Abstract
At least 600000 individuals worldwide annually die of hepatitis B virus (HBV)-related diseases, such as chronic hepatitis B (CHB), liver cirrhosis (LC), and hepatocellular carcinoma (HCC). Many viral factors, such as viral load, genotype, and specific viral mutations, are known to affect disease progression. HBV reverse transcriptase does not have a proofreading function, therefore, many HBV genotypes, sub-genotypes, mutants, and recombinants emerge. Differences between genotypes in response to antiviral treatment have been determined. To date, 10 HBV genotypes, scattered across different geographical regions, have been identified. For example, genotype A has a tendency for chronicity, whereas viral mutations are frequently encountered in genotype C. Both chronicity and mutation frequency are common in genotype D. LC and progression to HCC are more commonly encountered with genotypes C and D than the other genotypes. Pathogenic differences between HBV genotypes explain disease intensity, progression to LC, and HCC. In conclusion, genotype determination in CHB infection is important in estimating disease progression and planning optimal antiviral treatment.
Keywords: Hepatitis B virus, Genotypes, Chronic hepatitis B, Anti-viral therapy, Viral mutation
Core tip: Hepatitis B virus (HBV) infection is the leading cause of chronic liver disease and death worldwide. The clinical course and consequences of HBV infection are affected by several factors such as viral load, mutation, host, environment, and viral genotypes. Different HBV genotypes are associated with different mutations in the HBV precore and core promoter gene regions. HBV genotypes are closely related with optimal treatment strategy for chronic hepatitis B patients and clinical outcomes.
INTRODUCTION
HBV is an enveloped, hepatotropic, non-cytopathic virus that can cause acute and chronic hepatitis[1]. Although there is currently a safe vaccine against hepatitis B virus (HBV), it remains a severe public health issue, especially in Asia, Africa and South America, and may result in death. HBV infection may lead to a variety of clinical pictures, ranging from asymptomatic carrier state to acute hepatitis, fulminant hepatitis, chronic hepatitis, liver cirrhosis (LC) and hepatocellular carcinoma (HCC). Progression of chronic hepatitis B (CHB) disease to severe liver diseases, such as LC and HCC, is determined by the genetic characteristics of the host, as well as by viral and environmental factors[2,3].
GLOBAL DISTRIBUTION OF HBV GENOTYPES AND SUBGENOTYPES
HBV is differentiated into many genotypes, according to genome sequence. To date, eight well-known genotypes (A-H) of the HBV genome have been defined. Moreover, two new genotypes, I and J, have also been identified. Some HBV genotypes are further classified as sub-genotypes. HBV sequence is characterized by > 8% nucleotide differences for genotype, and 4%-8% nucleotide differences for sub-genotype. Over 30 related sub-genotypes belonging to HBV genotypes have been determined to date, but the mechanisms of different pathogenic characteristics of HBV genotypes are not known for certain. Many studies have reported that different genotypes and sub-genotypes show different geographical distribution, and are related to disease progression, clinical progression, response to antiviral treatment, and prognosis. A-D and F genotypes are divided into various sub-genotypes; no sub-genotypes have been defined for E, G and H genotypes[1,3-5]. Genotype A is widespread in sub-Saharan Africa, Northern Europe, and Western Africa; genotypes B and C are common in Asia; genotype C is primarily observed in Southeast Asia; genotype D is dominant in Africa, Europe, Mediterranean countries, and India; genotype G is reported in France, Germany, and the United States; and genotype H is commonly encountered in Central and South America. Genotype I has recently been reported in Vietnam and Laos. The newest HBV genotype, genotype J, has been identified in the Ryukyu Islands in Japan. Geographic distribution of HBV genotypes may be related to route of exposure. For example, genotypes B and C are more common in high-endemic regions of perinatal or vertical exposure, which plays an important role in viral transmission. Other genotypes are primarily observed in regions of horizontal exposure[6-9]. Therefore, genotyping provides an epidemiological clue in the investigation of acquisition, because this lies in the geographical distribution of HBV. Figure 1 shows genotype distribution across the world.
Figure 1.
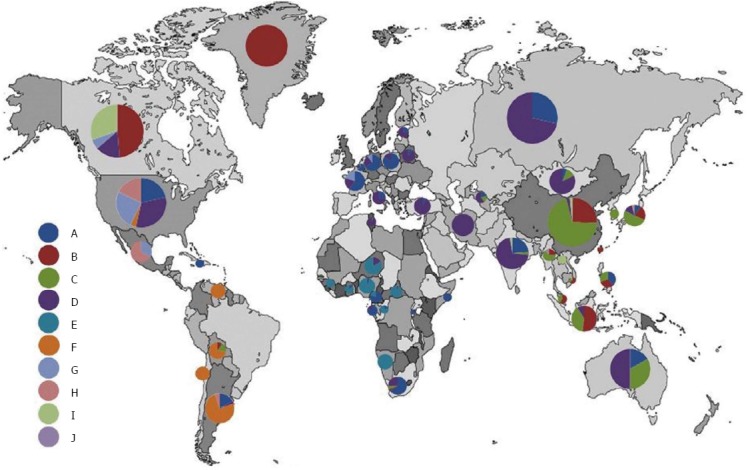
Geographic distribution of hepatitis B virus genotypes worldwide. Permission for Figure 1 has been granted by the Publisher, from Shi et al[4].
Identification of HBV genotype is important for many reasons. An epidemiological study conducted in China showed that genotype B was primarily distributed in the south and genotype C in the north of the country. Clear HBV genotype-related associations exist between clinical outcomes and treatment efficacy in patients with CHB[10]. Genotype G was initially identified in studies conducted in France in 2000. It was found during co-infection with other genotypes, especially HBV/A2. HBV/C and H genotypes have also been reported with co-infection[11]. The most important characteristics of HBV epidemiology in India are the dominance of genotype D and the increased frequency of hepatitis B e antigen (HBeAg)-negative chronic infection. To date, nine (D1-D9) sub-genotypes of genotype D have been identified. However, A/D recombinant species of HBV have been identified in chronic HBV patients in Northern India[12].
