Abstract
AIM: To identify potential biomarkers of primary gallbladder cancer (PGC).
METHODS: Fresh PGC, cholecystitis and normal gallbladder tissue specimens collected from 10 patients, respectively, were subjected to comparative proteomic analysis. The proteomic patterns of PGC were compared with those of cholecystitis and normal gallbladder tissues using two-dimensional gel electrophoresis (2-DE). The differentially expressed proteins were then identified using a MALDI-TOF mass spectrometer (MS) and database searches. To further validate these proteins, 20 samples of PGC tissues and normal tumor-adjacent tissues were collected for Western blot, quantitative real-time PCR, and immunohistochemical staining assay.
RESULTS: Seven differentially expressed protein spots were detected by 2-ED analysis by comparing the average maps of PGC, cholecystitis and normal gallbladder tissues. Six of the seven differentially expressed proteins were identified using MALDI-TOF MS, with three overexpressed and three underexpressed in PGC tissue. Protein levels of annexin A4 (ANXA4) were significantly elevated, and heat shock protein 90-beta (Hsp90β) and dynein cytoplasmic 1 heavy chain 1 (Dync1h1) were decreased in PGC tissues relative to the normal tumor-adjacent tissues as shown by Western blot analysis. However, levels of actin, aortic smooth muscle and gamma-actin were unchanged. In addition, the mRNA levels of all 5 proteins showed similar changes to those of the protein levels (P < 0.01). Further validation by immunohistochemical analysis showed the upregulated expression of ANXA4 and decreased expression of Hsp90β and Dync1h1 in the cytoplasm of PGC tissues relative to the normal tumor-adjacent tissues.
CONCLUSION: Three proteins are identified as potential biomarkers of PGC using proteomic analysis. The functions of these proteins in the carcinogenesis of PGC remain to be studied.
Keywords: Primary gallbladder cancer, Proteomic analysis, Annexin A4, Dynein cytoplasmic 1 heavy chain 1, Heat shock protein 90-beta, Biomarker
Core tip: A new comparative proteomic study using human primary gallbladder cancer (PGC), cholecystitis and normal gallbladder tissues to identify biomarkers of PGC was performed. Three new potential biomarkers of PGC were validated and may be informative in understanding the carcinogenic mechanisms and monitoring the treatment of PGC.
INTRODUCTION
Primary gallbladder carcinoma (PGC) is the most frequent biliary tract malignancy and the fifth most common malignant neoplasm involving the gastrointestinal tract. PGC has a high morbidity and poor prognosis as it often shows early metastasis and invasiveness[1]. The incidence of PGC varies globally, with the highest incidence in Poland and Northern India, and in Native American and South American populations. Each year 17000-18000 new cases of PGC are diagnosed in India with a comparable annual death rate. Despite recent advances in the diagnosis and management of gastrointestinal cancers, PGC remains a challenge with a poor overall prognosis. Many PGC tumors are not resectable at the time of presentation, and the 5-year survival rate in patients with PGC is < 10% in most reported series[2,3]. An investigation of the carcinogenesis and molecular mechanisms of PGC development has the potential to improve clinical strategies for treatment and disease outcomes. Biomarkers which are involved in the development and progression of PGC are needed. To date, no reliable molecular markers have been established for PGC, although some genes show promise[4]. However, studies on candidate genes for PGC susceptibility have not yet confirmed any associations, and data are limited on the consistent molecular changes associated with gallbladder carcinogenesis and disease progression[2].
Completion of the human genome project has allowed the identification of genes involved in the appearance, progression, and treatment of cancer, as well as information on the functions of specific genes, interactions among genes in communication networks, and the activities of their protein products in molecular pathways. In most cases, however, although studies on differential mRNA expression are informative, they do not always correlate with protein concentrations as proteins are often subjected to proteolytic cleavage or post-translational modifications such as phosphorylation or glycosylation.
Proteomics is a complementary method to genetic profiling. In addition to genetic and epigenetic alterations, other molecular changes in translation, post-translational modification and intracellular mislocalization are involved in tumor initiation and growth. Factors involved in these changes cannot be detected by measuring amounts of RNA or detecting nucleotide sequence variations[5]. Thus, discovery strategies for cancer biomarkers often target expressed proteins.
Proteomics analysis has been applied to many types of tumors including cancers of the breast, prostate, liver, esophagus, and stomach[6-10]. Proteomics can overcome some limitations of the approaches used to elucidate the molecular mechanisms of PGC. Although proteomic studies on PGC have included cancer cell lines and blood samples[11-13], few studies have applied a proteomic approach to tumor tissues from PGC patients. Compared to proteomic studies using cancer cell lines and blood samples, studies using tumor tissue should be more informative in terms of carcinogenic mechanisms, prognostic evaluation and therapy for PGC. Therefore, we aimed to identify biomarkers in human PGC tissue using proteomics as a high throughput method. The results presented here provide information for further studies on the carcinogenic mechanisms, diagnosis, and therapy of PGC.
