Abstract
Lipoxygenases (LOXs) and cyclooxygenases (COXs) metabolize poly-unsaturated fatty acids into inflammatory signaling molecules. Modulation of the activity of these enzymes may provide new approaches for therapy of inflammatory diseases. In this study, we screened novel anacardic acid derivatives as modulators of human 5-LOX and COX-2 activity. Interestingly, a novel salicylate derivative 23a was identified as a surprisingly potent activator of human 5-LOX. This compound showed both non-competitive activation towards the human 5-LOX activator adenosine triphosphate (ATP) and non-essential mixed type activation against the substrate linoleic acid, while having no effect on the conversion of the substrate arachidonic acid. The kinetic analysis demonstrated a non-essential activation of the linoleic acid conversion with a KA of 8.65 μM, αKA of 0.38 μM and a β value of 1.76. It is also of interest that a comparable derivative 23d showed a mixed type inhibition for linoleic acid conversion. These observations indicate the presence of an allosteric binding site in human 5-LOX distinct from the ATP binding site. The activatory and inhibitory behavior of 23a and 23d on the conversion of linoleic compared to arachidonic acid are rationalized by docking studies, which suggest that the activator 23a stabilizes linoleic acid, whereas the larger inhibitor 23d blocks the enzyme active site.
Keywords: Human 5-lipoxygenase, Anacardic acid, Cyclooxygenase-2, Enzyme kinetics, Allosteric binding
1. Introduction
The lipoxygenases (LOXs) and the cyclooxygenases (COXs) are enzymes that play key roles in the major metabolic pathways of poly-unsaturated fatty acids (PUFAs). These pathways convert PUFAs into inflammatory mediators such as lipoxins, leukotrienes, and prostaglandins, which play a regulatory role in numerous inflammatory and proliferative diseases including asthma, arthritis and cancer [1]. The biological roles of LOXs and COXs in metabolic pathways towards inflammatory mediators and signaling molecules demonstrate the utility of these enzymes as therapeutic targets.
Lipoxygenases are non-heme iron containing enzymes. There are four types of lipoxygenases with different positional specificities for arachidonic acid oxidation present in mammalian tissues; 5-LOX, 8-LOX, 12-LOX and 15-LOX. In general, lipoxygenases comprise of two domains in which the N-terminal domain is the regulatory domain and the C-terminal domain is the catalytic domain [2]. Although, human lipoxygenases show about 60% sequence similarities [3], their regulatory mechanisms are variable. Unlike other LOXs, 5-LOX activity requires the presence of activators such as Ca2+ and adenosine triphosphate (ATP). In addition, different types of LOXs have been identified based on their distribution over different tissues. For example, 12-LOX is subdivided in platelet 12-LOX (p12-LOX) and 12R-LOX. As its name implies, platelet 12-LOX (p12-LOX) is mostly found in platelets, whereas 12R-LOX is most frequently found in skin cells [4,5]. Similarly, 15-LOX is subdivided in 15-LOX-1 and 15-LOX-2. 15-LOX-1 is highly expressed in leukocytes and airways endothelial cells [6,7], whereas 15-LOX-2 is expressed in multiple tissues such as prostate, lung, cornea, liver, colon, kidney, and brain but not in leukocytes [8]. Moreover, stimulation of cells by interleukin (IL)-4 and IL-13 shows selective enhancement of the 15-LOX-1 expression and not 15-LOX-2 expression [9]. In addition, 15-LOX-1 and 15-LOX-2 lack similarity at the primary sequence level, which also hints at distinct biological roles and the possibility to develop selective inhibitors or activators [10].
The overexpression of certain lipoxygenases has been reported in numerous inflammatory diseases. For example, increased 15-LOX-1 expression in bronchial epithelium, which is accompanied by elevated concentrations of 15- hydroxyeicosatetraenoic acid (15-HETE), has been observed in patients with asthma and chronic bronchitis [11,12]. In addition, activity of other LOXs isoenzymes has been reported to be significantly increased in disease conditions. For example, upregulation of 12R-LOX activity has been observed in the development of skin tumor and melanoma [13,14]. In addition, overexpression of 5-LOX has been reported to contribute to airway inflammation in asthma and to the growth of some tumors in, for example, lung, prostate, brain and colon [15–19]. In contrast, lipoxins, another type of arachidonic acid metabolites, have been reported to have important roles as anti-inflammatory and pro-resolution mediators [20], which play an important role in the termination of immune responses. Production of lipoxins upon inflammatory stimulation involves two main lipoxygenase-mediated pathways. The first pathway involving 15-LOX and 5-LOX occurs in mucosal tissues, such as the respiratory tract, gastrointestinal tract and the oral cavity. The second pathway involves 5-LOX and p12-LOX, in which 5-LOX produces the Leukotriene A4 (LTA4) and p12-LOX converts the LTA4 to lipoxin A4 (LXA4) [21]. Lipoxin A4 has important roles in stopping neutrophil migration, stimulation of monocyte and macrophage activation, and inhibition of leukotriene B4 formation [20]. Induction of 15-LOX-1 by IL-13 in synovial tissues of patients with rheumatoid arthritis enhances the level of LX4 in a negative feedback loop to limit inflammatory responses induced by pro-inflammatory mediators such as Leukotriene B4 [22]. Another study showed that prior to induction with peroxisome proliferator-activated receptors gamma (PPARγ) agonists in brain, the 5-LOX expression was enhanced in association with the increase of cerebral LX4 production and inhibition of Leukotriene B4 production [23]. The association of 5-lipoxygenase activity in the production of both pro- and anti-inflammatory mediators indicates its crucial role in pathophysiological processes of inflammation. Based on these findings we speculate that activators of 5-lipoxygenase might also have beneficial effects in inflammation by triggering the termination of immune responses. In addition, it might be speculated that the redirection of lipoxygenase metabolism by 5-LOX activators towards other substrates might also lead to the formation of less harmful pro-inflammatory products. Enzyme activation as a concept in drug discovery has been shown to be successful for glucokinase activators [24]. We stress, however, that lipoxygenase activation by small molecules with the aim to limit inflammation is a novel concept and lacks experimental proof.
Prostaglandin endoperoxide H synthases (PGHSs), are also known as cyclooxygenases (COXs), catalyze the formation of prostaglandins (PGs). COXs are comprised of two main sites, a heme site with peroxidase activity and a bis-oxygenase site. COXs convert arachidonic acid into prostaglandin endoperoxide H2 (PGH2) in a two steps process. The first step is enzymatic catalysis of the insertion of two molecules O2 into arachidonic acid to generate prostaglandin G2 (PGG2). The second step is the peroxidase reaction in which the PGG2 is oxidized to PGH2 by a two electron abstraction. The peroxidase reaction occurs at a heme-containing active site located near the protein surface, whereas the oxygenation reaction occurs in a hydrophobic channel in the core of the enzyme [25]. The COX isoenzymes COX-1 and COX-2 are differentially expressed in various tissues. Under normal conditions, COX-1 is expressed in most mammalian cells, whereas COX-2 is not expressed. The COX-2 expression is increased under inflammatory conditions. Animal models of inflammatory arthritis indicate that overexpression of COX-2 is responsible for the increase of PGs in joint tissue and cause acute and chronic inflammation [26]. Elevated levels of PGs were observed in inflammatory bowel diseases (IBD) [27–28]. A similar behavior was observed in cancers. For example, the expression of COX-2, but not COX-1, was observed in various regions of the colon in human colorectal cancer [29,30]. Therefore, COX-2 inhibition is considered to be a relevant therapeutic strategy for treatment of cancer and inflammatory diseases.
Anacardic acid is a naturally occurring 6-alkyl substituted salicylate that is identified from cashew nutshells. Previously, we found that this salicylate and its derivatives are allosteric regulators of lipoxygenase activity. We identified an allosteric inhibitor of soybean lipoxygenase-1 (SLO-1) (19) and an allosteric activator of potato 5-lipoxygenase (5-LOXs) (21) [31]. It should, however, be noted that the lack of similarity between the human and plant enzymes prohibits extrapolation or these data towards biological effects in cell-based studies [32]. Therefore, we continue our previous studies by exploring the inhibitory potency of a salicylate-based compound collection for inhibition of human 5-lipoxygenase and cyclooxygenase-2. In this study, we describe the synthesis of a novel focused collection of salicylate-based compounds and we investigate their influence on the enzyme activity of human 5-LOX and human COX-2. This enabled the identification of a novel activator 23a and a mixed inhibitor 23d of the conversion of the linoleic acid by h-5-LOX. Interestingly, no effect on the arachidonic acid conversion was observed. Enzyme kinetic investigations demonstrated that activator 23a binds to an allosteric binding site on h-5-LOX that is different from the ATP regulatory site. Modeling studies suggest that 23a binds close to the substrate linoleic acid, thereby enhancing the Km and kcat for its conversion.
2. Results and discussion
2.1. Synthesis
Our salicylate-based compound collection that is inspired by anacardic acid (14) comprises of the previously published salicylates 14–19, and 21 and the newly synthesized salicylates 20, 22, 23a–d, 24b–c and benzoxazole 25. The synthesis of anacardic acid (14) and its derivatives 15–19, and 21 were described previously by Ghizzoni et. al. [33, 34]. This collection includes the previously described compounds that were reported as inhibitors or activator of soybean lipoxygenase-1 (SLO1) (19) and potato 5-LOX (21) [31]. The newly synthesized compounds were inspired by compound 19 and 21, which contain either a 6-phenylethyl substitution or a 6-alkoxy substitution. This was merged into a para-alkoxybenzyloxy substitution, which can be synthesized using convenient methods.
The starting materials, acetonide 1 (Scheme 1) and triflate 10 (Scheme 2), were synthesized as described by Uchiyama et al. [35]. 2-hydroxy-6-(nonyloxy)benzoic acid (22) was synthesized from commercially available 1-bromononane and acetonide 1 following synthetic procedure described by Ghizzoni et. al. [33] (Scheme 1). p-alkyloxybenzylbromides (3a–d) and m-alkyloxybenzylbromides (5b–c) were prepared in three steps from p-hydroxybenzaldehyde or m-hydroxybenzaldehyde. The first step involves the coupling of alkylbromides with the hydroxybenzaldehydes to give the corresponding ethers. Subsequently, the benzaldehydes were reduced to the corresponding benzyl alcohols 2a–d and 4b–c [36] in high yields. Direct coupling of benzyl alcohols to phenol 1 by Mitsunobu reaction did not provide the desired ethers. Therefore, the benzylalcohols 2a–d and 4b–c were brominated using PBr3 to give the corresponding benzylbromides 3a–d and 4b–c. The benzylbromides were coupled to acetonide 1 to give the corresponding products 12a–d and 13b–c in high yields (>70%). Subsequently, the acetonide protecting group was cleaved using potassium hydroxide in THF to provide the final products 23a–d as potassium salts (Scheme 1). Isolation of salicylates 23a–d as salicylate salts proved to be necessary to avoid degradation of the p-alkoxybenzylethers by the strongly acidic salicylic acids. Acidification results a slow hydrolysis of the product to give 2,6-dihydroxybenzoate and the corresponding (4-(alkyloxy)benzylium cation, which is stabilized by the electrondonating alkyloxy substituent on the para- postion. As expected, for the meta-substituted benzyl substituents 24b–c this problem was not observed.
