, Muhlebach MS. Review: Staphylococcus aureus and MRSA in cystic fibrosis. J Cyst Fibros. 2011;10:298–306. doi: 10.1016/j.jcf.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong DS, Grimwood K, Carlin JB, et al. Lower airway inflammation in infants and young children with cystic fibrosis. Am J Respir Crit Care Med. 1997;156:1197–1204. doi: 10.1164/ajrccm.156.4.96-11058. [DOI] [PubMed] [Google Scholar]
- 3.Cystic Fibrosis Foundation Patient Registry. 2011 Annual Data Report. Bethesda, MD: Cystic Fibrosis Foundation; 2012. [Google Scholar]
- 4.Otter JA, French GL. Community-associated meticillin-resistant Staphylococcus aureus strains as a cause of healthcare-associated infection. J Hosp Infect. 2011;79:189–193. doi: 10.1016/j.jhin.2011.04.028. [DOI] [PubMed] [Google Scholar]
- 5.Boye K, Bartels MD, Andersen IS, Moller JA, Westh H. A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I–V. Clin Microbiol Infect. 2007;13:725–727. doi: 10.1111/j.1469-0691.2007.01720.x. [DOI] [PubMed] [Google Scholar]
- 6.Gostev VV, Sidorenko SV. Staphylococcal cassette chromosome mec, evolution and genetic lines of methicillin-resistant Staphylococcus aureus. Antibiot Khimioter. 2012;57:38–46. [PubMed] [Google Scholar]
- 7.Elizur A, Orscheln RC, Ferkol TW, et al. Panton-Valentine Leukocidin-positive methicillin-resistant Staphylococcus aureus lung infection in patients with cystic fibrosis. Chest. 2007;131:1718–1725. doi: 10.1378/chest.06-2756. [DOI] [PubMed] [Google Scholar]
- 8.Kidd TJ, Grimwood K, Ramsay KA, Rainey PB, Bell SC. Comparison of three molecular techniques for typing Pseudomonas aeruginosa isolates in sputum samples from patients with cystic fibrosis. J Clin Microbiol. 2011;49:263–268. doi: 10.1128/JCM.01421-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silbert S, Barth AL, Sader HS. Heterogeneity of Pseudomonas aeruginosa in Brazilian cystic fibrosis patients. J Clin Microbiol. 2001;39:3976–3981. doi: 10.1128/JCM.39.11.3976-3981.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boukadida J, De Montalembert M, Lenoir G, Scheinmann P, Veron M, Berche P. Molecular epidemiology of chronic pulmonary colonisation by Pseudomonas aeruginosa in cystic fibrosis. J Med Microbiol. 1993;38:29–33. doi: 10.1099/00222615-38-1-29. [DOI] [PubMed] [Google Scholar]
- 11.Solis A, Brown D, Hughes J, Van Saene HK, Heaf DP. Methicillin-resistant Staphylococcus aureus in children with cystic fibrosis: An eradication protocol. Pediatr Pulmonol. 2003;36:189–195. doi: 10.1002/ppul.10231. [DOI] [PubMed] [Google Scholar]
- 12.Vanderhelst E, De Wachter E, Willekens J, et al. Eradication of chronic methicillin-resistant Staphylococcus aureus infection in cystic fibrosis patients. An observational prospective cohort study of 11 patients. J Cyst Fibros. 2013 doi: 10.1016/j.jcf.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Vanderhelst E, De Meirleir L, Verbanck S, Pierard D, Vincken W, Malfroot A. Prevalence and impact on FEV(1) decline of chronic methicillin-resistant Staphylococcus aureus (MRSA) colonization in patients with cystic fibrosis. A single-center, case control study of 165 patients. J Cyst Fibros. 2012;11:2–7. doi: 10.1016/j.jcf.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Miall LS, McGinley NT, Brownlee KG, Conway SP. Methicillin-resistant Staphylococcus aureus (MRSA) infection in cystic fibrosis. Arch Dis Child. 2001;84:160–162. doi: 10.1136/adc.84.2.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dasenbrook EC, Checkley W, Merlo CA, Konstan MW, Lechtzin N, Boyle MP. Association between respiratory tract methicillin-resistant Staphylococcus aureus and survival in cystic fibrosis. JAMA. 2010;303:2386–2392. doi: 10.1001/jama.2010.791. [DOI] [PubMed] [Google Scholar]
- 16.Taccetti G, Neri AS, Festini F, Galici V, Cocchi P, Campana S. Methicillin-resistant Staphylococcus aureus in cystic fibrosis. Pediatr Pulmonol. 2008;43:309. doi: 10.1002/ppul.20774. [DOI] [PubMed] [Google Scholar]
- 17.Ren CL, Morgan WJ, Konstan MW, et al. Presence of methicillin resistant Staphylococcus aureus in respiratory cultures from cystic fibrosis patients is associated with lower lung function. Pediatr Pulmonol. 2007;42:513–518. doi: 10.1002/ppul.20604. [DOI] [PubMed] [Google Scholar]
- 18.Al-Zubeidi D, Burnham CD, Hogan GP, Collins R, Hunstad DA, Fritz SA. Molecular Epidemiology of Recurrent Cutaneous Methicillin-Resistant Staphylococcus aureus Infections in Children. JPIDS. 2013 doi: 10.1093/jpids/pit046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twenty-third informational supplement; M100-S23. Wayne, PA: Clinical and Laboratory Standards Institute; 2013. Jan, Update. [Google Scholar]