MATERIALS AND METHODS
Materials
Tissue specimens were collected from patients at Shanghai Chang Zheng Hospital Affiliated to the Second Military Medical University, in accordance with human subject guidelines approved by the Scientific and Ethical Committee of the Second Military Medical University, Shanghai, China. All samples were obtained by experienced surgeons and examined by experienced pathologists. For proteomics analysis, fresh PGC, cholecystitis and normal gallbladder tissues were obtained from 10 patients, respectively, from 2006 to 2008. Normal gallbladder tissues were obtained from patients undergoing liver transplantation. Informed consent was obtained from all patients or their relatives for the use of tissues in experimental procedures. All specimens, which were from surgical resections, were immediately frozen in liquid nitrogen and stored at -80 °C until use. PGC patients did not receive antineoplastic therapy prior to surgery.
An IEF system (IPGphor), Immobiline Dry Strips (18 cm, pH 4-7 NL) and CLEAN-UP Kits were from Amersham Biosciences (Piscataway, NJ, United States). Iodoacetamide was from Sigma-Aldrich (St. Louis, MO, United States). Experiments were carried out using an ABI 4700 MALDI-TOF/TOF mass spectrometer (MS) (Applied Biosystems, Framingham, MA, United States). Sequence-grade trypsin was from Promega (Madison, WI, United States), and the standard peptide mixture used for calibration was from Applied Biosystems (Foster City, CA, United States). All buffers were prepared with Milli-Q water.
Sample preparation
Tissue samples were crushed and ground in a mortar containing liquid nitrogen. The resulting powder was immediately suspended in a lysis buffer (7 mol/L urea, 2 mol/L thiourea, 4% CHAPS, 65 mmol/L DTT, 2% IPG buffer, and trace cocktail protease inhibitor). After vigorous stirring for 1 h, cell debris and insoluble substances were removed by centrifugation at 100000 g for 45 min. Protein concentration was determined using a modified Bradford method as described previously[14]. Supernatants were aliquoted and stored at -80 °C.
Two-dimensional electrophoresis
Two-dimensional electrophoresis (2-DE) was performed as described previously with minor modifications[15]. Briefly, 100 μg of protein sample was diluted to 170 μL with a rehydration solution (7 mol/L urea, 2 mol/L thiourea, 2 g/L DTT, 5 mL/L pH 4-7 IPG buffer and trace bromphenol blue) and applied to IPG strips (18 cm, pH 4-7 NL) for 14 h rehydration. Proteins were focused on an IPGphor IEF system (Amersham Biosciences) for 61 kVh. Strips were equilibrated in equilibration buffer (25 mmol/L Tris-HCl, pH 8.8, 6 mol/L urea, 200 mL/L glycerol, 20 g/L SDS, and 130 mmol/L DTT) for 15 min, followed by 15 min in the same buffer containing 200 mmol/L iodoacetamide instead of DTT. For 2-DE, 120 mL/L SDS-PAGE gels were used and stained using CBB R-350 (Merck, Germany) according to the supplier’s protocol. Each sample was run in triplicate.
Image analysis
2-DE maps were obtained by scanning the gels using an Imagescanner. Gel analysis was performed using PDQuest analysis software and included background subtraction, spot detection, volume normalization and the establishment of a reference gel. Proteins were considered differentially expressed if: (1) the spot was observed on 2D images in six samples; (2) intensities of the corresponding spots showed a difference of > 2-fold variation in PGC tissues as compared with normal gallbladder and cholecystitis tissues; and (3) intensities of the corresponding spots in cholecystitis tissues were between the intensities in normal gallbladder and PGC tissues.
In-gel digestion and MS
Selected protein spots were excised from the gels and destained. In-gel digestion was carried out with 0.01 g/L trypsin (Promega, Madison, WI, United States) for 15 h at 37 °C. Tryptic peptides were extracted from the gels and dried by centrifugal lyophilization. Peptide mixtures were redissolved in 5 mL/L TFA. Finally, the digested peptide samples were co-crystallized with an equal volume of saturated matrix solution [CHCA in 1 mL/L TFA in H2O/ACN (2:1)] on MALDI sample target plates. Peptide mass spectra were obtained with a MALDI-TOF/TOF MS (4700 Proteomics Analyzer, Applied Biosystems, Foster City, CA, United States).
Protein spots were identified by MS and MS/MS as described by Jeong et al[16]. Before sample acquisition, six external standards (mass standard kit for the 4700 proteomics analyzer calibration mixture, Applied Biosystems) were used to calibrate each spectrum to a mass accuracy of within 5 ppm for MS Reflector Positive Operating Mode or within 10 ppm for MS-MS 1 KV Positive Operating Mode. A combined database search of MS and MS/MS measurements was performed using GPS Explorer™ software (Version 3.6; Applied Biosystems, Foster City, CA, United States) and Mascot software (Version 2.0; Matrix Science, London, United Kingdom). Searches were performed with carbamidomethylation of cysteine and oxidation of methionine as variable modifications. One trypsin miscleavage was allowed. Peptide mass tolerance and fragment mass tolerance were set to 50 ppm and ± 0.3 Da, respectively. Peptide mixtures that yielded statistically significant search scores (> 95%CI: equivalent to Mascot expected value < 0.05) and accounted for the majority of ions present in the mass spectrum were defined as positive identifications. Redundant proteins that appeared in the database under different names and accession numbers were eliminated. If more than one protein was identified for a single spot, the protein member with the highest protein score was selected.