Scheme 1.
Reagents and conditions: a) SOCl2, DMAP, DME, Acetone, 0 °C for 1 h, then R.T. overnight; b) 1-bromo alkane, K2CO3, DMF, R.T. overnight; c) NaBH4, MeOH, 0 °C for 1 h, then R.T. for 30 min; d) PBr3, CH2Cl2, 0 °C for 1.5 h; e) 1-bromononane, K2CO3, DMF, R.T. overnight; f) compound 1, K2CO3, DMF, R.T. overnight; g) 5 M KOH, THF, 60 °C overnight.
Scheme 2.

Reagents and conditions: a) Decanoylchloride, AlCl3, CH2Cl2, 60 °C for 1 h; b) NH2NH2.H2O, KOH, 1-octanol, reflux, 3h; c) Trimethylsilyl-acetylene, CuI, PdCl2(PPh3)2, Et2NH, PPh3, CH3CN, (MW, 120 °C, 95 W, 35 min), d) TBAF, THF, 0 °C, 10 min; e) CuI, PdCl2(PPh3)2, Et2NH, triflate 10, CH3CN (MW, 120 °C, 70 W, 35 min); f) H2, Pd/C, MeOH, 45 °C, 24 h; g) 2-aminophenol, polyphosphatic acid, 180 °C for 6.5 h.
p-decylphenyl bromide (8) (Scheme 2) was prepared from bromobenzene 6 and decanoylchloride following two reaction steps. The first step was the formation of 1-(4-bromophenyl)decan-1-one 7 through Friedel-Craft acylation using AlCl3 [37]. The second step was the Wolff-Kishner reduction of 1-(4-bromophenyl)decan-1-one 7 using hydrazine monohydrate and potassium hydroxide to give product 8 [37]. Subsequently, arylbromide 8 was converted to alkyne 9 by coupling with trimethyl-silyl (TMS) acetylene by a Sonogashira reaction and subsequent removal of the TMS protection using tetra butyl ammonium fluoride (TBAF). The resulting alkyne 9 was coupled to triflate 10 [38] to give the product in high yield (80%), which was subsequently converted into product 20 by catalytic hydrogenation.
2.2. Enzyme inhibition
LOXs are non-heme iron dependent enzymes. Benzoxazole 25 was designed based on the presumption that iron-binding is a key property for the inhibition of lipoxygenases and cyclooxygenases. Benzoxazole 25 was synthesized from salicylic acid and 2-amino phenol through direct cyclization using polyphosphoric acid (Scheme 2) [39]. Compound 25 did not inhibit h-5-LOX, however it gives 50% COX-2 inhibition at 50 μM inhibitor concentration. This indicates that 25 is a potential starting point for development of selective inhibitors of COX-2 (Figure 1).
Figure 1.
Residual enzyme activity that was observed for the screening of a salicylate-based compound collection for inhibition of h-5-LOX and COX-2 activity in the presence of 50 μM of the respective compounds. The h-5-LOX activity was determined in presence of 100 μM linoleic acid as a substrate. The COX-2 activity was determined in presence of 2 mM arachidonic acid as a substrate. The results presented were the average of three independent experiments and the standard deviations are shown.
The influence of the salicylates (Table 1) on the enzyme activity of human 5-lipoxygenase and/or cyclooxygenase-2 was investigated. The h-5-LOX activity was monitored in real time by detection of the formation of the UV absorbance of the conjugated diene 13-hydroperoxy-cis-9-trans-11-octa decanoic acid (13-HPOD) from linoleic acid [40,41]. The residual h-5-LOX activity was monitored after 10 minutes pre-incubation with 50 μM of the compound of interest. The COX-2 activity was determined by measuring the conversion or arachidonic acid, which conversion is monitored by the oxidation of N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD) [42]. The residual enzyme activity was measured after 5 minutes pre-incubation in the presence of 50 μM of the compound of interest. The enzyme activity without inhibitor present was taken as control and set to 100% and the activity without enzyme present was set to 0%. The residual enzyme activities of h-5-LOX and COX-2 in the presence of different salicylates are shown in Figure 1.
Table 1.
Focused compound collection
| Compounds | R1 | R2 | R3 | |
|---|---|---|---|---|

|
14 | H | H |
|
| 15 | H | H |
|
|
| 16 | H | H |
|
|
| 17 | H | OH |
|
|
| 18 | H | H |
|
|
| 19 | H | OH | ||
| 20 | H | OH |
|
|
| 21 | H | H |
|
|
| 22 | H | H |
|
|
| 23a (PK131) | K | H |
|
|
| 23b | K | H |
|
|
| 23c | K | H |
|
|
| 23d (PK147) | K | H |
|
|
| 24b | H | H |
|
|
| 24c | H | H |
|
|
| 25 |

|
|||
Anacardic acid 14, compounds 15, and 16 show little inhibition of both h-5-LOX and COX-2 at 50 μM. In comparison to anacardic acid 14, a slight improvement in the inhibitory potency of h-5-LOX was observed for compound 17. Importantly, neither inhibition nor activation was observed from compound 21, which in our previous report [31] shows a strong activation on potato 5-LOX, which demonstrates the difference between plant and human enzymes.
Compounds 23a–d with alkoxy substituents in the para- positions of the benzylethers, show an interesting structure activity relationship for modulation of h-5-LOX activity. Compound 23a shows strong activation of h-5-LOX at 50 μM, whereas compound 23b and 23c with a longer side chain show no or little inhibition of h-5-LOX. Furthermore, compound 23d with a branched side chain provides almost 50% inhibitions of h-5-LOX at the same concentration. The compound is racemic at its asymmetric carbon
The data in Figure 1 demonstrate that the salicylate-based compounds do not inhibit COX-2 activity more than 50% at 50 μM, which indicates that this type of compounds has a tendency for selective modulation of h-5-LOX compared to COX-2. Interestingly, benzoxazole 25 inhibits the COX-2 activity by about 50%, whereas no inhibition was observed on h-5-LOX, which indicates that further optimization of this class of compound may be feasible.
In order to assure that the assays are performed in homogenous solutions the critical micelle concentrations (CMCs) of the different compounds were determined and the assays were performed at concentration below the CMC. Homogenous solutions are required for proper analysis of the enzyme kinetics. The CMCs for 23a and 23d at the assay conditions (50 mM Tris buffer, 2 mM EDTA and 2 mM CaCl2, pH 7.5, R.T.) are, respectively, 628 μM and 395 μM (Figure 2B,C) which indicate that micelle formation does not occur at concentrations employed for inhibition or activation of h-5-LOX. In addition, we found a CMC value for linoleic acid of 185 μM (Figure 2A) [43], which demonstrates that the substrate concentration in the inhibition studies (100 μM) was below the CMC.
Figure 2.



Surface tensions of (A) Anacardic acid 14 and (B) compound 23a (C) compound 23d against the logarithm of concentration. CMC values (A–C) were measured in human 5-LOX assay conditions, Tris buffer (50 mM), 2 mM EDTA and 2 mM CaCl2, pH 7.5, R.T.(t = 19°C).
2.3. Enzyme kinetic studies for human 5-lipoxygenase
A steady-state enzyme kinetic analysis of h-5-LOX in the presence of the activator 23a was performed in order to unravel the activation mechanism. The kinetic characterization of h-5-LOX activity versus linoleic acid in the presence of activator 23a shows a concentration dependent activation (Figure 3). The activation was determined in the presence of various concentrations of the natural substrate linoleic acid or the h-5-LOX activator ATP. The initial velocities of h-5-LOX were determined at various concentration of ATP, various concentrations 23a (0, 12.5 and 25 μM) and a fixed concentration of linoleic acid (100 μM). The velocities were plotted in the Michaelis-Menten and the Lineweaver-Burk plot as shown in Figure 4.
Figure 3.
Concentration dependent activation of the linoleic acid conversion by h-5-LOX in the presence of various concentrations of compound 23a and in its absence (control). The results were the average of three independent experiments with error bars (±S.D)
Figure 4.
Steady-state kinetic characterization of the linoleic acid conversion versus the ATP concentration of h-5-LOX activator 23a. (a) the Michaelis-Menten plots and (b) the Lineweaver-Burk plots show the relation of h-5-LOX activity versus the ATP concentration at three selected concentration of 23a (■) 0 μM, (●) 12.5 μM, and (▲) 25 μM in the presence of 100 μM linoleic acid substrate.
The Michaelis-Menten data show that activator 23a causes an increase in Vmax, whereas the Km remains constant (Table 2). Lineweaver-Burk analysis provided similar values. The values demonstrate non-competitive activation of linoleic acid conversion by h-5-LOX by compound 23a compared to ATP. The fact that binding of 23a does not influence the binding constant of ATP to h-5-LOX indicates that 23a does not compete for binding in the ATP binding pocket that allosterically regulates h-5-LOX activity. This indicates the presence of an allosteric binding pocket that is distinct from the ATP binding pocket and regulates the h-5-LOX enzyme activity.
Table 2.
Enzyme kinetic parameter for activation of h-5-LOX by activator 23a versus ATP
| 23a (μM) | Kmapp (μM) | Vmaxapp (nM/s) | R2 |
|---|---|---|---|
| 0 | 27.9 ± 1.7 | 230.9 ± 4.2 | 0.998 |
| 12.5 | 28.3 ± 1.7 | 386.1 ± 7.3 | 0.999 |
| 25 | 28.6 ± 2.3 | 523.6 ± 13.7 | 0.999 |
The enzyme kinetics of 23a were further investigated through the Michaelis-Menten and the Lineweaver-Burk plots (Figure 5). The initial velocities of h-5-LOX at different concentration of linoleic acid and a fixed concentration of ATP (100 μM), and at several selected concentrations of 23a (0, 12.5 and 25 μM) were measured. Activator 23a causes an increase in Vmax and a decrease in Km (Table 3), which indicates non-essential mixed type activation. This indicates that the activator can bind to the free enzyme (E, Scheme 3) and also to the substrate bound enzyme (ES, Scheme 3). In this model binding of the substrate and 23a mutually influence each other. According to this model the activation of h-5-LOX by 23a can be described by equation 1 (Figure 7).