Genotype distribution shows variations between countries, and even between geographical regions within countries. The HBV genotype and sub-genotype distributions between countries are shown in Table 1.
Table 1.
Distribution of genotypes and sub-genotypes among countries
| Country | Genotypes | Sub-genotypes | Ref. |
| China | B, C | B2, C1, C2 | Lin et al[10] |
| Indonesia | C, B | C1, B3, B7, C10, B9 and C8 | Siburian et al[13] |
| Prasetyo et al[14] | |||
| Tunisia | D, F | - | Ayari et al[15] |
| Turkey | D | D2, D1, D3 | Sunbul et al[16] |
| Brazil | A, F | A1, F2a, A2, F4 | Moura et al[3] |
| Nabuco et al[17] | |||
| Vietnam | B, C, I | B2-5, C5-16 | Shi[18] |
| Taiwan | B | B2,B5 | Kao[19] |
| South Korea | C | - | Kao[19] |
| Hong Kong | C,B | - | Kao[19] |
| Gambia, Nigeria, Haiti, Congo, Rwanda, Cameroon | A | A4, A5, A6, A7 | Shi[18] |
| Japan | A, C | C1, C2, C3 | Sakai et al[20] |
| Kobayashi et al[21] | |||
| Philippines | A, B, C | A1, B5, C5 | Sakamoto et al[7] |
| India | A, C, D | - | Biswas et al[2] |
| Canada | C, B, A, D | Congly et al[22] | |
| Central African | A, D, E | A1,D4 | Komas et al[23] |
| Republic | |||
| Saudi Arabia | D, E | D1 | Khan et al[24] |
| Iran | D | D1 | Geramizadeh et al[25] |
| Norouzi et al[26] | |||
| Mongolia | D | - | Oyunsuren et al[27] |
| South Africa | D | D3 | Yousif et al[28] |
| Thailand | C, B | C1-5 | Louisirirotchanakul et al[29] |
| Italy | D | - | Lampertico et al[30] |
| Morocco | D, A | D1, D7, A2 | Baha et al[31] |
| Argentina | F | F1, F2, F4 | Torres et al[32] |
| Egypt | D | D1 | Ragheb et al[33] |
| Pakistan | D | - | Ali et al[34] |
| Australia | C, D | C4, D4 | Davies et al[35] |
| Sugauchi et al[36] | |||
| Spain | A, D, F | - | Buti et al[37] |
Pathogenic differences between various HBV genotypes are now partially understood. Intracellular levels of HBV DNA and extracellular levels of HBV DNA and HBeAg have been revealed as higher in genotypes B and C than in genotypes A and D. HBV DNA and intracellular accumulation of viral antigens may play a role in the development of cellular damage in hepatocytes. In addition, the high replication capacity of genotype C may be the reason for increased genotype-related severe hepatic damage[19]. The following findings were reported from an in vitro study: (1) when a pre-core (PC) or basal core promoter (BCP) region mutation affected HBeAg expression in genotype C, intracellular HBV core protein expression was increased; (2) In PC wild-type HBV genotype C patients, intracellular HBV surface protein expression was lower than in HBV genotype B patients; (3) Extracellular HBV DNA was lower in PC-mutant patients; (4) there was less hepatitis B surface antigen (HBsAg) formation in HBV genotype C than in genotype B; and (5) there was less secretion of HBeAg in HBV genotype B than in genotype C[38].
CLINICAL IMPORTANCE OF HBV GENOTYPES
A greater understanding of the relationship between HBV genotypes, progression of hepatitis B disease, and clinical outcomes has developed over time. Clinical outcomes of chronic HBV infections are variable, and many viral factors, such as host factors, HBV genotype, specific viral mutations, viral load, and quantitative HBsAg levels are important in their prediction. HBV genotypes in viral factors are not only predictive of clinical progression, but are also related to interferon (IFN)-α treatment response[6]. In a study comparing genotypes B and C, alanine aminotransferase (ALT) levels were higher in patients with genotype C. However, the reason for this is not yet known[39]. The primary clinical and virological features among HBV genotypes are shown in Table 2[9].
Table 2.
Comparison of clinical and virological features among hepatitis B virus genotypes
| Genotype | B | C | A | D | E-J |
| Clinical characteristics | |||||
| Modes of transmission | Perinatal/vertical | Perinatal/vertical | Horizontal | Horizontal | Horizontal |
| Tendency of chronicity | Lower | Higher | Higher | Lower | ND |
| Positivity of HBeAg | Lower | Higher | Higher | Lower | ND |
| HBeAg Seroconversion | Earlier | Later | Earlier | Later | ND |
| HBsAg seroclearance | More | Less | More | Less | ND |
| Histological activity | Lower | Higher | Lower | Higher | ND |
| Clinical outcomes (LC, HCC) | Better | Worse | Better | Worse | Worse in genotype F |
| Response to INF-α | Higher | Lower | Higher | Lower | Lower in genotype G |
| Response to nucleos(t)ide analogs | No significant differences among genotypes A to D | ND | |||
| Virological characteristics | |||||
| Serum HBV DNA level | Lower | Higher | ND | ND | ND |
| Frequency of PC A1896 mutation | Higher | Lower | Lower | Higher | ND |
| Frequency of basal core promoterT1762/A1764 mutation | Lower | Higher | Higher | Lower | ND |
| Frequency of preS deletion mutation | Lower | Higher | ND | ND | ND |
ND: No data available; HBsAg: Hepatitis B surface antigen; HBeAg: Hepatitis B e antigen; LC: Liver cirrhosis; HCC: Hepatocellular carcinoma; HBV: Hepatitis B virus.