- 20.Fritz SA, Hogan PG, Camins BC, et al. Mupirocin and chlorhexidine resistance in Staphylococcus aureus in patients with community-onset skin and soft tissue infections. Antimicrob Agents Chemother. 2013;57:559–568. doi: 10.1128/AAC.01633-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Feghaly RE, Stamm JE, Fritz SA, Burnham CA. Presence of the bla(Z) beta-lactamase gene in isolates of Staphylococcus aureus that appear penicillin susceptible by conventional phenotypic methods. Diagn Microbiol Infect Dis. 2012;74:388–393. doi: 10.1016/j.diagmicrobio.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 22.Glikman D, Siegel JD, David MZ, et al. Complex molecular epidemiology of methicillin-resistant Staphylococcus aureus isolates from children with cystic fibrosis in the era of epidemic community-associated methicillin-resistant. S. aureus. Chest. 2008;133:1381–1387. doi: 10.1378/chest.07-2437. [DOI] [PubMed] [Google Scholar]
- 23.Goodrich JS, Sutton-Shields TN, Kerr A, Wedd JP, Miller MB, Gilligan PH. Prevalence of community-associated methicillin-resistant Staphylococcus aureus in patients with cystic fibrosis. J Clin Microbiol. 2009;47:1231–1233. doi: 10.1128/JCM.00255-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stone A, Quittell L, Zhou J, et al. Staphylococcus aureus nasal colonization among pediatric cystic fibrosis patients and their household contacts. Pediatr Infect Dis J. 2009;28:895–899. doi: 10.1097/inf.0b013e3181a3ad0a. [DOI] [PubMed] [Google Scholar]
- 25.Popovich KJ, Weinstein RA, Hota B. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin Infect Dis. 2008;46:787–794. doi: 10.1086/528716. [DOI] [PubMed] [Google Scholar]
- 26.Liu C, Graber CJ, Karr M, et al. A population-based study of the incidence and molecular epidemiology of methicillin-resistant Staphylococcus aureus disease in San Francisco, 2004–2005. Clin Infect Dis. 2008;46:1637–1646. doi: 10.1086/587893. [DOI] [PubMed] [Google Scholar]
- 27.Saiman L, O'Keefe M, Graham PL, et al. Hospital transmission of community-acquired methicillin-resistant Staphylococcus aureus among postpartum women. Clin Infect Dis. 2003;37:1313–1319. doi: 10.1086/379022. [DOI] [PubMed] [Google Scholar]
- 28.Centers for Disease Control and Prevention. Community-associated methicillin-resistant Staphylococcus aureus infection among healthy newborns--Chicago and Los Angeles County, 2004. MMWR Morb Mortal Wkly Rep. 2006;55:329–332. [PubMed] [Google Scholar]
- 29.Hulten KG, Kaplan SL, Lamberth LB, et al. Hospital-acquired Staphylococcus aureus infections at Texas Children's Hospital, 2001–2007. Infect Control Hosp Epidemiol. 2010;31:183–190. doi: 10.1086/649793. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez BE, Rueda AM, Shelburne SA, Musher DM, Hamill RJ, Hulten KG. Community-associated strains of methicillin-resistant Staphylococccus aureus as the cause of healthcare-associated infection. Infect Control Hosp Epidemiol. 2006;27:1051–1056. doi: 10.1086/507923. [DOI] [PubMed] [Google Scholar]
- 31.Healy CM, Hulten KG, Palazzi DL, Campbell JR, Baker CJ. Emergence of new strains of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. Clin Infect Dis. 2004;39:1460–1466. doi: 10.1086/425321. [DOI] [PubMed] [Google Scholar]
- 32.Cocchi P, Cariani L, Favari F, et al. Molecular epidemiology of meticillin-resistant Staphylococcus aureus in Italian cystic fibrosis patients: a national overview. J Cyst Fibros. 2011;10:407–411. doi: 10.1016/j.jcf.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Cramer N, Wiehlmann L, Ciofu O, Tamm S, Hoiby N, Tummler B. Molecular epidemiology of chronic Pseudomonas aeruginosa airway infections in cystic fibrosis. PLoS One. 2012;7:e50731. doi: 10.1371/journal.pone.0050731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saiman L, Siegel J. Cystic Fibrosis Foundation. Infection control recommendations for patients with cystic fibrosis: microbiology, important pathogens, and infection control practices to prevent patient-to-patient transmission. Infect Control Hosp Epidemiol. 2003;24:S6–S52. doi: 10.1086/503485. [DOI] [PubMed] [Google Scholar]
- 35.Goerke C, Wolz C. Adaptation of Staphylococcus aureus to the cystic fibrosis lung. International Journal of Medical Microbiology. 2010;300:520–525. doi: 10.1016/j.ijmm.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell G, Grondin G, Bilodeau G, Cantin AM, Malouin F. Infection of polarized airway epithelial cells by normal and small-colony variant strains of Staphylococcus aureus is increased in cells with abnormal cystic fibrosis transmembrane conductance regulator function and is influenced by NF-kappaB. Infect Immun. 2011;79:3541–3551. doi: 10.1128/IAI.00078-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yagci S, Hascelik G, Dogru D, Ozcelik U, Sener B. Prevalence and genetic diversity of Staphylococcus aureus small-colony variants in cystic fibrosis patients. Clin Microbiol Infect. 2013;19:77–84. doi: 10.1111/j.1469-0691.2011.03742.x. [DOI] [PubMed] [Google Scholar]
- 38.Dasenbrook EC. Update on methicillin-resistant Staphylococcus aureus in cystic fibrosis. Curr Opin Pulm Med. 2011;17:437–441. doi: 10.1097/MCP.0b013e32834b95ed. [DOI] [PubMed] [Google Scholar]
- 39.Wolter DJ, Emerson JC, McNamara S, et al. Staphylococcus aureus small-colony variants are independently associated with worse lung disease in children with cystic fibrosis. Clin Infect Dis. 2013;57:384–391. doi: 10.1093/cid/cit270. [DOI] [PMC free article] [PubMed] [Google Scholar]