Figure 5.
Steady-state kinetic characterization of the linoleic acid conversion by h-5-LOX activation by activator 23a. (a) Michaelis-Menten plots and (b) Lineweaver-Burk plots show the relation of h-5-LOX activity versus linoleic acid concentrations at three selected concentrations of 23a (■) 0 μM, (●) 12.5 μM, and (▲) 25 μM.
Table 3.
Enzyme kinetic parameters for activation of h-5-LOX by activator 23a versus linoleic acid.
| 23a (μM) | Kmapp (mM) | Vmaxapp (μM/s) | R2 |
|---|---|---|---|
| 0 | 0.590 ± 0.080 | 2.14 ± 0.21 | 0.999 |
| 12.5 | 0.438 ± 0.020 | 2.39 ± 0.07 | 0.999 |
| 25 | 0.383 ± 0.011 | 2.70 ± 0.05 | 0.999 |
Scheme 3.

Kinetic model for non-essential activation.
Figure 7.

Equation 1 for the enzyme kinetics according to the model in Scheme 2.44 v is the reaction velocity, Vmax is the maximal reaction velocity, [S] is the substrate concentration and Km is the Michaelis-Menten constant, [A] is the activator concentration. α and β, respectively, are the parameters to describe the change in the affinity of substrate binding and the change in the catalytic constant.
The enzyme kinetic parameters were derived as described in the material and methods using methods from Leskovac [44]. The activator dissociation constant (KA), the change in the affinity of substrate binding (α) and the change in the catalytic constant (β) were determined by re-plotting the slopes and the y-interceptions derived from the Lineweaver-Burk plot as a function of activator 23a concentration (Figure 6) [44] The KA value was determined from the intersection of 1/Δslopes plot and 1/Δy-intercept plot. The intersection of 1/Δy-intercept plot with the ordinate is defined as β.Vmax/(β-1) from which the β value was derived. Subsequently, the resulting β value was substituted to the equation β.Vmax/Km(β-α), which is the intersection of 1/Δslopes plot with the ordinate, to acquire the α value.
Figure 6.
Re-plot of 1/Δslopes and 1/Δy-intercept versus concentration of 23a.
The analysis shows that α is 0.0438, β is 1.76 and KA is 8.65 μM. The α value, which indicates the change in substrate binding, gives about 50-fold affinity enhancement of the substrate binding to the activator bound enzyme compared to the free enzyme. Compound 23a shows a high potency as an activator of h-5-LOX with the KA value in the micromolar range. Furthermore, the affinity of 23a to the substrate bound enzyme is about 50-fold enhanced, which indicates that activation of h-5-LOX at high substrate concentrations (> 10 μM) takes place at low activator 23a concentrations (<10 μM) (Figure 3). The catalytic constant for substrate conversion reaction was enhanced by 1.76 fold in the presence of the activator 23a compared to the normal reaction conditions. In conclusion, compound 23a gives a non-essential mixed-type activation of h-5-LOX and binds with high affinity (αKA = 390nM) to the substrate bound enzyme and enhances the conversion of the substrate linoleic acid.
The observed results from the enzyme kinetic studies that give non-essential mixed-type activation versus linoleic acid and non-essential non-competitive activation versus ATP demonstrate that there is an allosteric regulatory site in h-5-LOX that is distinct from the ATP binding site. In order to get a better understanding of this allosteric site in the regulation of the enzyme activity, we also studied the inhibition mechanism of h-5-LOX by compound 23d.
Interestingly, compound 23d has a structure that is closely related to 23a and this compound inhibits h-5-LOX at 50 μM. We analyzed the underlying enzyme kinetic parameters. The Michaelis-Menten plots and the Lineweaver-Burk plots show an increase of the Kmapp and reduction of the Vmaxapp (Table 4), which is a mixed-type inhibition of h-5-LOX (Figure 8). This indicates that inhibitor 23d can bind to the free enzyme as well as to the substrate bound enzyme following the inhibition model in Scheme 4 from which the kinetic parameters Vmax and Km can be derived by non-linear curve fitting of equation 2 (Figure 9). Furthermore, the changes in the Km and Vmax are α/α′. Km and Vmax/α′ are used to determine the Ki and Ki′ values for inhibitor 23d using equation 3 and 4 (Figure 9). Ki and Ki′ values for inhibitor 23d respectively are 70.3 μM and 140 μM. The fact that inhibitor 23d shows a mixed type inhibition also indicates the presence of an allosteric site in human 5-lipoxygenase. We presume that compound 23d binds to the same allosteric binding pocket as 23a but that the difference in the alkyl chain causes a difference in affinity as well as a change from activation to inhibition.
Table 4.
Enzyme kinetic parameter for inhibition of h-5-LOX by inhibitor 23d versus linoleic acid
| 23d (μM) | Kmapp (mM) | Vmaxapp (μM/s) | R2 |
|---|---|---|---|
| 0 | 0.624 ± 0.085 | 2.24 ± 0.23 | 0.999 |
| 25 | 0.666 ± 0.121 | 1.84 ± 0.25 | 0.998 |
| 50 | 0.690 ± 0.101 | 1.47 ± 0.16 | 0.999 |
Figure 8.

Steady-state kinetic characterization of linoleic acid conversion by h-5-LOX by inhibitor 23d. (a) Michaelis-Menten plots and (b) Lineweaver-Burk plots show the relation of h-5-LOX activity versus linoleic acid concentration at three selected concentrations of 23d (■) 0 μM, (●) 25 μM, and (▲) 50 μM.
Scheme 4.

Kinetic model for mixed type enzyme inhibition.
Figure 9.

Equation 2 (a), Equation 3 (b) and equation 4 (c) for the enzyme kinetics according to the model in Scheme 2. v is the reaction velocity, Vmax is the maximal reaction velocity, [S] is the substrate concentration and Km is the Michaelis-Menten constant. α and α′, respectively, are the parameters to describe the change of substrate binding affinity to the enzyme and the change of the maximum velocities. Ki is the dissociation constant of the inhibitor to the free enzyme and Ki′ is the dissociation constant of the inhibitor to the enzyme-substrate complex.
Taking this together, the kinetic studies on activation or inhibition of human 5-lipoxygenase by either 23a or 23d indicate the presence of an allosteric binding site that influences the enzyme activity. In addition, both activation and inhibition obey a comparable kinetic model, which suggests that both effects originate from the same binding site in the enzyme. Presumably, depending on the structure of the compounds that bind to this allosteric site the enzyme would either be activated or inhibited. This is in line with the literature where the presence of an allosteric binding site in h-5-LOX that regulates its activity has been described frequently [45–48]. We conclude that anacardic acid derivatives can be used as tools to modulate the h-5-LOX activity toward linoleic acid and that these compounds modulate the activity of this enzyme by binding to an allosteric binding site.
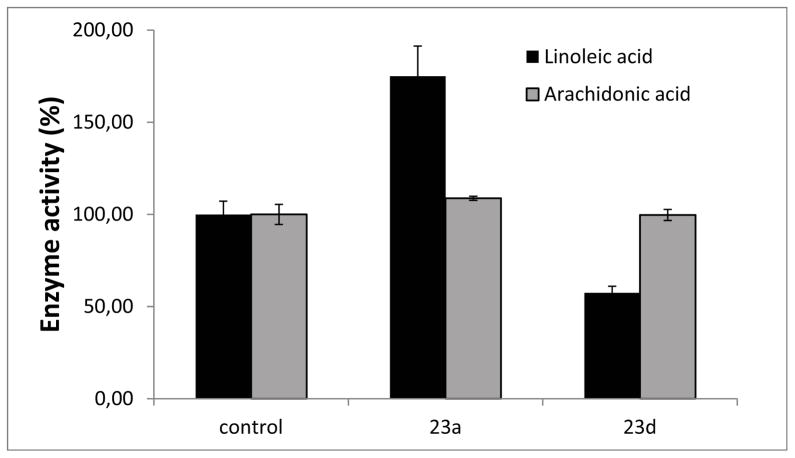
We continued our studies with investigation of the modulation of 23a and 23d of the h-5-LOX activity for arachidonic acid (Figure 10). Interestingly, no modulation of the h-5-LOX activity toward arachidonic acid was observed in presence of 23a or 23d. These results demonstrate that 23a and 23d are selective modulators of linoleic acid conversion and not arachidonic acid conversion by h-5-LOX.
Figure 10.
Residual enzyme activity that was observed for compounds 23a and 23d for inhibition of h-5-LOX in the presence of 50 μM of the respective compounds with linoleic acid or arachidonic acid as a substrate both at final concentrations of 100 μM. The results presented were the average of three independent experiments and the standard deviations are shown.
2.4. Molecular modeling
These results justify the hypothesis that the allosteric regulators 23a and 23d bind to an allosteric regulatory site in h-5-LOX that is close to the active site. The hypothesis is supported by a previous report, which stated that h-5-LOX has a wide active site compared to the others LOXs, which could harbor two substrate molecules [49]. The observation that 23a and 23d modulate the conversion of linoleic acid, which has 18 carbon atoms, and not arachidonic acid, which has 20 carbon atoms, is in line with this hypothesis, because it is an indication for steric interactions between the substrate and 23a or 23d. Therefore, we hypothesize that the substrates together with either 23a or 23d bind in the active site, thus resulting in a different binding constant and a different turnover rate of the substrate linoleic acid. We employed this presumption as a basis for the modeling studies.
The binding models of the activator 23a and the inhibitor 23d in the linoleic acid bound as well as the arachidonic acid bound h-5-LOX active site were made based on the recently published crystal structure of h-5-LOX (PDB code 3V99) [50]. The model of h-5-LOX was further refined using the related PDB structures 3V98 and 3V92 to model Ile673 and its carboxylic acid of the C-terminal (missing in 3V99), which coordinates the catalytic iron in the active site. We note that the model is not meant to capture the role of the phosphomimic S663D, but only as the structural basis for substrate binding. Additionally, the active site is in an open configuration accessible to a second small molecule. The above notwithstanding the structure is nearly indistinguishable from the wild type structure [50].