A study conducted in China investigated the reasons for the longer immune clearance period in HBV patients infected with genotype C compared with genotype B; higher level of viral replication; high hepatic histological activity, recurrent or persistently high ALT levels and IFN, nucleos(t)ide analogs; and low response to treatment. The possible relationship among genotypes B and C and peripheral blood follicular helper T (Tfh) cells in CHB patients under treatment was investigated. Tfh cells play a major role in spreading signals that affect cellular division; help with activation of B cells; and regulate the humoral response. In addition, Tfh cells secrete specific cytotoxic T lymphocyte (CTL) interleukin (IL)-21 in order to sustain long-acting, effective, antiviral immunity in chronic infection. High serum HBV DNA and ALT ratios in patients with genotype C might be related to lower peripheral blood Tfh cell levels, which would cause low IL-21 levels, when compared to genotype B. It has been reported that HBV-specific CTL levels are lower[40].
TENDENCY TO CHRONICITY DEVELOPMENT AFTER ACUTE HBV INFECTION
There are differences in date obtained from studies that reviewed genotypes and chronicity. Some recent studies showed that progression to chronic infection was increased in individuals with acute infection due to HBV genotype A[41,42]. However, a study conducted in China reported that chronic infection developed more frequently in patients with C2 sub-genotype than in those with sub-genotype B2, and genotype C2 was an independent risk factor for chronicity development[43]. Studies using a limited number of patients concluded that in those with genotypes A and D, chronicity ratios were higher than in patients with genotypes B and C[21,44]. In a Japanese study, the ratio of persistent HBV infection development after acute hepatitis B infection was higher in patients with genotype A than in those with genotypes B and C. It was also reported that chronicity ratio after HBV infection was relatively higher in patients with genotype D[6]. In addition, chronicity of HBV infection after acute hepatitis B infection was explained by genotype, as well as multifactorial reasons, such as the amount of viral inoculum, route of acquisition, and different interactions between host and virus[45].
HBeAg SEROCONVERSION AND HBsAg SEROCLEARANCE
HBeAg seroconversion and HBsAg seroclearance are the most important steps in the natural progression of HBV infection, and the annual incidence of these are 12% and 2%, respectively. Earlier HBeAg seroconversion is generally accepted as a positive outcome. Conversely, delayed seroconversion, or its absence, after recurrent hepatitis attacks may indicate progression of chronic hepatitis to LC. Following their study in Taiwanese patients, Lin et al[45] reported that spontaneous HBeAg seroconversion in patients with genotype C was lower than that in genotype B patients (27% vs 47%, P < 0.025). HBeAg seroconversion rate in genotype B patients is lower than genotype C patients, and a more lengthy persistence of HBV replication explains why LC and HCC development was found in patients with this genotype.
HBV GENOTYPES AND ANTIVIRAL TREATMENT
IFN-based therapy
Predictive parameters for response to IFN treatment are age, HBV DNA level, sex, ALT, hepatic inflammatory activity index, HBeAg status, and genotype. Many studies have indicated the role of HBV genotypes in response to IFN treatment, and response is greater with regard to genotypes A and B than genotypes C and D, with the worst response to IFN being observed with genotype D[3,18,46]. An experimental study investigating the effect of IFN showed that IFN/Peg-IFN was more effective in genotypes A or B than in genotypes C, D and I[47]. In a separate study, different serological and virological results were obtained for the varying HBV genotypes. In addition, it was determined that HBsAg and HBV DNA kinetics were specific for genotype. During IFN treatment, the most rapid decrease in quantitative HBsAg level was observed in genotypes A and B, and the best serological responses were obtained after treatment discontinuation in these genotypes. It was indicated that genotype C patients receiving IFN treatment reached HBV DNA negativity the earliest. In the same study, it was reported that genotype E was the most difficult genotype to treat, and that a longer period of time was required for treatment[46].
Biochemical and serological response rates in patients with genotypes A and B were significantly higher than in patients with genotypes C and D at 6-12 mo after IFN treatment was discontinued. Similarly, it was reported that persistent HBeAg clearance frequency was higher in patients with genotypes A and B, compared to patients with genotypes C and D 3 years after treatment was discontinued. During longer follow-up periods, the HBsAg clearance ratio in HBeAg-negative patients with genotype A who were treated with IFN was markedly higher than in the patients with the other genotypes. Moreover, there were differences between HBV genotypes in HBsAg clearance kinetics during IFN treatment. For example, the mean decrease in HBsAg levels at the end of treatment was the highest in patients with genotype A, was moderate in those with genotypes B and D, and was lowest in patients with genotypes C and E. Although decreases in serum HBsAg levels continued in patients with HBV genotypes A and D during the follow-up period, HBsAg rebound was observed in those with genotypes B and E. In two large global studies, ALT levels were higher in IFN-treated, HBeAg-positive patients with high ALT levels, or in genotype A patients with low HBV DNA levels. In addition, there was a possibility of a persistent response with IFN treatment in genotype B and C patients with lower HBV DNA, and it was concluded that IFN should be assessed for its therapeutic use. Conversely, HBV genotype D patients had the lowest persistent response independently of ALT and HBV DNA levels, so IFN treatment should be recommended for these patients[19,48]. In conclusion, identification of HBV genotype provides the clinician with very important clues with regard to the treatment response of disease and disease progression.