Western blot
All proteins to be validated were separated by SDS-PAGE with 20 μg protein per lane and transferred to PVDF membranes (Millipore, Billerica, MA, United States) in transfer buffer [pH = 11.0, 25 mmol/L Tris, 0.2 mol/L glycine, 20% (v/v) methanol] for 45 min at 1.5 mA/cm2 on a semidry electroblotter (Bio-Rad Laboratories, Richmond, VA, United States). After blocking with 1% skim milk for 1 h at 25 °C, the membranes were probed with primary antibody against annexin A4 (ANXA4), heat shock protein 90-beta (Hsp90β), dynein cytoplasmic 1 heavy chain 1 (Dync1h1), gamma-actin (ACTG), actin, aortic smooth muscle actin (ACTA2), or α-tubulin (Santa Cruz Biotechnology, CA, United States) at 1:300 dilution overnight at 4 °C. Horseradish peroxidase-conjugated rabbit anti-goat IgG at 1:3000 dilution (AMS Biotechnology, Oxon, United Kingdom) was used as a secondary antibody. The immunoblots were developed using ECL detection reagent (Pierce Chemical, Rockford, IL, United States).
Immunohistochemistry assay
Sections were subjected to routine deparaffination and rehydration. Antigen retrieval was achieved by microwaving in 0.01 mol/L citrate buffer for 10 min and then cooling for 30 min. The endogenous peroxidase activity was inhibited by incubation with 30 mL/L hydrogen peroxide in methanol for 20 min and nonspecific binding was blocked by incubation with 50 mL/L bovine serum albumin at room temperature. After washing, the specimens were reacted overnight at 4 °C with primary antibody against ANXA4, Hsp90β or Dync1h1 (Santa Cruz Biotechnology). After incubation with horseradish peroxidase-conjugated IgG, the signal was developed with 3,30-diaminobenzidine tetrahydrochloride in Tris-HCl buffer (pH = 7.6) containing 0.2 mL/L hydrogen peroxide. The sections were then counterstained with hematoxylin and mounted. Negative controls were prepared by replacing the primary antibody with nonspecific IgG at the same concentration.
Quantitative real-time polymerase chain reaction
Total RNA was extracted from tissues or cells at indicated time points and was subsequently reverse-transcribed using the AMV Reverse Transcription System (Takara, Shiga, Japan). Quantitative real-time polymerase chain reaction (PCR) was performed using SYBR Green PCR mix on an ABI Prism 7900HT (Applied Biosystems, Foster City, CA, United States). Thermocycler conditions included 2 min incubation at 50 °C, then 95 °C for 10 min; this was followed by a 2-step PCR program, as follows: 95 °C for 15 s and 60 °C for 60 s for 40 cycles. β-actin was used as an internal control to normalize differences in the amount of total RNA in each sample. The primers are listed in Table 1.
Table 1.
Quantitative real-time Polymerase chain reaction primers
| Name | Sequence |
| β-actin forward | 5’-CTCCATCCTGGCCTCGCTGT-3’ |
| β-actin reverse | 5’-GCTGTCACCTTCACCGTTCC-3’ |
| ANXA4 forward | 5’-GAGCACCATCGGCAGGGACT-3’ |
| ANXA4 reverse | 5’-TCATACAGCACCGTGGGCGT-3’ |
| Hsp90β forward | 5’-AGAAGGTTGAGAAGGTGACAA-3’ |
| Hsp90β reverse | 5’-AAGAGTAGAGAGGGAATGGG-3’ |
| Dync1h1 forward | 5’-AGTTGGTGGAATGTGGGTTG-3’ |
| Dync1h1 reverse | 5’-TGATTGATCTGGGTGATCTGA-3’ |
| ACTG forward | 5’-GGCATGGAGTCAGCTGGAATT-3’ |
| ACTG reverse | 5’-TCTGCATCCTGTCAGCAATCC-3’ |
| ACTA2 forward | 5’-CCTCCCTTGAGAAGAGTTACG-3’ |
| ACTA2 reverse | 5’-GAGCAGGAAAGTGTTTTAGAA-3’ |
ANXA4: Antibody against annexin A4; ACTG: Gamma-actin.
Statistical analysis
Statistical analysis was performed using the two-tailed Student’s t test, and P < 0.05 was considered statistically significant. Data are expressed as mean ± SE of triplicate samples, and reproducibility was confirmed in three separate experiments.