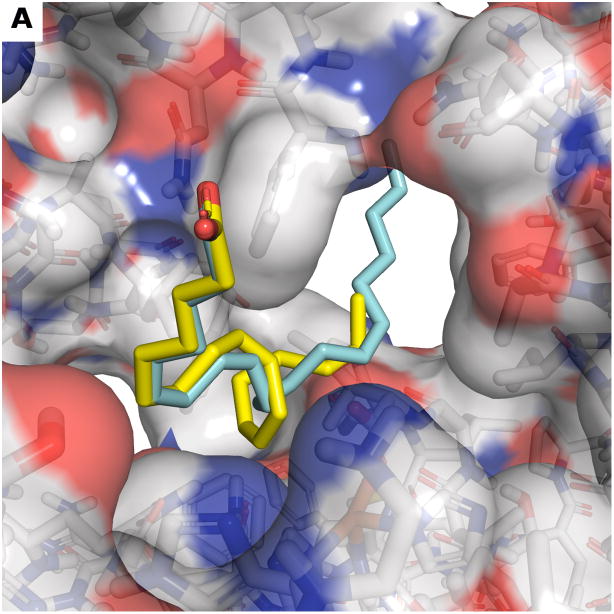
The arachidonic acid substrate molecule lacks visible specific interactions with the protein except for the interaction of the cis double bond at carbon 11 to the catalytic iron. This lack of visible specific interactions renders the virtual re-docking of arachidonic acid and de novo docking of linoleic acid in the h-5-LOX enzyme active site futile. Therefore, we modeled a bound structure of linoleic acid by manually threading the lipid into the crystal structure of arachidonic acid using PyMOL [51]. The threaded linoleic acid was then minimized using default settings in UCSF Chimera (version 1.6) [52] to produce the final model of linoleic acid bound to the h-5-LOX active site as shown in Figure 11a. This model is justified by the high similarity of linoleic acid to arachidonic acid, which is present in the crystal structure. This model demonstrates that arachidonic acid protrudes further into the active site then linoleic acid, which support the idea that steric interaction between 23a and 23d with the lipid substrate plays a role in the observed enzyme kinetics.
Figure 11. Docking models of Compounds 23 a,b,c,d in a model of linoleic acid bound to h-5-LOX.
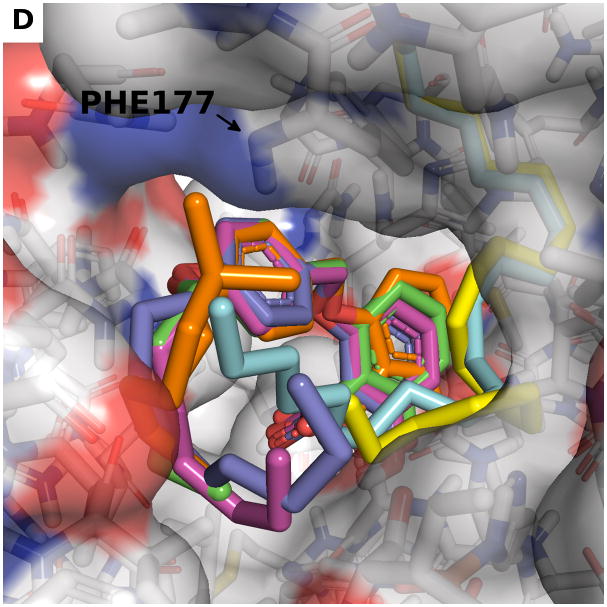
(a) Model of bound linoleic acid (yellow) based on a superposition to the configuration of arachidonic acid in the crystal structure (blue). Front (b) and back (c) view of the highest ranking binding mode of compound 23a (green) and compound 23d (orange) in the linoleic acid bound active site (a). Yellow dashed lines indicate the hydrogen bond between the ether oxygen and the backbone nitrogen of Phe177. Black dashed lines indicate hydrophobic interactions. (d) Superposition of highest ranked docked configurations of compounds 23a (green), 23b (purple), 23c (dark blue), 23d (orange) show that larger carbon tails block larger portions of the entrance to the catalytic site. Linoleic acid (yellow) and arachidonic acid (blue) are also shown as references.
Compound 23a and 23d were docked in the free enzyme, which can be justified by the observations that their affinity for the enzyme is higher (23a KA 8.6 μM, 23d Ki 70 μM) than the substrate linoleic acid (Km 600 μM). Using the structure of h-5-LOX with the arachidonic acid substrate removed, we modeled the binding mode of 23a and 23d by molecular docking using smina [55] and the resulting poses were reranked using the vina scoring function. Despite the large binding site (relative to the size of the compound), we observe a common binding mode for compounds 23a and 23d (as well as 23b and 23c, not shown). Subsequently, the substrate linoleic acid or arachidonic acid were put into the enzyme as described above. This binding mode of 23a and 23d is characterized by a strong pi-pi stacking interaction between the central phenyl ring of the compound and Phe177 on one side and the hydrophobic side chain of Ala410 on the other (Figure 11b). The buried polar functionalities of the planar salicylate head group may be stabilized by a hydrogen bond between the carboxylic -OH group of the head group and the side chain of His372. Also of importance is the hydrogen bond between the backbone nitrogen of Phe177 and the ether oxygen of the hydrophobic tail (Figure 11c). The dominant docking poses of compounds 23a–d share a similar geometry. A few of the low score poses have the salicylate head group rotated by 180 degrees. Compounds 23a–d are identical except for the length of their alkoxy tails (Table 1). The only differences in the full series of docked conformations is in the long flexible alkoxy tails of 23c–d that can fit in a variety of conformations with almost identical energy scores (Figure 11d).
The models of 23a–d (Figure 11d) are consistent with our experimental observations. Figure 1 and Table 1 demonstrate that the h-5-LOX enzymatic activity decreases with an increase in the alkoxy tail length. In addition, it was observed that 23a and 23d influence the linoleic acid conversion and not the arachidonic acid conversion by h-5-LOX (Figure 10).
Our docking experiments suggest that 23a binds alongside the substrate linoleic acid and stabilizes its binding and enhances the enzyme activity. Compound 23d has a longer tail with multiple binding modes that are likely to block the end of the binding site, hindering the entry of linoleic acid and thus reducing enzymatic activity (Figure 11c). Thus, the shorter linoleic acid substrate leaves space for the alkoxy tails of 23a and 23d (Figure 11c). This model would explain the high affinity of 23a for the linoleic acid bound enzyme (αKA 0.38 μM) compared to the free enzyme (KA 8.7 μM) and it is also in line with a higher conversion rate (β 1.76). Perhaps more interestingly, the model of 23d indicates that an increase in ‘bulk’ for the alkoxy tail provides inhibition of the enzyme activity for linoleic acid in contrast to the observed activation by 23a. Namely, the increased ‘bulk’ in 23d must interfere with the enzymatic activity for linoleic acid by disrupting the necessary binding pose or by blocking the entrance to the active site, which limits the access of linoleic acid. Conversely, the larger arachidonic acid might not permit binding of 23a or 23d and thus is its enzymatic activity is unaffected.
Taking all these data together we propose a model to explain our observations. We conclude that compound 23a is an activator for linoleic acid conversion by h-5-LOX, presumably because it stabilizes the substrate-enzyme complex without blocking the entrance to the active site. On the other hand, compound 23d could bind in a comparable configuration but its larger carbon tail, presumably, blocks one end of the active site which interferes with the diffusion of the substrate and molecular oxygen thus overriding the stabilization effect. Although these models provide plausible explanations for the observed behavior of this compound class, we realize crystallography studies would provide the ultimate experimental proof for the mode of action of these molecules.
3. Conclusion
Anacardic acid derivatives show interesting structure activity relationships that demonstrate that these compounds can either activate or inhibit the activity of human-5-LOX. Compound 23a and 23d either activate or inhibit the activity of human 5-LOX toward linoleic acid as a substrate depending on the substitution in para- position of the 2-(benzyloxy)-6-hydroxybenzoic acid core. Compound 23a was identified as a non-essential activator of human 5-lipoxygenase with KA is 8.65 μM, α is 0.0438, and β is 1.76. The affinity of activator 23a to the substrate bound enzyme (αKA) is 0.38 μM, which is 50 times higher than to the free enzyme. Furthermore, activator 23a acts as non-competitive activator against ATP, which indicates the presence of an allosteric site that is different from the ATP binding site. Kinetic studies on inhibitor 23d show a mixed type inhibition, which indicates the presence of an allosteric regulatory site for human 5-LOX activity. Both modulators 23a and 23d demonstrated no effect on the conversion of the substrate arachidonic acid by h-5-LOX. Molecular modeling studies indicate that 23a and 23d bind close to the substrate linoleic acid in the active site of h-5-LOX, but that these modulators do not fit simultaneously in the active site with the arachidonic acid, which is slightly larger than linoleic acid.
4. Experimental
4.1. Chemistry
4.1.1. General
All reagents and solvents were purchased from commercial suppliers (Fluka, Sigma-Aldrich, Acros Organics) and were used without further purification. Dichloromethane was distilled over CaH2 before use. Merck silica gel 60 F254 plates were used for analytical thin layer chromatography (TLC) and spots were detected by UV light, or stained using KMnO4 solution. Column chromatography was performed with MP Ecochrom silica gel 32–63, 60Ǻ using the flash chromatography technique. 1H (200 MHz) and 13C (50 MHz) NMR spectra were recorded on a Varian Gemini 200 spectrometer. 13C NMR spectra were recorded using the attached proton test (APT). Chemical shifts are reported in ppm (δ) relative to the solvent. Atmospheric Pressure Photoionization mass spectra (APPI-MS) and electrospray ionization (ESI) were recorded on an Applied Biosystems/SCIEX API3000-triple quadrupole mass spectrometer. High-resolution mass spectra (HR-MS) were recorded using a flow injection method on a LTQ-Orbitrap XL mass spectrometer (Thermo Electron, Bremen, Germany) with a resolution of 60,000 at m/z 400. Protonated testosterone (lock mass m/z = 289.2162) was used for internal recalibration in real time.
N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD) hydrochloride, human 5-lipoxygenase enzyme, and COX-2 (human recombinant) were obtained from Cayman Chemicals. Adenosine tri-phosphate (ATP), arachidonic acid, and linoleic acid were obtained from Sigma-Aldrich. Raw Blue cells assay was obtained from InvivoGen. The MTS assay kit was obtained from Promega. Interferon γ (IFN-γ) was obtained from PeproTech.
4.1.2 Synthetic Procedure 1: Synthesis of alkoxy benzyl alcohol
1-bromo alkane (13 mmol) was added to a light yellow suspension of hydroxybenzaldehyde (1.1 g, 9.0 mmol) and K2CO3 (5.2 g, 37 mmol) in 45 mL DMF under nitrogen atmosphere. The suspension was stirred overnight at room temperature. The reaction mixture was diluted with demiwater (100 mL) and extracted with ethylacetate (3 × 80 mL). The combined organic layers were washed with demiwater (4 × 80 mL), 0.1 M aqueous HCl (100 mL), brine (2 × 30 mL), dried with MgSO4, filtered and concentrated in vacuo.