IFN-α shows antiviral, immunomodulatory, antiproliferative, and gene induction activities through binding to type I IFN receptor (IFNAR). In another study conducted in China, hepatic expression of type I IFN-α receptor β subunit (IFNAR2) in CHB infection in response to IFN treatment and its relationships with HBV genotypes was investigated. It was reported that IFNAR2 was expressed at high rates in liver in genotype B patients, and better response rates were obtained with IFN treatment when compared to patients with other HBV genotypes[48].
In a similar manner to chronic hepatitis C patients, correlation between IFN treatment and IL28B polymorphism has been investigated in CHB patients in recent years. Lampertico et al[49] studied HBeAg-negative CHB patients, who are known to be difficult to treat, investigating the correlation between IFN and IL28B polymorphisms, and HBsAg clearance was accepted as the response at the end of treatment. It was shown that HBsAg seroclearance and persistent viral responses were increased in patients with the IL28B CC genotype. Therefore, IL28B polymorphism might be an additional predictor in HBeAg-negative genotype D patients with regard to the optimization and discontinuation of the treatment.
Nucleos(t)-ide analog therapy
Treatment responses similar to lamivudine, adefovir, entecavir and telbivudine have been shown among different HBV genotypes in many clinical studies[45,50-52]. A recent meta-analysis showed that there was no difference in the response between HBV genotypes and nucleos(t)-ide analogs[53].
PROGRESSION TO LC OR HCC
LC and HCC are the most severe complications of hepatitis B. Therefore, studies conducted in the past decade have focused particularly on the correlation between HCC and HBV variants, and correlations between two important mutations of the HBV virus, PC mutant in nucleotide 1896 and BCP mutants in nucleotides 1762 and 1764, as well as clinical intensity of the disease, were investigated in detail. A study conducted in China, on HBeAg-positive patients infected by the HBV/C1 sub-genotype, reported that, in addition toT1762/A1764 BCP mutations, V1753 or/and A1768 mutations were closely correlated with HCC. It has been shown that increased HCC risk caused by BCP variants was partially based on modifications of biological functions of HBx protein[54]. In a study conducted in Turkish patients, a correlation between T1773 and T1764/G1766 mutations and high viral load was observed, but a definite correlation between BCP, PC, and/or core region mutations and disease prognosis could not be determined[55]. Many similar studies have indicated that genotype C caused more common severe liver diseases, such as LC and HCC, than the other genotypes caused. Concomitant presence of high serum HBV DNA levels, mutations, such as 1653T, 1753V and A1762T/G1764A, as well as acute hepatic failure, LC and HCC have also been noted. In addition, subsequent studies similarly showed that HBV genotype C infection caused mutations more frequently than did genotype B infection. It is currently accepted that there is a positive correlation between HBeAg expression and HBV DNA level, which is the HBV replication indicator. Some prospective studies have also indicated that HCC risk is increased as basal serum HBV DNA levels are increased[18,56,57].
In a meta-analysis investigating the correlation between HBV genotypes and HCC, HCC detection was 12% for genotype B, and 25% for genotype C (OR = 2.05, 95%CI: 1.52-2.76, P < 0.001). Moreover, a correlation between genotype C patients and more severe liver diseases has been reported. Conversely, genotype A (14%) and genotype C (11%) have a similar HCC risk. In accordance with previous studies, another meta-analysis showed that HCC development was associated more with genotype C patients than with patients with the other HBV genotypes[58].
In a study conducted in India, Ghosh et al[59] showed a correlation between advanced clinical stage and mutation frequency in genotype D patients with HBeAg-negative chronic HBV infection. Many of these mutations were localized in regions that regulated transcription (BCP/EnhII/NRE/SP1). According to the results, deletion at the preS region was the most important predictor of LC. Moreover, the only clinically important mutation was identified as S183P at the HBV core protein C terminus, which was out of the ectopic region, and this caused HBV genotype D to shift from an inactive carrier state to progression to CHB and LC. It was also reported that G1896A and G1899A PC mutants in HBV were correlated with more aggressive liver diseases, as has previously been reported in several studies. Therefore, it was indicated that high-risk patients could be identified before disease progression by the use of viral genomic markers, and thereby incidences of LC and HCC decreased. Conversely, an investigation of correlation between sub-genotypes and severe liver diseases indicated that sub-genotype B1 was related to fulminant hepatitis B infection in Japan, whereas sub-genotype B2 was related to HCC and HCC recurrence in East Asia. A separate study reported that risk of HCC was high in sub-genotype C2 infections[18,56,57].
A study carried out in South Korea showed that HBV mutations were significantly related to HCC in CHB patients infected by genotype C, especially in major histocompatibility complex class II-restricted regions. In the prec/C region, there are six (preC-W28*, C-P5H/L/T, C-E83D,C-I97F/L, C-L100I and C-Q182K/*) and seven (preC-W28*, preC-G29D, C-D32N/H, CE43K, C-P50A/H/Y, C-A131G/N/P and C-S181H/P) types of mutations that are related to HCC, and these define HBeAg serostatus. The results showed that HBV variants in the C region could lead to HCC progression in chronic patients infected by genotype C by the immune escape pathway against the CD4 T-cell-mediated immune response. Therefore, it was concluded that HCC-related hepatitis B core antigen mutations and HBeAg serostatus could be used as diagnostic markers for early diagnosis of liver disease progression, including HCC[60]. Interestingly, many studies reported that HCC development was observed in patients at an advanced age with genotype C, whereas this occurred earlier in genotype B patients[61-63].
Conversely, in the study conducted in India, it was noted that subtypes D1 and D3 were accompanied by chronic and occult infections, respectively[4,12]. However, a different study reported that there was a correlation between occult hepatitis and genotype C[64].