RESULTS
2-DE profiling of proteins from PGC, cholecystitis and normal gallbladder tissues
Fresh PGC, cholecystitis and normal gallbladder tissues from 10 patients, respectively (mean age, 59 ± 7.4 years for PGC; 48.8 ± 10.6 years for cholecystitis; and 46.4 ± 14.3 years for normal gallbladder), were analyzed in triplicate by 2-DE. Image analysis confirmed that the 2-DE maps were well resolved and reproducible. Coomassie staining of 2D gels detected 1483 ± 76 protein spots from PGC samples, 1460 ± 62 from cholecystitis samples, and 1514 ± 73 from normal gallbladder tissues in the pH range of 4-7 (Figure 1). An average electrophoresis map was constructed for human PGC, cholecystitis and normal gallbladder tissues by combining ten 2-DE maps using PDQuest 7.1.1 2-DE gel analysis software. Seven differentially expressed protein spots were detected by comparing the average maps of PGC, cholecystitis and normal gallbladder tissues. Of these spots, three were significantly upregulated and four were downregulated in PGC tissues (Figure 1).
Figure 1.
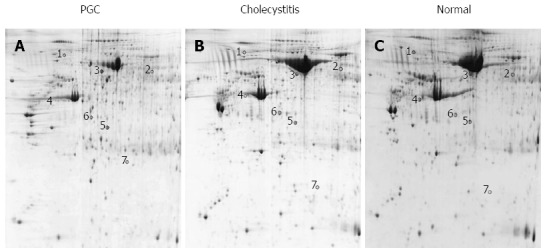
Representative two-dimensional gel electrophoresis protein profiles. Two-dimensional gel electrophoresis (2-DE) profiles of Primary gallbladder carcinoma (PGC) (A), cholecystitis (B) and normal gallbladder (C) samples. Proteins were separated by pI (X-axis) and molecular mass (Y-axis). Black circles indicate 7 differentially expressed protein spots identified by MALDI-TOF/TOF MS.
Protein identification by MALDI-TOF MS or tandem MS
Six of the seven differentially expressed proteins were identified using MALDI-TOF MS or tandem MS. Compared with cholecystitis and normal tissues, the overexpressed proteins in PGC tissues were serum albumin, ANXA4 and ACTG and the underexpressed proteins were Hsp90β, ACTA2 and Dync1h1. The identified proteins and their gene number, theoretical molecular weight, P value, Mascot score, and sequence coverage (%) are shown in Table 1.
Confirmation of biomarkers associated with PGC
To investigate whether the differentially expressed proteins would be useful diagnostic markers and to confirm the 2-DE results, additional samples were examined by Western blot, quantitative real-time PCR and IHC assay. Given that serum albumin is an abundant soluble protein, is approximately one-half of the protein content in serum and could lead to a false positive diagnosis, we studied the other five proteins. Consistent with the 2-DE data, the protein levels of ANXA4 were significantly elevated, and both Hsp90β and Dync1h1 were decreased in PGC tissues relative to the normal tumor-adjacent tissues in three paired tissue samples (Figure 2). However, Western blot analysis showed that the levels of ACTG and ACTA2 were unchanged (Figure 2). In addition, the mRNA levels of ANXA4, Hsp90β and Dync1h1 showed similar changes to those of the protein levels, suggesting that the expression of these proteins was affected by transcriptional factors (Figure 3).
Figure 2.
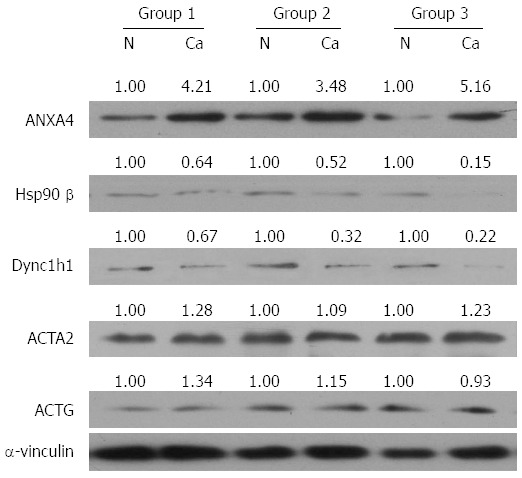
Western blot analysis of proteins. Expression of ANXA4, Hsp90β, Dync1h1, ACTG, ACTA2 and α-vinculin in paired gallbladder cancer tissues (Ca) and normal tumor-adjacent tissues (N). Quantification was performed by densitometry of Western blot bands. Densities were normalized to α-vinculin, and relative folds were normalized to expression levels in normal tumor-adjacent tissues. The data are representative of 20 paired groups. ANXA4: Antibody against annexin A4; ACTG: Gamma-actin; ACTA2: Aortic smooth muscle actin; Dync1h1: Dynein cytoplasmic 1 heavy chain 1; Hsp90β: Heat shock protein 90-beta.
Figure 3.
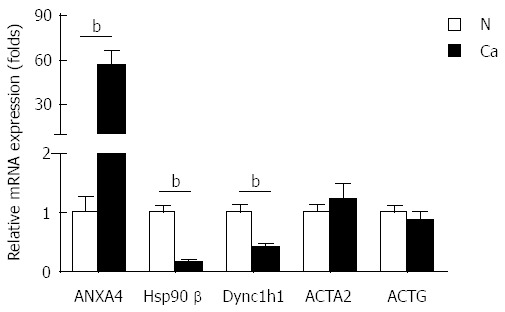
mRNA expression levels of biomarkers. The mRNA levels of ANXA4, Hsp90β, Dync1h1, ACTG and ACTA2 in normal tumor-adjacent tissue (N) and cancer tissue (Ca) were measured by quantitative real-time PCR. Data are normalized to expression levels of normal tumor-adjacent tissue. Results are mean ± SE (n = 6). bP < 0.01. ANXA4: Antibody against annexin A4; ACTG: Gamma-actin; ACTA2: Aortic smooth muscle actin; Dync1h1: Dynein cytoplasmic 1 heavy chain 1; Hsp90β: Heat shock protein 90-beta.