NaBH4 (0.17 g, 4.5 mmol) was added to a solution of benzaldehyde (8.0 mmol) in 30 mL methanol at 0 °C under nitrogen atmosphere. The reaction mixture was stirred for one hour at 0 °C and then warmed to room temperature. The reaction mixture was diluted with demiwater (80 mL) and extracted with ethylacetate (2 × 60 mL). The combined organic layers were washed with demiwater (2 × 40 mL), brine (2 × 40mL), dried with MgSO4, filtered and concentrated in vacuo. The product was obtained after crystallization or purification using column chromatography.
4.1.3. Synthetic procedure 2: Bromination of alkyloxy benzyl alcohol
Phosphorus tribromide (PBr3) (0.34 mL, 3.6 mmol) was added slowly to a suspension of alkyloxy benzyl alcohol (2.5 mmol) in 10 mL CH2Cl2 under nitrogen atmosphere at 0 °C to give an orange suspension. The reaction mixture was stirred for 1.5 hours at 0 °C. The reaction mixture was poured into 80 mL ice water, and extracted with ethylacetate (3 × 50 mL). The combined organic layers were washed with demiwater (50 mL), 0.1 M aqueous HCl (2 × 30 mL), brine (50 mL), dried with MgSO4, filtered and concentrated in vacuo. The aryl bromide product was obtained after purification using column chromatography.
4.1.4. Synthetic Procedure 3: Williamson ether synthesis coupling of aryl bromide
5-hydroxy-2,2-dimethyl-4H-benzo[d][1,3]dioxin-4-one 1 (0.22 g, 1.1 mmol) and K2CO3 (0.63 g, 4.6 mmol) were suspended in 8 mL DMF under nitrogen atmosphere. A suspension of aryl bromide in 7 mL DMF was added to the suspension. The mixture was stirred overnight at room temperature and was diluted with demiwater (60 mL) and extracted with ethylacetate (3 × 45 mL). The combined organic layers were washed with demiwater (2 × 40 mL), 0.1 M aqueous HCl (40 mL), brine (2 × 20 mL), dried over MgSO4, filtered and concentrated in vacuo. The product was obtained after purification using column chromatography.
4.1.5. Synthetic procedure 4: saponification of the acetonide
5 M KOH (0.16 mL, 0.80 mmol) was added to a solution of 2,2-dimethyl-5-((4-(alkyloxy)benzyl)oxy)-4H-benzo[d][1,3]dioxin-4-one (0.15 g, 0.40 mmol) in 10 mL THF. The reaction mixture was heated till 60 °C and stirred overnight affording an orange suspension. The reaction mixture was concentrated in vacuo yielding the product as potassium salt.
4.1.6. Synthetic procedure 5: hydrolysis of acetonide
5 M KOH (1.2 mL, 6 mmol) was added to a colorless solution of acetonide (0.60 mmol) in 10 mL THF. The reaction mixture was stirred for 24 hours at 62 °C. The reaction mixture was diluted and acidified with 1 N aqueous HCl (2.6 mL) and extracted with ethylacetate (2 × 40 mL). The combined organic layers were washed with water (50 mL), brine (2 × 30 mL), dried over MgSO4, filtered and concentrated in vacuo.
4.1.7. (4-(pentyloxy)phenyl)methanol (2a)
The product was obtained using Williamson ether synthesis followed by aldehyde reduction of 1-bromopentane, and 4-hydroxybenzaldehyde using synthetic procedure 1. The product was purified using column chromatography with heptane:EtOAc 5:1 (v/v) as an eluent. Yield 73%. Light orange solid. Rf=0.41 (heptane:EtOAc 1:1). 1H NMR (200 MHz, CDCl3) δ 7.40 – 7.18 (m, 2H), 6.97 – 6.79 (m, 2H), 4.59 (s, 2H), 3.95 (t, J = 6.6, 2H), 1.87 (s, 1H), 1.86 – 1.72 (m, 2H), 1.46 – 1.34 (m, 4H), 0.94 (t, J = 7.0, 3H). 13 C NMR (50 MHz, CDCl3) δ 158.47, 133.10, 128.80, 114.73, 68.24, 65.20, 29.14, 28.38, 22.65, 14.20. MS (APPI): m/z 194.2 [M+.].
4.1.8. 1-(bromomethyl)-4-(pentyloxy)benzene (3a)
The product was obtained from (4-(pentyloxy)phenyl)methanol 2a using synthetic procedure 2. The product was obtained in high purity and no further purification was required. Yield 97%. Yellow solid. Rf = 0.72 (heptane:EtOAc 1:1).1H NMR (200 MHz, CDCl3) δ 7.38 – 7.22 (m, 2H), 6.94 – 6.74 (m, 2H), 4.51 (s, 2H), 3.94 (t, J = 6.2, 2H), 1.91 – 1.65 (m, 2H), 1.59 – 1.21 (m, 4H), 0.94 (t, J = 7.1, 3H). 13 C NMR (50 MHz, CDCl3) δ 159.47, 130.60, 129.88, 114.94, 68.26, 34.30, 29.11, 28.38, 22.65, 14.22. MS (ESI): m/z 177.1 [M-Br]+.
4.1.9. 2,2-dimethyl-5-((4-(pentyloxy)benzyl)oxy)-4H-benzo[d][1,3]dioxin-4-one (12a)
The product was obtained from 5-hydroxy-2,2-dimethyl-4H-benzo[d][1,3]dioxin-4-one 1, and 1-(bromomethyl)-4-(pentyloxy)benzene 3a using synthetic procedure 3. The product was crystallized from iso-propanol. Yield 70%. White solid. Rf = 0.6 (heptane:EtOAc 1:1).1H NMR (200 MHz, CDCl3) δ 7.50 – 7.32 (m, 3H), 6.90 (d, J = 8.3, 2H), 6.65 (d, J = 8.5, 1H), 6.54 (d, J = 8.2, 1H), 5.18 (s, 2H), 3.95 (t, J = 6.6, 2H), 1.81 – 1.70 (m, 8H), 1.46 – 1.40 (m, 4H), 0.93 (t, J = 7.1, 3H). 13C NMR (50 MHz, CDCl3) δ 160.69, 159.10, 157.99, 136.40, 128.53, 128.26, 114.82, 109.57, 107.62, 105.41, 105.22, 104.17, 70.89, 68.23, 29.17, 28.41, 25.86, 22.67, 14.23. MS (APPI): m/z 370.2 [M+.].
4.1.10. potassium 2-hydroxy-6-((4-(pentyloxy)benzyl)oxy)benzoate (23a)
The product was obtained from 2,2-dimethyl-5-((4-(pentyloxy)benzyl)oxy)-4H-benzo[d][1,3]dioxin-4-one 12a using synthetic procedure 4. Yield quantitatively determined from the final yield of product in excess of KOH. Orange gum. Rf = 0.16 (heptane:EtOAc 1:1 containing 1% acetic acid). The product was more than 95% pure as judged from TLC and NMR. 1H NMR (200 MHz, D2O) δ 7.15 (d, J = 8.5, 2H), 6.89 (t, J = 8.3, 1H), 6.68 (d, J = 8.5, 2H), 6.34 (d, J = 8.3, 1H), 6.27 (d, J = 8.3, 1H), 4.91 (s, 2H), 3.68 (t, J = 6.5, 2H), 1.50 – 1.47 (m, 2H), 1.14 – 1.11 (m, 4H), 0.70 (t, J = 7.0, 3H).13C NMR (50 MHz, D2O) δ 195.90, 159.11, 158.08, 157.85, 131.54, 129.50, 128.81, 114.48, 111.31, 109.15, 105.52, 70.26, 68.25, 28.21, 27.56, 21.89, 13.38. HRMS: m/z [M–H]− calcd for C19H21O5 329.13945, found 329.13995.
4.1.11. (4-(heptyloxy)phenyl)methanol (2b)
The product was obtained using Williamson ether synthesis followed by aldehyde reduction of 1-bromoheptane, and 4-hydroxybenzaldehyde using synthetic procedure 1. The product was crystallized from heptane. Yield 87%. Colorless to white crystal. Rf = 0.44 (heptane:EtOAc 1:1). 1H NMR (200 MHz, CDCl3) δ 7.49 – 7.16 (m, 2H), 6.99 – 6.79 (m, 2H), 4.59 (s, 2H), 3.95 (t, J = 6.6, 2H), 1.83 (s, 1H), 1.81 – 1.71 (m, 2H), 1.55 – 1.19 (m, 8H), 0.89 (t, J = 6.4, 3H).13C NMR (50 MHz, CDCl3) δ 158.97, 133.09, 128.81, 114.75, 68.27, 65.22, 31.98, 29.46, 29.26, 26.20, 22.80, 14.28. MS (APPI): m/z 222.2 [M+.].
4.1.12. 1-(bromomethyl)-4-(heptyloxy)benzene (3b)
The product was obtained from (4-(heptyloxy)phenyl)methanol 2b using synthetic procedure 2. The product was obtained in high purity and no further purification was required. Yield 97%. Yellow oil. Rf = 0.71 (heptane:EtOAc 1:1).1H NMR (200 MHz, CDCl3) δ 7.37 – 7.27 (m, 2H), 6.91 – 6.75 (m, 2H), 4.51 (s, 2H), 3.95 (t, J = 6.5, 2H), 1.91 – 1.68 (m, 2H), 1.57 – 1.18 (m, 8H), 0.91 (t, J = 6.5, 3H).13C NMR (50 MHz, CDCl3) δ 159.47, 130.60, 129.87, 114.94, 68.27, 34.30, 31.98, 29.42, 29.25, 26.19, 22.81, 14.29. MS (ESI): m/z 205.1 [M-Br]+.
4.1.13. 5-((4-(heptyloxy)benzyl)oxy)-2,2-dimethyl-4H-benzo[d][1,3]dioxin-4-one (12b)
The product was obtained from 5-hydroxy-2,2-dimethyl-4H-benzo[d][1,3]dioxin-4-one 1, and 1-(bromomethyl)-4-(heptyloxy)benzene 3b using synthetic procedure 3. The product was purified using column chromatography with heptane:EtOAc 11:1 (v/v) as an eluent. Yield 78%. White solid. Rf = 0.41 (heptane:EtOAc 3:1).1H NMR (200 MHz, CDCl3) δ 7.50 – 7.32 (m, 3H), 6.90 (d, J = 8.6, 2H), 6.90 (d, J = 8.4, 1H), 6.65 (d, J = 8.4, 1H), 5.18 (s, 2H), 3.94 (t, J = 6.5, 2H), 1.85 – 1.65 (m, 8H), 1.40 – 1.30 (m, 8H), 0.89 (t, J = 6.6, 3H). 13C NMR (50 MHz, CDCl3) δ 160.68, 159.11, 158.23, 157.99, 136.40, 128.53, 128.25, 114.82, 109.57, 107.61, 105.41, 104.17, 70.88, 68.25, 31.99, 29.48, 29.27, 26.21, 25.85, 22.82, 14.29. MS (APPI): m/z 398.2 [M+.].