CONCLUSION
Nowadays, There is a large accumulation of knowledge regarding HBV, as well as hepatitis B disease and treatment. HBV genotype varies according to countries and ethnic backgrounds. Chronicity of the disease, response to antiviral treatment, and progression of LC and HCC differ according to genotypes, which also enables the physician to individualize the treatment and to identify disease-related risks. In addition to the existent potent antivirals currently used in treatment, the introduction of new antivirals, such as core inhibitors, which inhibit covalently closed circular DNA activity, may make it possible to achieve more definite treatment outcomes in the near future. The positive outcomes of a routine neonatal vaccination program will be observed more closely in the coming years. However, much remains unknown about HBV disease, so further studies, especially into its complex immunopathogenesis, are required for clarification and understanding.
Footnotes
P- Reviewers: Kietzmann T, Koch TR, Rostami-Nejad M, Yalniz M S- Editor: Qi Y L- Editor: Kerr C E- Editor: Ma S
References
- 1.Huang CC, Kuo TM, Yeh CT, Hu CP, Chen YL, Tsai YL, Chen ML, Chou YC, Chang C. One single nucleotide difference alters the differential expression of spliced RNAs between HBV genotypes A and D. Virus Res. 2013;174:18–26. doi: 10.1016/j.virusres.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Biswas A, Panigrahi R, Pal M, Chakraborty S, Bhattacharya P, Chakrabarti S, Chakravarty R. Shift in the hepatitis B virus genotype distribution in the last decade among the HBV carriers from eastern India: possible effects on the disease status and HBV epidemiology. J Med Virol. 2013;85:1340–1347. doi: 10.1002/jmv.23628. [DOI] [PubMed] [Google Scholar]
- 3.Moura IF, Lopes EP, Alvarado-Mora MV, Pinho JR, Carrilho FJ. Phylogenetic analysis and subgenotypic distribution of the hepatitis B virus in Recife, Brazil. Infect Genet Evol. 2013;14:195–199. doi: 10.1016/j.meegid.2012.11.022. [DOI] [PubMed] [Google Scholar]
- 4.Shi W, Zhang Z, Ling C, Zheng W, Zhu C, Carr MJ, Higgins DG. Hepatitis B virus subgenotyping: history, effects of recombination, misclassifications, and corrections. Infect Genet Evol. 2013;16:355–361. doi: 10.1016/j.meegid.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 5.Cooksley WG. Do we need to determine viral genotype in treating chronic hepatitis B? J Viral Hepat. 2010;17:601–610. doi: 10.1111/j.1365-2893.2010.01326.x. [DOI] [PubMed] [Google Scholar]
- 6.Liu CJ, Kao JH. Global perspective on the natural history of chronic hepatitis B: role of hepatitis B virus genotypes A to J. Semin Liver Dis. 2013;33:97–102. doi: 10.1055/s-0033-1345716. [DOI] [PubMed] [Google Scholar]
- 7.Sakamoto T, Tanaka Y, Orito E, Co J, Clavio J, Sugauchi F, Ito K, Ozasa A, Quino A, Ueda R, et al. Novel subtypes (subgenotypes) of hepatitis B virus genotypes B and C among chronic liver disease patients in the Philippines. J Gen Virol. 2006;87:1873–1882. doi: 10.1099/vir.0.81714-0. [DOI] [PubMed] [Google Scholar]
- 8.Schaefer S. Hepatitis B virus taxonomy and hepatitis B virus genotypes. World J Gastroenterol. 2007;13:14–21. doi: 10.3748/wjg.v13.i1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allain JP. Epidemiology of Hepatitis B virus and genotype. J Clin Virol. 2006;36 Suppl 1:S12–S17. doi: 10.1016/s1386-6532(06)80003-x. [DOI] [PubMed] [Google Scholar]
- 10.Lin S, Liu C, Shang H, Chen H, Yang B, Chen J, Chen Y, Chen D, Ou Q. HBV serum markers of 49164 patients and their relationships to HBV genotype in Fujian Province of China. J Clin Lab Anal. 2013;27:130–136. doi: 10.1002/jcla.21573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakamoto T, Tanaka Y, Watanabe T, Iijima S, Kani S, Sugiyama M, Murakami S, Matsuura K, Kusakabe A, Shinkai N, et al. Mechanism of the dependence of hepatitis B virus genotype G on co-infection with other genotypes for viral replication. J Viral Hepat. 2013;20:e27–e36. doi: 10.1111/jvh.12022. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh S, Banerjee P, Deny P, Mondal RK, Nandi M, Roychoudhury A, Das K, Banerjee S, Santra A, Zoulim F, et al. New HBV subgenotype D9, a novel D/C recombinant, identified in patients with chronic HBeAg-negative infection in Eastern India. J Viral Hepat. 2013;20:209–218. doi: 10.1111/j.1365-2893.2012.01655.x. [DOI] [PubMed] [Google Scholar]
- 13.Siburian MD, Utama A, Dhenni R, Arnelis N, Fanany I, Intan MD, Kurniasih TS, Andriani F, Afadlal S, Julianto EB, et al. High prevalence of hepatitis B virus genotype C/C1 in the Minangkabau ethnic group in Indonesia. Virol J. 2013;10:27. doi: 10.1186/1743-422X-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prasetyo AA, Dirgahayu P, Sari Y, Hudiyono H, Kageyama S. Molecular epidemiology of HIV, HBV, HCV, and HTLV-1/2 in drug abuser inmates in central Javan prisons, Indonesia. J Infect Dev Ctries. 2013;7:453–467. doi: 10.3855/jidc.2965. [DOI] [PubMed] [Google Scholar]