To further investigate these potential PGC biomarkers, IHC was performed to determine the expression of ANXA4, Hsp90β and Dync1h1. As shown in Figure 4, ANXA4 expression was markedly upregulated, while the expression of Hsp90β and Dync1h1 was decreased in the cytoplasm of PGC tissues relative to normal tumor-adjacent tissues (Figure 4). These results suggested that these three proteins, ANXA4, Hsp90β and Dync1h1, may be used as diagnostic biomarkers of PGC.
Figure 4.
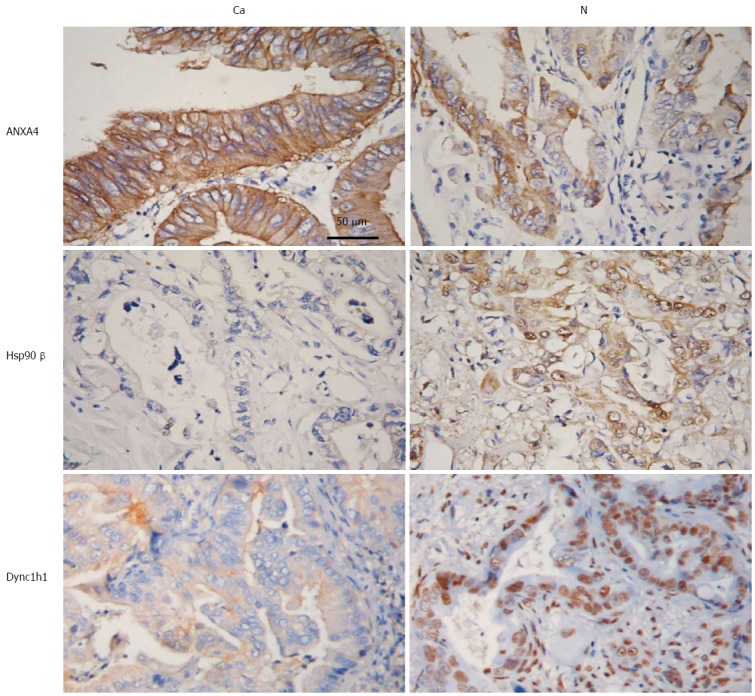
Immunohistochemical assay of proteins. Representative photographs of the expression levels of ANXA4, Hsp90β and Dync1h1 in paired gallbladder cancer tissues (Ca) and normal tumor-adjacent tissues (N). Scale bar: 100 μm. ANXA4: Antibody against annexin A4; Dync1h1: Dynein cytoplasmic 1 heavy chain 1; Hsp90β: Heat shock protein 90-beta.
DISCUSSION
The mortality rate of PGC has remained high for many years. A poor understanding of the risk factors and molecular pathogenesis of PGC and a lack of chemotherapeutic agents for this cancer have hampered our ability to devise strategies for this disease. An investigation of the carcinogenic and molecular mechanisms of PGC could improve clinical strategies against this disease and outcomes. Epidemiological data demonstrated that gallstones and cholecystitis are significantly associated with PGC; gallstones and cholecystitis are considered precancerous lesions of PGC[17]. In this study, comparative proteomics was performed using human PGC, cholecystitis and normal gallbladder tissues to advance the study of the carcinogenic mechanisms, diagnosis, and treatment of PGC, as well as the development of new drugs. Six non-redundant proteins were identified by MALDI-TOF/TOF as differentially expressed in human PGC tissues compared with cholecystitis and normal gallbladder tissues. Three of these proteins were upregulated (SA, ANXA4, and ACTG) and three were downregulated (Hsp90β, ACTA2 and Dync1h1).
In previous PGC proteomics studies, Tan et al[12] demonstrated that overexpression of annexinA3 correlated significantly with those cases with a lower histological grade, lymph node or distant metastasis, or a shorter survival time after operation, suggesting that annexinA3 might be a potential biomarker in the initiation and progression of human gallbladder cancer. Their subsequent study identified 24 differentially expressed serum proteins between gallbladder cancer and healthy cancer-free control patients, with 12 upregulated proteins such as splicing factor 3B subunit 5, cystatin-B, S100A10 protein, histone H2B type 2-E, profilin-1, and eukaryotic translation initiation factor 1A, and 12 downregulated proteins such as apolipoprotein A-I precursor, myosin regulatory light chain 2, proteasome subunit β type-2, superoxide dismutase, isoform 1 of Ficolin-3 precursor, and HP haptoglobin isoform 2 preproprotein. Tan et al[13] suggested that these differentially expressed proteins were potential serum biomarkers for the early diagnosis of gallbladder cancer. Wang et al[11] identified 15 proteins that were differentially expressed between the highly metastatic PGC-SD18H cell line and the poorly metastatic PGC-SD18L cell line, including CLIC1, WDR1, ezrin, vimentin, ANXA3, GDIR, TPI, PSME3, GDI2, GLUD1, TCP-1, THF synthase, hnRNP-K, PGK1 and PCD8. The present study is a new comparative proteomic study using human PGC, cholecystitis and normal gallbladder tissues and may be more informative in understanding the carcinogenic mechanisms and monitoring the treatment of PGC compared to the previous two studies.