4.1.14. Potassium 2-((4-(heptyloxy)benzyl)oxy)-6-hydroxybenzoate (23b)
The product was obtained from 5-((4-(heptyloxy)benzyl)oxy)-2,2-dimethyl-4H-benzo[d][1,3]dioxin-4-one 12b using synthetic procedure 4. Yield quantitatively determined from the final yield of product in excess of KOH. Orange solid. Rf = 0.49 (heptane:EtOAc 2:1 containing 1% acetic acid). The product was more than 95% pure as judged from TLC and NMR. 1H NMR (200 MHz, D2O) δ 6.93 (m, 2H), 6.60 (m, 1H), 6.42 (m, 2H), 6.18 (m, 1H), 5.94 (m, 1H), 4.74 (s, 2H), 3.38 (m, 2H), 1.31 – 1.06 (m, 10H), 0.68 (t, J = 6.6, 3H). 13C NMR (50 MHz, CD3OD) δ 202.2, 179.61, 161.89, 161.16, 160.88, 133.19, 131.99, 130.65, 116.15, 111.26, 106.62, 105.29, 72.48, 69.85, 33.85, 31.30, 31.08, 28.01, 24.53, 15.27. HRMS: m/z [M–H]− calcd for C21H25O5357.17075, found 357.17109.
4.1.15. (4-(nonyloxy)phenyl)methanol (2c)
The product was obtained using Williamson ether synthesis followed by aldehyde reduction of 1-bromononane, and 4-hydroxybenzaldehyde using synthetic procedure 1. The product was crystallized from heptane. Yield 73%. White crystal. Rf = 0.44 (heptane:ethylacetate 1:1).1H NMR (200 MHz, CDCl3) δ 7.35 – 7.20 (m, 2H), 6.95 – 6.82 (m, 2H), 4.60 (s, 2H), 3.95 (t, J = 6.6, 2H), 1.85 – 1.74 (m, 2H), 1.71 (s, 1H), 1.42 – 1.29 (m, 12H), 0.89 (t, J = 7.0, 3H).13C NMR (50 MHz, CDCl3) δ 158.99, 133.09, 128.82, 114.76, 68.28, 65.27, 32.08, 29.74, 29.61, 29.47, 26.24, 22.87, 14.31. MS (APPI): m/z 250.2 [M+.].
4.1.16. 1-(bromomethyl)-4-(nonyloxy)benzene (3c)
The product was obtained from (4-(nonyloxy)phenyl)methanol 2c using synthetic procedure 2. The product was obtained in high purity and no further purification was required. Yield 94%. Light yellow oil. Rf = 0.74 (heptane:EtOAc 1:1).1H NMR (200 MHz, CDCl3) δ 7.39 – 7.20 (m, 2H), 6.94 – 6.76 (m, 2H), 4.50 (s, 2H), 3.95 (t, J = 6.5, 2H), 1.88 – 1.66 (m, 2H), 1.54 – 1.14 (m, 12H), 0.89 (t, J = 6.4, 3H).13C NMR (50 MHz, CDCl3) δ 159.48, 130.61, 129.88, 114.96, 68.29, 34.31, 32.09, 29.74, 29.59, 29.47, 29.42, 26.23, 22.88, 14.32. MS (ESI): m/z 233.1 [M-Br]+.
4.1.17. 2,2-dimethyl-5-((4-(nonyloxy)benzyl)oxy)-4H-benzo[d][1,3]dioxin-4-one (12c)
The product was obtained from 5, -hydroxy-2,2-dimethyl-4H-benzo[d][1,3]dioxin-4-one 1, and 1-(bromomethyl)-4-(nonyloxy)benzene 3c using synthetic procedure 3. The product was purified using column chromatography with heptane:EtOAc 15:1 (v/v) as an eluent. Yield 85%. White solid. Rf = 0.48 (heptane:EtOAc 3:1). 1H NMR (200 MHz, CDCl3) δ 7.53 – 7.29 (m, 3H), 6.90 (d, J = 8.4, 2H), 6.65 (d, J = 8.4, 1H), 6.54 (d, J = 8.4, 1H), 5.18 (s, 2H), 3.94 (t, J = 6.5, 2H), 1.88 – 1.64 (m, 8H), 1.36 (d, J = 30.5, 12H), 0.88 (t, J = 6.2, 3H).13C NMR (50 MHz, CDCl3) δ 202.3, 160.67, 159.09, 158.20, 157.98, 136.39, 128.52, 128.24, 114.81, 109.56, 107.60, 105.40, 102.26, 70.87, 68.24, 32.08, 29.74, 29.61, 29.47, 26.25, 25.85, 22.87, 14.35, 14.31. MS (APPI): m/z 426.2 [M+.].
4.1.18. Potassium 2-hydroxy-6-((4-(nonyloxy)benzyl)oxy)benzoate (23c)
The product was obtained from 2,2-dimethyl-5-((4-(nonyloxy)benzyl)oxy)-4H-benzo[d][1,3]dioxin-4-one 12c using synthetic procedure 4. Yield quantitatively determined from the final yield of product in excess of KOH. Rf = 0.18 (heptane:EtOAc 1:1 containing 1% acetic acid). The product was more than 95% pure as judged from TLC and NMR. 1H NMR (200 MHz, D2O) δ 6.95 (d, J = 8.3, 2H), 6.57 (t, J = 8.5, 1H), 6.43 (d, J = 8.3, 2H), 6.16 (d, J = 8.5, 1H), 5.91 (d, J = 8.5, 1H), 4.75 (s, 2H), 3.38 (m, 2H), 1.34 (m, 2H), 1.06 (m, 12H), 0.71 (t, J = 7.0, 3H). 13C NMR (50 MHz, CD3OD) δ 202.2, 164.24, 160.76, 156.88, 133.10, 131.98, 130.67, 116.19, 111.48, 109.33, 106.36, 72.41, 69.89, 61.77, 33.83, 31.47, 31.31, 31.18, 28.10, 27.95, 24.52, 14.22. HRMS: m/z [M–H]− calcd for C23H29O5 385.20205, found 385.20248.
4.1.19. (4-((3,7-dimethyloctyl)oxy)phenyl)methanol (2d)
The product was obtained using Williamson ether synthesis followed by aldehyde reduction of 1-1-bromo-3,7-dimethyloctane, and 4-hydroxybenzaldehyde using synthetic procedure 1. The product was used for the next reaction step without further purification. Yield 92%. Yellow oil. Rf=0.48 (heptane:EtOAc 1:1). 1H NMR (200 MHz, CDCl3) δ 7.27 (d, J = 8.6, 2H), 6.88 (d, J = 8.6, 2H), 4.59 (s, 2H), 3.98 (t, J = 6.5, 2H), 1.88 – 1.46 (m, 5H), 1.32 – 1.14 (m, 6H), 0.93 (d, J = 6.2, 3H),0.87 (d, J = 6.5, 6H). 13C NMR (50 MHz, CDCl3) δ 158.98, 133.10, 128.82, 114.77, 66.59, 65.27, 39.45, 37.49, 36.40, 30.06, 28.17, 24.86, 22.91, 22.81, 19.86. MS (APPI): m/z 264.1 [M+.].
4.1.20. 1-(bromomethyl)-4-((3,7-dimethyloctyl)oxy)benzene (3d)
The product was obtained from (4-((3,7-dimethyloctyl)oxy)phenyl)methanol 2d using synthetic procedure 2. The product was used for the next reaction step without further purification. Yield quantitative. Light yellow oil. Rf = 0.75 (heptane:EtOAc 1:1). 1H NMR (200 MHz, CDCl3) δ 7.30 (d, J = 8.6, 2H), 6.85 (d, J = 8.6, 2H), 4.49 (s, 2H), 3.98 (t, J = 6.5, 2H), 1.87 – 1.46 (m, 4H), 1.32 – 1.10 (m, 6H), 0.93 (d, J = 6.2, 3H), 0.87 (d, J = 6.4, 6H).13C NMR (50 MHz, CDCl3) δ 159.46, 130.60, 129.88, 114.95, 66.59, 39.45, 37.47, 36.35, 34.30, 30.04, 28.18, 24.86, 22.92, 22.82, 19.85. MS (ESI): m/z 247.1 [M-Br]+.
4.1.21. 5-((4-((3,7-dimethyloctyl)oxy)benzyl)oxy)-2,2-dimethyl-4H-benzo[d][1,3]dioxin-4-one (12d)
The product was obtained from 5-hydroxy-2,2-dimethyl-4H-benzo[d][1,3]dioxin-4-one 1, and 1-(bromomethyl)-4-((3,7-dimethyloctyl)oxy)benzene 3d using synthetic procedure 3. The product was purified using column chromatography with heptane:EtOAc 19:1 (v/v) as eluent. Yield 68%. White solid. Rf = 0.35 (heptane:EtOAc 4:1). 1H NMR (200 MHz, CDCl3) δ 7.46 – 7.26 (m, 3H), 6.92 (d, J = 8.6, 2H), 6.65 (d, J = 8.5, 1H), 6.54 (d, J = 8.5, 1 H), 5.18 (s, 2H), 3.98 (t, J = 6.5, 2H), 1.84 – 1.5 (m, 10H), 1.32 – 1.14 (m, 6H), 0.93 (d, J = 6.2, 3H), 0.87 (d, J = 6.4, 6H). 13C NMR (50 MHz, CDCl3) δ 160.69, 159.10, 158.23, 157.99, 136.40, 128.53, 128.25, 114.82, 109.57, 107.61, 105.41, 70.89, 66.56, 39.45, 37.49, 36.40, 30.05, 28.17, 25.85, 24.85, 22.91, 22.81, 19.86. MS (APPI): m/z 440.4 [M+.].