- 15.Ayari R, Lakhoua-Gorgi Y, Bouslama L, Safar I, Kchouk FH, Aouadi H, Jendoubi-Ayed S, Najjar T, Ayed K, Abdallah TB. Investigation of DNA sequence in the Basal core promoter, precore, and core regions of hepatitis B virus from Tunisia shows a shift in genotype prevalence. Hepat Mon. 2012;12:e6191. doi: 10.5812/hepatmon.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sunbul M, Leblebicioglu H. Distribution of hepatitis B virus genotypes in patients with chronic hepatitis B in Turkey. World J Gastroenterol. 2005;11:1976–1980. doi: 10.3748/wjg.v11.i13.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nabuco LC, Mello FC, Gomes Sde A, Perez RM, Soares JA, Coelho HS, Nogueira CA. Hepatitis B virus genotypes in Southeast Brazil and its relationship with histological features. Mem Inst Oswaldo Cruz. 2012;107:785–789. [PubMed] [Google Scholar]
- 18.Shi YH. Correlation between hepatitis B virus genotypes and clinical outcomes. Jpn J Infect Dis. 2012;65:476–482. doi: 10.7883/yoken.65.476. [DOI] [PubMed] [Google Scholar]
- 19.Kao JH. Molecular epidemiology of hepatitis B virus. Korean J Intern Med. 2011;26:255–261. doi: 10.3904/kjim.2011.26.3.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakai T, Shiraki K, Inoue H, Okano H, Deguchi M, Sugimoto K, Ohmori S, Murata K, Fujioka H, Takase K, et al. HBV subtype as a marker of the clinical course of chronic HBV infection in Japanese patients. J Med Virol. 2002;68:175–181. doi: 10.1002/jmv.10180. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi M, Suzuki F, Arase Y, Akuta N, Suzuki Y, Hosaka T, Saitoh S, Kobayashi M, Tsubota A, Someya T, et al. Infection with hepatitis B virus genotype A in Tokyo, Japan during 1976 through 2001. J Gastroenterol. 2004;39:844–850. doi: 10.1007/s00535-004-1400-3. [DOI] [PubMed] [Google Scholar]
- 22.Congly SE, Wong P, Al-Busafi SA, Doucette K, Fung SK, Ghali P, Fonseca K, Myers RP, Osiowy C, Coffin CS. Characterization of hepatitis B virus genotypes and quantitative hepatitis B surface antigen titres in North American tertiary referral liver centres. Liver Int. 2013;33:1363–1369. doi: 10.1111/liv.12222. [DOI] [PubMed] [Google Scholar]
- 23.Komas NP, Vickos U, Hübschen JM, Béré A, Manirakiza A, Muller CP, Le Faou A. Cross-sectional study of hepatitis B virus infection in rural communities, Central African Republic. BMC Infect Dis. 2013;13:286. doi: 10.1186/1471-2334-13-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan A, Al Balwi MA, Tanaka Y, Hajeer A, Sanai FM, Al Abdulkarim I, Al Ayyar L, Badri M, Saudi D, Tamimi W, et al. Novel point mutations and mutational complexes in the enhancer II, core promoter and precore regions of hepatitis B virus genotype D1 associated with hepatocellular carcinoma in Saudi Arabia. Int J Cancer. 2013;133:2864–2871. doi: 10.1002/ijc.28307. [DOI] [PubMed] [Google Scholar]
- 25.Geramizadeh B, Nikeghbalian S, Kazemi K, Shamsaifar A, Bahador A, Salahi H, Malekhosseini SA, Kashtkar Jahromi M, Bakhshai Dehkordi A. Hepatocellular carcinoma in explanted livers of patients with genotype d HBV cirrhosis: report of the first experience from Iran. Arch Iran Med. 2013;16:348–350. [PubMed] [Google Scholar]
- 26.Norouzi M, Ghorashi S, Abedi F, Nejatizadeh A, Ataei B, Malekzadeh R, Alavian S, Judaki M, Ghamari S, Namazi A, et al. Identification of Hepatitis B Virus Surface Antigen (HBsAg) Genotypes and Variations in Chronic Carriers from Isfahan Province, Iran. Iran J Public Health. 2012;41:104–111. [PMC free article] [PubMed] [Google Scholar]
- 27.Oyunsuren T, Kurbanov F, Tanaka Y, Elkady A, Sanduijav R, Khajidsuren O, Dagvadorj B, Mizokami M. High frequency of hepatocellular carcinoma in Mongolia; association with mono-, or co-infection with hepatitis C, B, and delta viruses. J Med Virol. 2006;78:1688–1695. doi: 10.1002/jmv.20755. [DOI] [PubMed] [Google Scholar]
- 28.Yousif M, Kramvis A. Genotype D of hepatitis B virus and its subgenotypes: An update. Hepatol Res. 2013;43:355–364. doi: 10.1111/j.1872-034X.2012.01090.x. [DOI] [PubMed] [Google Scholar]
- 29.Louisirirotchanakul S, Olinger CM, Arunkaewchaemsri P, Poovorawan Y, Kanoksinsombat C, Thongme C, Sa-Nguanmoo P, Krasae S, Theamboonlert A, Oota S, et al. The distribution of hepatitis B virus genotypes in Thailand. J Med Virol. 2012;84:1541–1547. doi: 10.1002/jmv.23363. [DOI] [PubMed] [Google Scholar]
- 30.Lampertico P, Viganò M, Di Costanzo GG, Sagnelli E, Fasano M, Di Marco V, Boninsegna S, Farci P, Fargion S, Giuberti T, et al. Randomised study comparing 48 and 96 weeks peginterferon α-2a therapy in genotype D HBeAg-negative chronic hepatitis B. Gut. 2013;62:290–298. doi: 10.1136/gutjnl-2011-301430. [DOI] [PubMed] [Google Scholar]