Among the three identified candidate proteins, ANXA4 is a member of the annexins, which are found on membrane surfaces and may act as regulators of membrane fusion and possess the structural properties to form ion channels[18]. These properties suggest that annexins could be involved in processes such as growth, differentiation, and transformation[14]. Upregulation of ANXA4 has been observed in colorectal carcinoma, clear cell carcinoma of the ovary, renal clear cell carcinoma, pancreatic tumor, gastric cancer with Helicobacter pylori infection, and human penile squamous cell carcinoma. Elevated expression of ANXA4 is associated with tumor stage in colorectal cancer and lymph node metastasis of human penile squamous cell carcinoma[15,16,19-22].
Hsp90 is a molecular chaperone required for the stability and function of a number of conditionally expressed signaling proteins[23]. In cancer, Hsp90 is involved in the proteostatic maintenance of oncoproteins that promote tumor cell growth, survival or metastasis[24]. Thiel et al[25] using MALDI-TOF analysis, demonstrated that Hsp90β discriminated significantly between cancerous and noncancerous oral squamous cells. The SELDI-TOF MS platform is extremely useful for detecting and quantifying proteins associated with cancer due to its high sensitivity, convenience, and ability to profile a large number of clinical samples rapidly. This platform has been mainly used for serum or plasma samples.
The Dync1h1 protein is a large (> 530 ku), crucial subunit of the cytoplasmic dynein complex responsible for retrograde axonal transport in neurons[26]. Several studies have reported a correlation between missense mutations of Dync1h1 and Charcot-Marie-Tooth disease, severe intellectual disability, or other variable neuronal migration defects, suggesting that Dync1h1 could have broad effects on nervous system development and maintenance[27]. Recently, several studies reported that Dync1h1 is somatically mutated in pancreatic neuroendocrine neoplasms, ovarian cancer, and glioblastoma multiforme[28,29]. However, whether altered expression of Dync1h1 is involved in PGC is largely unknown. In this study, we used SELDI-TOF MS to analyze and validate Hsp90β and Dync1h1 as potential biomarkers in patients with PGC.
ACTA2 is a smooth muscle α-actin whose expression is transformation sensitive to growth signals in normal cells. Heterozygous missense mutations in ACTA2 cause predisposition to a variety of vascular diseases, including thoracic aortic aneurysm and dissection, early onset coronary artery disease and stroke[30,31]. ACTG mutations are associated with hearing loss[32,33]. Neither ACTG nor ACTA2 is known to be correlated with PGC.
In conclusion, three proteins were identified as potential biomarkers of PGC using proteomic analysis. These proteins may participate in the progression of malignant growth or the maintenance of normal gallbladder conditions. However, it is still too early to conclude that the three possible biomarkers reported here are novel biomarkers for PGC diagnosis. The functions of these proteins in the carcinogenesis of PGC remain to be studied. We hope that our results will provide useful information for studies on the early detection and therapy of PGC.
COMMENTS
Background
Primary gallbladder carcinoma (PGC) is the most frequent biliary tract malignancy with high morbidity and poor prognosis. Despite recent advances in the diagnosis and management of gastrointestinal cancers, PGC remains a challenge with a poor overall prognosis. To date, no reliable molecular markers have been established for PGC. Proteomic analysis has been applied to many types of tumors and can overcome some limitations of the approaches used to elucidate the molecular mechanisms of PGC. However, few studies have applied this method to tumor tissues from PGC patients.
Research frontiers
In most cases, although studies on differential mRNA expression are informative, they do not always correlate with protein concentrations as proteins are often subjected to proteolytic cleavage or post-translational modifications. Proteomics is a complementary method to genetic profiling. It is widely used to investigate molecular changes in translation, post-translational modification and intracellular mislocalization involved in tumor initiation and growth.
Innovations and breakthroughs
Although proteomic studies on PGC have used cancer cell lines and blood samples, few studies have applied a proteomic approach to tumor tissues from PGC patients. Studies using tumor tissue should be more informative in terms of carcinogenic mechanisms, prognostic evaluation and therapy for PGC. This is a new comparative proteomic study using human PGC, cholecystitis and normal gallbladder tissues, and three new potential biomarkers of PGC were validated.
Applications
This study may be informative in understanding the carcinogenic mechanisms and monitoring the treatment of PGC.
Terminology
Proteomics is a blend of protein and genome, and the large-scale study of proteins, particularly their structures and functions. It is often used specifically for protein purification and mass spectrometry.