4.1.22. Potassium 2-((4-((3,7-dimethyloctyl)oxy)benzyl)oxy)-6-hydroxybenzoate (23d)
The product was obtained from 5-((4-((3,7-dimethyloctyl)oxy)benzyl)oxy)-2,2-dimethyl-4H-benzo[d][1,3]dioxin-4-one 12d using synthetic procedure 4. The compound is racemic at its asymmetric carbon. Yield quantitatively determined from the final yield of product in excess of KOH. Orange solid. Rf = 0.54 (heptane:EtOAc 1:1 containing 1% acetic acid). The product was more than 95% pure as judged from TLC and NMR. 1H NMR (200 MHz, CD3OD) δ 7.42 (d, J = 8.7, 2H), 7.05 (t, J = 8.2, 1H), 6.95 – 6.85 (m, 2H), 6.43 – 6.38 (m, 2H), 5.03 (s, 2H), 3.99 (t, J = 6.7 2H), 1.92 – 1.48 (m, 4H), 1.40 – 1.16 (m, 6H), 0.95 (d, J = 6.2, 3H), 0.89 (d, J = 6.5, 6H). 13C NMR (50 MHz, CD3OD) δ 202.3, 164.42, 161.42, 160.86, 132.74, 132.06, 130.69, 116.17, 111.76, 105.95, 72.47, 68.11, 41.28, 39.28, 38.27, 31.89, 29.99, 26.67, 23.95, 23.86, 20.90. HRMS: m/z [M–H]− calcd for C24H31O5 399.2177, found 399.21844.
4.1.23. (3-(heptyloxy)phenyl)methanol (4b)
The product was obtained using Williamson ether synthesis followed by aldehyde reduction of 1-bromoheptane, and 3-hydroxybenzaldehyde using synthetic procedure 1. The product was used for the next reaction step without further purification. Yield 94%. Brown solid. Rf = 0.48 (heptane:EtOAc 1:1). 1H NMR (200 MHz, CDCl3) δ 7.26 – 7.18 (m, 1H), 6.96 – 6.75 (m, 3H), 4.64 (s, 2H), 3.95 (t, J = 6.5, 2H), 1.88 – 1.67 (m, 3H), 1.55 – 1.17 (m, 8H), 0.89 (t, J = 6.5, 3H). 13C NMR (50 MHz, CDCl3) δ 159.62, 142.67, 129.74, 119.09, 114.01, 113.09, 68.18, 65.48, 31.99, 29.49, 29.27, 26.22, 22.82, 14.34. MS (APPI): m/z 222.2 [M+.].
4.1.24. 1-(bromomethyl)-3-(heptyloxy)benzene (5b)
The product was obtained from (3-(heptyloxy)phenyl)methanol 4b using synthetic procedure 2. Therefore product was used for the next reaction step without further purification. Yield 86%. Brown solid. Rf = 0.71 (heptane:EtOAc 1:1). 1H NMR (200 MHz, CDCl3)7.32 – 6.27 (m, 4H), 4.76 (s, 2H), 3.92 (t, J = 6.4, 2H), 1.72–1.65(m, 2H), 1.34 – 1.15 (m, 8H), 0.89 (t, J = 6.5, 3H). 13C NMR (50 MHz, CDCl3) δ 159.41, 129.63, 119.81, 114.36, 105.23, 68.05, 32.65, 32.01, 29.53, 29.36, 26.24, 22.84, 14.31. MS (ESI): m/z 285.0 [M+.].
4.1.25. 5-((3-(heptyloxy)benzyl)oxy)-2,2-dimethyl-4H-benzo[d][1,3]dioxin-4-one (13b)
The product was obtained from 5-hydroxy-2,2-dimethyl-4H-benzo[d][1,3]dioxin-4-one 1, and 1-(bromomethyl)-3-(heptyloxy)benzene 5b using synthetic procedure 3. The product was purified using column chromatography with heptane:EtOAc 16:1 (v/v) as eluent. Yield 58%. White solid. Rf = 0.29 (heptane:EtOAc 5:1).1H NMR (200 MHz, CDCl3) δ 7.42 – 7.35 (m, 2H), 7.25 (d, J = 7.5, 1H), 7.04 (d, J = 7.5, 1H), 6.83 (d, J = 7.9, 1H), 6.70 – 6.49 (m, 2H), 5.22 (s, 2H), 3.99 (t, J = 6.4, 2H), 1.87 – 1.68 (m, 2H) 1.56 (s, 6H), 1.42 – 1.12 (m, 8H), 0.89 (t, J = 6.5, 3H). 13C NMR (50 MHz, CDCl3) δ 160.53, 159.77, 158.23, 158.01, 138.02, 136.48, 129.73, 118.57, 114.61, 112.43, 109.67, 107.36, 105.46, 70.74, 68.19, 32.00, 29.50, 29.29, 26.26, 25.85, 22.82, 14.30. MS (APPI): m/z 399.2 [M+H]+.
4.1.26. 2-((3-(heptyloxy)benzyl)oxy)-6-hydroxybenzoic acid (24b)
The product was obtained from 5-((3-(heptyloxy)benzyl)oxy)-2,2-dimethyl-4H-benzo[d][1,3]dioxin-4-one 13b using synthetic procedure 5. Yield quantitative. Yellow oil. Rf = 0.42 (heptane:EtOAc 1:1 containing 1% acetic acid). The product was more than 95% pure as judged from TLC and NMR. 1H NMR (200 MHz, DMSO-d6) δ 7.26 – 7.19 (m, 2H), 7.08 (d, J = 7.5, 1H), 7.04 (d, J = 7.5, 1H), 6.79 (d, J = 8.11H), 6.31 (d, J = 8.2, 2H), 5.04 (s, 2H), 3.95 (t, J = 6.4, 2H), 1.73 – 1.63 (m, 2H), 1.36 – 1.18 (m, 8H), 0.87 (t, J = 7.0, 3H). 13C NMR (50 MHz, DMSO-d6) δ 170.28, 164.46, 159.34, 158.66, 139.68, 130.82, 129.05, 118.77, 113.03, 110.05, 109.85, 102.77, 69.61, 67.26, 31.26, 28.75, 28.48, 25.55, 22.07, 13.96. HRMS: m/z [M–H]− calcd for C21H25O5 357.17075, found 357.17114.
4.1.27. (3-(nonyloxy)phenyl)methanol (4c)
The product was obtained using Williamson ether synthesis followed by aldehyde reduction of 1-bromoheptane, and 3-hydroxybenzaldehyde using synthetic procedure 1. The product was used for the next reaction step without further purification. Yield 89%. Rf = 0.54 (heptane:EtOAc 1:1). 1H NMR (200 MHz, CDCl3) δ 7.22 (t, J = 7.8, 1H), 6.89 (s, 1H), 6.85 – 6.72 (m, 2H), 4.59 (s, 2H), 3.92 (t, J = 6.5, 2H), 2.32 (s, 1H), 1.79 – 1.69 (m, 2H), 1.42 – 1.28 (m, 12H), 0.88 (t, J = 6.3, 3H). 13C NMR (50 MHz, CDCl3) δ 159.52, 142.67, 129.62, 119.04, 113.88, 113.02, 68.11, 65.25, 32.05, 29.72, 29.58, 29.44, 26.22, 22.84, 14.26. MS (APPI): m/z250.2 [M+.].
4.1.28. 1-(bromomethyl)-3-(nonyloxy)benzene (5c)
The product was obtained from (3-(nonyloxy)phenyl)methanol 4c using synthetic procedure 2. Therefore the product was used for the next reaction step without further purification. Yield 78%. Yellow oil. Rf = 0.71 (heptane:EtOAc 1:1).1H NMR (500 MHz, CDCl3) δ 7.22 (t, J = 7.8, 1H), 6.96 – 6.92 (m, 2H), 6.84 – 6.81 (m, 1H), 4.46 (s, 2H), 3.95 (t, J = 7.0, 2H), 1.87 – 1.75 (m, 2H), 1.47 – 1.28 (m, 12H), 0.89 (t, J = 6.8, 3H). 13C NMR (50 MHz, CDCl3) δ 159.33, 139.04, 129.73, 121,03, 115.03, 114.67, 68.02, 33.55, 31.86, 29.52, 29.38, 29.24, 26.02, 22.65, 14.09. MS (ESI): m/z 313.0 [M+.]
4.1.29. 2,2-dimethyl-5-((3-(nonyloxy)benzyl)oxy)-4H-benzo[d][1,3]dioxin-4-one (13c)
The product was obtained from 5-hydroxy-2,2-dimethyl-4H-benzo[d][1,3]dioxin-4-one 1, and 1-(bromomethyl)-3-(nonyloxy)benzene 5c using synthetic procedure 3. The product was purified using column chromatography with heptane:EtOAc 16:1 (v/v) as eluent. Yield 88%. Yellow oil. Rf = 0.29 (heptane:EtOAc 5:1). 1H NMR (500 MHz, CDCl3) δ 7.41–7.35 (m, 2H), 7.25 – 7.20 (m, 2H), 7.04 (d, J = 7.0, 1H), 6.82 (d, J = 6.9, 1H), 6.59 (d, J = 8.0, 1H), 5.21 (s, 2H), 3.99 (t, J = 6.0, 2H), 1.89 – 1.62 (m, 8H), 1.51 – 1.13 (m, 12H), 0.87 (d, J = 6.3, 3H). 13C NMR (50 MHz, CDCl3) δ 160.46, 159.71, 157.95, 137.97, 136.45, 129.66, 118.51, 114.55, 112.37, 109.62, 107.30, 105.39, 103.97, 70.68, 68.13, 32.04, 29.71, 29.60, 29.43, 26.26, 25.79, 22.83, 14.28. MS (ESI): m/z 427.3 [M+H]+.
4.1.30. 2-hydroxy-6-((3-(nonyloxy)benzyl)oxy)benzoic acid (24c)
The product was obtained from 5-((3-(heptyloxy)benzyl)oxy)-2,2-dimethyl-4H-benzo[d][1,3]dioxin-4-one 13c using synthetic procedure 5. The product was obtained in high purity and no further purification was required. Yield 58%. Yellow oil. Rf = 0.60 (heptane:EtOAc 1:1 containing 1% acetic acid). The product was more than 95% pure as judged from TLC and NMR. 1H NMR (200 MHz, CD3OD) δ 7.27 – 7.20 (m, 2H), 7.11 – 7.01 (m, 2H), 6.81 (d, J = 7.8, 1H), 6.49 (d, J = 7.5, 2H), 5.16 (s, 2H), 3.97 (t, J = 6.2, 2H), 1.75 – 1.71 (m, 2H), 1.41 – 1.23 (m, 12H), 0.90 (t, J = 6.5, 3H). 13C NMR (50 MHz, CD3OD) δ 185.96, 161.65, 156.96, 141.16, 131.25, 121.03, 115.76, 115.03, 107.29, 94.05, 72.58, 69.81, 33.92, 31.56, 31.43, 31.30, 28.07, 24.59, 15.30. HRMS: m/z [M–H]− calcd for C23H29O5 385.20205, found 385.20261.