- 31.Baha W, Ennaji MM, Lazar F, Melloul M, El Fahime E, El Malki A, Bennani A. HBV genotypes prevalence, precore and basal core mutants in Morocco. Infect Genet Evol. 2012;12:1157–1162. doi: 10.1016/j.meegid.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 32.Torres C, Piñeiro y Leone FG, Pezzano SC, Mbayed VA, Campos RH. New perspectives on the evolutionary history of hepatitis B virus genotype F. Mol Phylogenet Evol. 2011;59:114–122. doi: 10.1016/j.ympev.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 33.Ragheb M, Elkady A, Tanaka Y, Murakami S, Attia FM, Hassan AA, Hassan MF, Shedid MM, Abdel Reheem HB, Khan A, et al. Multiple intra-familial transmission patterns of hepatitis B virus genotype D in north-eastern Egypt. J Med Virol. 2012;84:587–595. doi: 10.1002/jmv.23234. [DOI] [PubMed] [Google Scholar]
- 34.Ali M, Idrees M, Ali L, Hussain A, Ur Rehman I, Saleem S, Afzal S, Butt S. Hepatitis B virus in Pakistan: a systematic review of prevalence, risk factors, awareness status and genotypes. Virol J. 2011;8:102. doi: 10.1186/1743-422X-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies J, Littlejohn M, Locarnini SA, Whiting S, Hajkowicz K, Cowie BC, Bowden DS, Tong SY, Davis JS. Molecular epidemiology of hepatitis B in the Indigenous people of northern Australia. J Gastroenterol Hepatol. 2013;28:1234–1241. doi: 10.1111/jgh.12177. [DOI] [PubMed] [Google Scholar]
- 36.Sugauchi F, Mizokami M, Orito E, Ohno T, Kato H, Suzuki S, Kimura Y, Ueda R, Butterworth LA, Cooksley WG. A novel variant genotype C of hepatitis B virus identified in isolates from Australian Aborigines: complete genome sequence and phylogenetic relatedness. J Gen Virol. 2001;82:883–892. doi: 10.1099/0022-1317-82-4-883. [DOI] [PubMed] [Google Scholar]
- 37.Buti M, Rodriguez-Frias F, Jardi R, Esteban R. Hepatitis B virus genome variability and disease progression: the impact of pre-core mutants and HBV genotypes. J Clin Virol. 2005;34 Suppl 1:S79–S82. doi: 10.1016/s1386-6532(05)80015-0. [DOI] [PubMed] [Google Scholar]
- 38.Chen RY, Bowden S, Desmond PV, Dean J, Locarnini SA. Effects of interferon alpha therapy on the catalytic domains of the polymerase gene and basal core promoter, precore and core regions of hepatitis B virus. J Gastroenterol Hepatol. 2003;18:630–637. doi: 10.1046/j.1440-1746.2003.03019.x. [DOI] [PubMed] [Google Scholar]
- 39.Xibing G, Xiaojuan Y, Juanhua W. PD-1 expression on CTL may be related to more severe liver damage in CHB patients with HBV genotype C than in those with genotype B infection. J Viral Hepat. 2013;20:e1–e2. doi: 10.1111/jvh.12009. [DOI] [PubMed] [Google Scholar]
- 40.Xibing G, Xiaojuan Y, Juanhua W, Zhong H. Relationship between HBV genotypes B, C and follicular helper T cells in patients with chronic hepatitis B and its significance. Hepat Mon. 2013;13:e6221. doi: 10.5812/hepatmon.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogawa M, Hasegawa K, Naritomi T, Torii N, Hayashi N. Clinical features and viral sequences of various genotypes of hepatitis B virus compared among patients with acute hepatitis B. Hepatol Res. 2002;23:167–177. doi: 10.1016/s1386-6346(01)00176-0. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki Y, Kobayashi M, Ikeda K, Suzuki F, Arfase Y, Akuta N, Hosaka T, Saitoh S, Kobayashi M, Someya T, et al. Persistence of acute infection with hepatitis B virus genotype A and treatment in Japan. J Med Virol. 2005;76:33–39. doi: 10.1002/jmv.20320. [DOI] [PubMed] [Google Scholar]
- 43.Zhang HW, Yin JH, Li YT, Li CZ, Ren H, Gu CY, Wu HY, Liang XS, Zhang P, Zhao JF, et al. Risk factors for acute hepatitis B and its progression to chronic hepatitis in Shanghai, China. Gut. 2008;57:1713–1720. doi: 10.1136/gut.2008.157149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wai CT, Fontana RJ, Polson J, Hussain M, Shakil AO, Han SH, Davern TJ, Lee WM, Lok AS. Clinical outcome and virological characteristics of hepatitis B-related acute liver failure in the United States. J Viral Hepat. 2005;12:192–198. doi: 10.1111/j.1365-2893.2005.00581.x. [DOI] [PubMed] [Google Scholar]
- 45.Lin CL, Kao JH. The clinical implications of hepatitis B virus genotype: Recent advances. J Gastroenterol Hepatol. 2011;26 Suppl 1:123–130. doi: 10.1111/j.1440-1746.2010.06541.x. [DOI] [PubMed] [Google Scholar]
- 46.Boglione L, D’Avolio A, Cariti G, Milia MG, Simiele M, De Nicolò A, Ghisetti V, Di Perri G. Sequential therapy with entecavir and PEG-INF in patients affected by chronic hepatitis B and high levels of HBV-DNA with non-D genotypes. J Viral Hepat. 2013;20:e11–e19. doi: 10.1111/jvh.12018. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, Wu Y, Ye S, Wang T, Zhao R, Chen F, Abe K, Jin X. The response to interferon is influenced by hepatitis B virus genotype in vitro and in vivo. Virus Res. 2013;171:65–70. doi: 10.1016/j.virusres.2012.10.027. [DOI] [PubMed] [Google Scholar]