Peer review
There are novel findings about PGC using proteomic approach. The authors found three potential biomarkers of PGC. The study was overall well designed and appropriately controlled. In particular, samples with cholecystitis were included in the initial 2D-gel profiling as “non-specific disease controls” and the protein candidates identified were further confirmed for their significance with multiple approaches including Western blot, qRT-PCR and IHC in a separate set of 20 samples from patients with PGC.
Footnotes
P- Reviewers: Cho JY, Chen WX S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Liu XM
References
- 1.Giang TH, Ngoc TT, Hassell LA. Carcinoma involving the gallbladder: a retrospective review of 23 cases - pitfalls in diagnosis of gallbladder carcinoma. Diagn Pathol. 2012;7:10. doi: 10.1186/1746-1596-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srivastava K, Srivastava A, Sharma KL, Mittal B. Candidate gene studies in gallbladder cancer: a systematic review and meta-analysis. Mutat Res. 2011;728:67–79. doi: 10.1016/j.mrrev.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dutta U. Gallbladder cancer: can newer insights improve the outcome? J Gastroenterol Hepatol. 2012;27:642–653. doi: 10.1111/j.1440-1746.2011.07048.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhang SB, Song SP, Li B, Zhou YS, Zhang YD. Expression of N-myc downstream-regulated gene 1 in primary gallbladder carcinoma and its correlation with clinicopathological features and clinical outcome. Med Oncol. 2012;29:1866–1872. doi: 10.1007/s12032-011-0017-7. [DOI] [PubMed] [Google Scholar]
- 5.Li Z, Zhao X, Bai S, Wang Z, Chen L, Wei Y, Huang C. Proteomics identification of cyclophilin a as a potential prognostic factor and therapeutic target in endometrial carcinoma. Mol Cell Proteomics. 2008;7:1810–1823. doi: 10.1074/mcp.M700544-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke CH, Buckley JA, Fung ET. SELDI-TOF-MS proteomics of breast cancer. Clin Chem Lab Med. 2005;43:1314–1320. doi: 10.1515/CCLM.2005.225. [DOI] [PubMed] [Google Scholar]
- 7.Lin JF, Xu J, Tian HY, Gao X, Chen QX, Gu Q, Xu GJ, Song JD, Zhao FK. Identification of candidate prostate cancer biomarkers in prostate needle biopsy specimens using proteomic analysis. Int J Cancer. 2007;121:2596–2605. doi: 10.1002/ijc.23016. [DOI] [PubMed] [Google Scholar]
- 8.Zinkin NT, Grall F, Bhaskar K, Otu HH, Spentzos D, Kalmowitz B, Wells M, Guerrero M, Asara JM, Libermann TA, et al. Serum proteomics and biomarkers in hepatocellular carcinoma and chronic liver disease. Clin Cancer Res. 2008;14:470–477. doi: 10.1158/1078-0432.CCR-07-0586. [DOI] [PubMed] [Google Scholar]
- 9.Qi YJ, He QY, Ma YF, Du YW, Liu GC, Li YJ, Tsao GS, Ngai SM, Chiu JF. Proteomic identification of malignant transformation-related proteins in esophageal squamous cell carcinoma. J Cell Biochem. 2008;104:1625–1635. doi: 10.1002/jcb.21727. [DOI] [PubMed] [Google Scholar]
- 10.Hao Y, Yu Y, Wang L, Yan M, Ji J, Qu Y, Zhang J, Liu B, Zhu Z. IPO-38 is identified as a novel serum biomarker of gastric cancer based on clinical proteomics technology. J Proteome Res. 2008;7:3668–3677. doi: 10.1021/pr700638k. [DOI] [PubMed] [Google Scholar]
- 11.Wang JW, Peng SY, Li JT, Wang Y, Zhang ZP, Cheng Y, Cheng DQ, Weng WH, Wu XS, Fei XZ, et al. Identification of metastasis-associated proteins involved in gallbladder carcinoma metastasis by proteomic analysis and functional exploration of chloride intracellular channel 1. Cancer Lett. 2009;281:71–81. doi: 10.1016/j.canlet.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 12.Tan Y, Meng HP, Wu Q, Wang FQ, Wu HR. [Proteomic study of gallbladder cancer, with special reference on the expression and significance of annexin A3] Zhonghua Bingli xue Zazhi. 2010;39:382–386. [PubMed] [Google Scholar]
- 13.Tan Y, Ma SY, Wang FQ, Meng HP, Mei C, Liu A, Wu HR. Proteomic-based analysis for identification of potential serum biomarkers in gallbladder cancer. Oncol Rep. 2011;26:853–859. doi: 10.3892/or.2011.1353. [DOI] [PubMed] [Google Scholar]
- 14.Hayes MJ, Moss SE. Annexins and disease. Biochem Biophys Res Commun. 2004;322:1166–1170. doi: 10.1016/j.bbrc.2004.07.124. [DOI] [PubMed] [Google Scholar]