- 48.Fan HB, Guo YB, Zhu YF, Chen AS, Zhou MX, Li Z, Xu LT, Ma XJ, Yan FM. Hepatitis B Virus Genotype B and High Expression of Interferon Alpha Receptor β Subunit are Associated With Better Response to Pegylated Interferon Alpha 2a in Chinese Patients With Chronic Hepatitis B Infection. Hepat Mon. 2012;12:333–338. doi: 10.5812/hepatmon.6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lampertico P, Viganò M, Cheroni C, Facchetti F, Invernizzi F, Valveri V, Soffredini R, Abrignani S, De Francesco R, Colombo M. IL28B polymorphisms predict interferon-related hepatitis B surface antigen seroclearance in genotype D hepatitis B e antigen-negative patients with chronic hepatitis B. Hepatology. 2013;57:890–896. doi: 10.1002/hep.25749. [DOI] [PubMed] [Google Scholar]
- 50.Liu CJ, Kao JH, Chen DS. Therapeutic implications of hepatitis B virus genotypes. Liver Int. 2005;25:1097–1107. doi: 10.1111/j.1478-3231.2005.01177.x. [DOI] [PubMed] [Google Scholar]
- 51.Kao JH. Hepatitis B viral genotypes: clinical relevance and molecular characteristics. J Gastroenterol Hepatol. 2002;17:643–650. doi: 10.1046/j.1440-1746.2002.02737.x. [DOI] [PubMed] [Google Scholar]
- 52.Liu CJ, Kao JH. Genetic variability of hepatitis B virus and response to antiviral therapy. Antivir Ther. 2008;13:613–624. [PubMed] [Google Scholar]
- 53.Wiegand J, Hasenclever D, Tillmann HL. Should treatment of hepatitis B depend on hepatitis B virus genotypes? A hypothesis generated from an explorative analysis of published evidence. Antivir Ther. 2008;13:211–220. [PubMed] [Google Scholar]
- 54.Li W, Chen G, Yu X, Shi Y, Peng M, Wei J. Accumulation of the mutations in basal core promoter of hepatitis B virus subgenotype C1 increase the risk of hepatocellular carcinoma in Southern China. Int J Clin Exp Pathol. 2013;6:1076–1085. [PMC free article] [PubMed] [Google Scholar]
- 55.Sunbul M, Sugiyama M, Kurbanov F, Leblebicioglu H, Khan A, Elkady A, Tanaka Y, Mizokami M. Specific mutations of basal core promoter are associated with chronic liver disease in hepatitis B virus subgenotype D1 prevalent in Turkey. Microbiol Immunol. 2013;57:122–129. doi: 10.1111/1348-0421.12011. [DOI] [PubMed] [Google Scholar]
- 56.Chen Y, Yu D, Zhang W, Qiu C, Xiang G, Dai W, Wu S, Wang X. HBV Subgenotype C2 Infection, A1762T/G1764A Mutations May Contribute To Hepatocellular Carcinoma with Cirrhosis in Southeast China. Iran J Public Health. 2012;41:10–18. [PMC free article] [PubMed] [Google Scholar]
- 57.Malmström S, Eilard A, Larsson SB, Hannoun C, Norkrans G, Lindh M. Genotype impact on long-term virological outcome of chronic hepatitis B virus infection. J Clin Virol. 2012;54:321–326. doi: 10.1016/j.jcv.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 58.Wong GL, Chan HL, Yiu KK, Lai JW, Chan VK, Cheung KK, Wong EW, Wong VW. Meta-analysis: The association of hepatitis B virus genotypes and hepatocellular carcinoma. Aliment Pharmacol Ther. 2013;37:517–526. doi: 10.1111/apt.12207. [DOI] [PubMed] [Google Scholar]
- 59.Ghosh S, Mondal RK, Banerjee P, Nandi M, Sarkar S, Das K, Santra A, Banerjee S, Chowdhury A, Datta S. Tracking the naturally occurring mutations across the full-length genome of hepatitis B virus of genotype D in different phases of chronic e-antigen-negative infection. Clin Microbiol Infect. 2012;18:E412–E418. doi: 10.1111/j.1469-0691.2012.03975.x. [DOI] [PubMed] [Google Scholar]
- 60.Kim DW, Lee SA, Hwang ES, Kook YH, Kim BJ. Naturally occurring precore/core region mutations of hepatitis B virus genotype C related to hepatocellular carcinoma. PLoS One. 2012;7:e47372. doi: 10.1371/journal.pone.0047372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yin J, Zhang H, Li C, Gao C, He Y, Zhai Y, Zhang P, Xu L, Tan X, Chen J, et al. Role of hepatitis B virus genotype mixture, subgenotypes C2 and B2 on hepatocellular carcinoma: compared with chronic hepatitis B and asymptomatic carrier state in the same area. Carcinogenesis. 2008;29:1685–1691. doi: 10.1093/carcin/bgm301. [DOI] [PubMed] [Google Scholar]
- 62.Ni YH, Chang MH, Wang KJ, Hsu HY, Chen HL, Kao JH, Yeh SH, Jeng YM, Tsai KS, Chen DS. Clinical relevance of hepatitis B virus genotype in children with chronic infection and hepatocellular carcinoma. Gastroenterology. 2004;127:1733–1738. doi: 10.1053/j.gastro.2004.09.048. [DOI] [PubMed] [Google Scholar]
- 63.Kao JH, Chen PJ, Lai MY, Chen DS. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology. 2000;118:554–559. doi: 10.1016/s0016-5085(00)70261-7. [DOI] [PubMed] [Google Scholar]
- 64.Kim H, Lee SA, Kim DW, Lee SH, Kim BJ. Naturally occurring mutations in large surface genes related to occult infection of hepatitis B virus genotype C. PLoS One. 2013;8:e54486. doi: 10.1371/journal.pone.0054486. [DOI] [PMC free article] [PubMed] [Google Scholar]