- 15.Duncan R, Carpenter B, Main LC, Telfer C, Murray GI. Characterisation and protein expression profiling of annexins in colorectal cancer. Br J Cancer. 2008;98:426–433. doi: 10.1038/sj.bjc.6604128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong J, Kim Y, Kyung Seong J, Lee KJ. Comprehensive identification of novel post-translational modifications in cellular peroxiredoxin 6. Proteomics. 2012;12:1452–1462. doi: 10.1002/pmic.201100558. [DOI] [PubMed] [Google Scholar]
- 17.Andreotti G, Liu E, Gao YT, Safaeian M, Rashid A, Shen MC, Wang BS, Deng J, Han TQ, Zhang BH, et al. Medical history and the risk of biliary tract cancers in Shanghai, China: implications for a role of inflammation. Cancer Causes Control. 2011;22:1289–1296. doi: 10.1007/s10552-011-9802-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerke V, Moss SE. Annexins: from structure to function. Physiol Rev. 2002;82:331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- 19.Zimmermann U, Balabanov S, Giebel J, Teller S, Junker H, Schmoll D, Protzel C, Scharf C, Kleist B, Walther R. Increased expression and altered location of annexin IV in renal clear cell carcinoma: a possible role in tumour dissemination. Cancer Lett. 2004;209:111–118. doi: 10.1016/j.canlet.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Sitek B, Sipos B, Alkatout I, Poschmann G, Stephan C, Schulenborg T, Marcus K, Lüttges J, Dittert DD, Baretton G, et al. Analysis of the pancreatic tumor progression by a quantitative proteomic approach and immunhistochemical validation. J Proteome Res. 2009;8:1647–1656. doi: 10.1021/pr800890j. [DOI] [PubMed] [Google Scholar]
- 21.Lin LL, Chen CN, Lin WC, Lee PH, Chang KJ, Lai YP, Wang JT, Juan HF. Annexin A4: A novel molecular marker for gastric cancer with Helicobacter pylori infection using proteomics approach. Proteomics Clin Appl. 2008;2:619–634. doi: 10.1002/prca.200780088. [DOI] [PubMed] [Google Scholar]
- 22.Protzel C, Richter M, Poetsch M, Kakies C, Zimmermann U, Woenckhaus C, Klebingat KJ, Hakenberg OW, Giebel J. The role of annexins I, II and IV in tumor development, progression and metastasis of human penile squamous cell carcinomas. World J Urol. 2011;29:393–398. doi: 10.1007/s00345-010-0575-x. [DOI] [PubMed] [Google Scholar]
- 23.Neckers L, Ivy SP. Heat shock protein 90. Curr Opin Oncol. 2003;15:419–424. doi: 10.1097/00001622-200311000-00003. [DOI] [PubMed] [Google Scholar]
- 24.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 25.Thiel UJ, Feltens R, Adryan B, Gieringer R, Brochhausen C, Schuon R, Fillies T, Grus F, Mann WJ, Brieger J. Analysis of differentially expressed proteins in oral squamous cell carcinoma by MALDI-TOF MS. J Oral Pathol Med. 2011;40:369–379. doi: 10.1111/j.1600-0714.2010.00982.x. [DOI] [PubMed] [Google Scholar]
- 26.Pfister KK, Shah PR, Hummerich H, Russ A, Cotton J, Annuar AA, King SM, Fisher EM. Genetic analysis of the cytoplasmic dynein subunit families. PLoS Genet. 2006;2:e1. doi: 10.1371/journal.pgen.0020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsurusaki Y, Saitoh S, Tomizawa K, Sudo A, Asahina N, Shiraishi H, Ito J, Tanaka H, Doi H, Saitsu H, et al. A DYNC1H1 mutation causes a dominant spinal muscular atrophy with lower extremity predominance. Neurogenetics. 2012;13:327–332. doi: 10.1007/s10048-012-0337-6. [DOI] [PubMed] [Google Scholar]
- 28.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milewicz DM, Østergaard JR, Ala-Kokko LM, Khan N, Grange DK, Mendoza-Londono R, Bradley TJ, Olney AH, Adès L, Maher JF, et al. De novo ACTA2 mutation causes a novel syndrome of multisystemic smooth muscle dysfunction. Am J Med Genet A. 2010;152A:2437–2443. doi: 10.1002/ajmg.a.33657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leong WF, Chow VT. Transcriptomic and proteomic analyses of rhabdomyosarcoma cells reveal differential cellular gene expression in response to enterovirus 71 infection. Cell Microbiol. 2006;8:565–580. doi: 10.1111/j.1462-5822.2005.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu M, Yang T, Wei S, DeWan AT, Morell RJ, Elfenbein JL, Fisher RA, Leal SM, Smith RJ, Friderici KH. Mutations in the gamma-actin gene (ACTG1) are associated with dominant progressive deafness (DFNA20/26) Am J Hum Genet. 2003;73:1082–1091. doi: 10.1086/379286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Wijk E, Krieger E, Kemperman MH, De Leenheer EM, Huygen PL, Cremers CW, Cremers FP, Kremer H. A mutation in the gamma actin 1 (ACTG1) gene causes autosomal dominant hearing loss (DFNA20/26) J Med Genet. 2003;40:879–884. doi: 10.1136/jmg.40.12.879. [DOI] [PMC free article] [PubMed] [Google Scholar]