4.1.31. 2,2-dimethyl-5-(nonyloxy)-4H-benzo[d][1,3]dioxin-4-one (11)
The product was obtained from 5-hydroxy-2,2-dimethyl-4H-benzo[d][1,3]dioxin-4-one 1, and 1-bromononane using synthetic procedure 3. The product was purified using column chromatography with heptane as eluent followed by heptane:EtOAc 50:1 (v/v) as eluent until the starting material was completely removed from the column and the product was obtained by eluting the column with EtOAc as eluent. Yield 53%. Light yellow solid. Rf = 0.64 (heptane:EtOAc 1:1). 1H NMR (200 MHz, CDCl3) δ 7.40 (t, J = 8.4, 1H), 6.59 (d, J =, 8.1, 1H), 6.51 (d, J =, 8.2, 1H) 4.06 (t, J = 6.7, 2H), 1.95 – 1.81 (m, 2H), 1.69 (s, 6H), 1.50 – 1.26 (m, 12H), 0.85 (t, J = 6.6, 3H). 13C NMR (50 MHz, CDCl3) δ 161.30, 158.20, 157.96, 136.41, 109.03, 106.62, 105.28, 103.67, 69.60, 32.07, 29.69, 29.55, 29.46, 29.14, 26.03, 25.84, 22.86, 14.31. MS (APPI): m/z 321.0 [M+H]+.
4.1.32. 2-hydroxy-6-(nonyloxy)benzoic acid (22)
The product was obtained from 5-((3-(heptyloxy)benzyl)oxy)-2,2-dimethyl-4H-benzo[d][1,3]dioxin-4-one 11 using synthetic procedure 5. The product was obtained in high purity and no further purification was required. Yield quantitative. Light brown solid. Rf = 0.60 (heptane:EtOAc 1:1 containing 1% acetic acid). The product was more than 95% pure as judged from TLC and NMR. 1H NMR (200 MHz, DMSO-d6) δ 7.02 (t, J = 8.2, 1H), 6.28 (d, J = 8.0, 1H), 6.24 (d, J = 8.1, 1H), 3.87 (t, J = 6.5, 2H), 1.68 – 1.58 (m, 2H), 1.40 – 1.17 (m, 12H), 0.86 (t, J = 6.5, 3H). 13C NMR (50 MHz, DMSO-d6) δ 164.31, 160.21, 131.19, 110.51, 109.82, 103.07, 69.00, 31.74, 29.48, 29.34, 29.14, 25.92, 22.54, 14.40. HRMS: m/z [M–H]− calcd for C16H23O4 279.16018, found 279.16061.
4.1.33. 1-(4-bromophenyl)decan-1-one (7)
The product was obtained using Friedel-Craft acylation of bromobenzene 6 with decanoylchloride. AlCl3 (0.81 g, 6.1 mmol) was added slowly to a solution of bromobenzene (0.54 mL, 5.1 mmol) and decanoylchloride (1.2 mL, 5.8 mmol) in 20 mL CH2Cl2 in a 50 mL flask at 0 °C. AlCl3 slowly dissolved in the mixture and the reaction mixture was stirred for 1 hour at 60 °C. The reaction mixture was poured into ice-water (25 mL) and extracted with CH2Cl2 (2 × 30 mL). The combined organic layers were dried over brine and concentrated in vacuo. The product was dissolved in CH2Cl2 and washed with 2 N aqueous HCl (2 × 15 mL) and dried over brine (15 mL) and concentrated in vacuo. The product was crystallized from heptane in a yield 62% (0.49 mg, 1.5 mmol) as a white solid. Rf = 0.69 (heptane:EtOAc 1:1). 1H NMR (200 MHz, CDCl3) δ 7.89 – 7.75 (m, 2H), 7.65 – 7.53 (m, 2H), 2.92 (t, J = 7.5, 2H), 1.75 – 1.64 (m, 2H), 1.32 – 1.27 (m, 12H), 0.87 (t, J = 6.4, 3H). 13C NMR (50 MHz, CDCl3) δ 199.72, 136.01, 132.06, 129.82, 128.19, 38.81, 32.08, 29.67 (2C), 29.54, 29.49, 24.50, 22.88, 14.32. MS (APPI): m/z 311.2 [M+].
4.1.34. 1-bromo-4-decylbenzene (8)
The product was obtained using a Wolff-Kishner reaction to reduce 1-(4-bromophenyl)decan-1-one 7. 1-(4-bromophenyl)decan-1-one 7 (1.3 g, 4.2 mmol), hydrazine monohydrate (1.2 mL, 16 mmol) and KOH (1.8 g, 32 mmol) were dissolved in 25 mL of 1-octanol to yield a green suspension in a two-necked 50 mL flask. The reaction mixture was stirred at reflux at 135 °C for 2¾ hours to give an almost colorless solution. The reaction mixture was then cooled to room temperature and diluted with ether (50 mL), washed with 1 N aqueous HCl (50 mL), 2 N aqueous HCl (20 mL), brine (2 times, 30 mL), dried over MgSO4, filtered, concentrated in vacuo. 1-octanol was removed by vacuum distillation to yield about 2.7 g of the crude product. The product was purified using column chromatography with heptane as eluent. Yield 40%. Colorless oil. Rf = 0.61 (100% heptane). 1H NMR (500 MHz, CDCl3) δ 7.37 (d, J = 8.3, 2H), 7.03 (d, J = 8.1, 2H), 2.54 (t, J = 7.7, 2H), 1.60 – 1.54 (m, 2H), 1.28 – 1.25 (m, 14H), 0.88 (t, J = 6.8, 3H). 13C NMR (50 MHz, CDCl3) δ 142.05, 131.46, 130.37, 119.47, 35.59, 32.14, 31.56, 29.85, 29.82, 29.71, 29.57, 29.43, 22.93, 14.34. MS (APPI): m/z 296.2 [M+].
4.1.35. ((4-decylphenyl)ethynyl)trimethylsilane
Into a dried 10–20 mL microwave vial, PdCl2(PPh3) (52 mg, 74 μmol), CuI (18 mg, 95 μmol), and PPh3 (80 mg, 0.30 mmol) were suspended in CH3CN (2 mL) under nitrogen atmosphere. Subsequently, 1-bromo-4-decylbenzene 8 (0.46 g, 1.5 mmol), diethyl amine (Et2NH) (2.4 mL, 23 mmol), and trimethylsilyl acetylene (TMSA) (0.24 mL, 1.7 mmol), were added to the mixture. The mixture was heated at 120 °C for 35 minutes via microwave irradiation (95 W). The reaction mixture was diluted with EtOAc (50 mL) and washed with water (3 × 50 mL). The water layer was washed with EtOAc (2 × 50 mL). The combined organic phases were extracted with brine (1 × 50 mL) and dried over Mg2SO4. The solvent was evaporated under reduced pressure and the residue was purified by column chromatography with heptane as eluent. Yield 65%. Brown oil. Rf = 0.41 (100% heptane). 1H NMR (500 MHz, CDCl3) δ 7.37 (d, J = 8.0, 2H), 7.10 (d, J = 7.9, 2H), 2.58 (t, J = 7.6, 2H), 1.58 – 1.54 (m, 2H), 1.28 – 1.25 (m, 14H), 0.88 (t, J = 6.8, 3H), 0.24 (s, 9H);13C NMR (50 MHz, CDCl3) δ 143.89, 132.08, 128.52, 120.44, 105.63, 93.45, 36.11, 32.12, 31.42, 29.82, 29.69 (2C), 29.55, 29.42, 22.91, 14.34, 0.26. MS (APPI): m/z 314.2 [M+].
4.1.36. 1-decyl-4-ethynylbenzene (9)
((4-decylphenyl)ethynyl)trimethylsilane (0.29 g, 0.91 mmol) was dissolved in THF (5 mL) and the solution was cooled to 0 °C. Tetra butyl ammonium fluoride (TBAF) (1.4 mL, 1.4 mmol) in THF (1 M, 2.6 mL) was added dropwise and the reaction mixture was stirred for 10 minutes. The reaction mixture was diluted with EtOAc (50 mL), extracted with water (4 × 50 mL) and washed with brine (2 × 50 mL). The organic phase was dried over MgSO4 and filtered. The solvent was evaporated and the product was used without further purification. Yield 93%. Brown oil. Rf = 0.45 (100% heptane). 1H NMR (500 MHz, CDCl3) δ 7.40 (d, J = 8.0, 2H), 7.13 (d, J = 7.8, 2H), 3.02 (s, 1H), 2.59 (t, J = 7.6, 2H), 1.59 – 1.55 (m, 2H), 1.29 – 1.26 (m, 14H), 0.88 (t, J = 6.8, 2H). 13C NMR (50 MHz, CDCl3) δ 144.22, 132.25, 128.63, 119.41, 105.22, 92.67, 36.12, 32.12, 31.45, 29.82, 29.79, 29.69, 29.55, 29.46, 22.91, 14.34. MS (APPI): m/z 242.2 [M+.].
4.1.37. Benzyl 2,4-bis(benzyloxy)-6-((4-decylphenyl)ethynyl)benzoate
The product was obtained using Sonogashira coupling reaction of 2,4-bis(benzyloxy)-6-(((trifluoromethyl)sulfonyl)oxy)benzoate 10, and 1-decyl-4-ethynylbenzene 9. Distilled diethylamine (0.11 mL, 1.05 mmol) and 1-decyl-4-ethynylbenzene 9 (0.18 g, 0.74 mmol) were subsequently added to a solution of the triflate 10 (0.36 g, 0.63 mmol), CuI (9.0 mg, 47 μmol), and PdCl2(PPh3)2 (18 mg, 0.026 mmol) in degassed anhydrous acetonitrile (1.0 mL) under nitrogen atmosphere. The mixture was subjected to microwave irradiation for 35 min at 120 °C (70 W). The reaction mixture was diluted with ether (50 mL) and washed with demi water (2 × 30 mL), brine (2 × 50 mL), dried over MgSO4 and filtered. The solvent was evaporated under reduced pressure, and the residue was purified by column chromatography with heptanes/EtOAc 5:1 (v/v) as eluent. Yield 80%. Brown oil. Rf = 0.40 (heptane:EtOAc 1:1). 1H NMR (500 MHz, CDCl3) δ 7.43 – 7.27 (m, 15H), 7.22 (d, J = 8.0, 2H), 7.12 (d, J = 7.9, 2H), 6.75 (s, 1H), 6.56 (s, 1H), 5.37 (s, 2H), 5.06 (s, 2H), 5.04 (s, 2H), 2.60 (t, J = 7.5, 2H), 1.60- 1.55 (m, 2H), 1.30- 1.26 (m, 14H), 0.88 (t, J = 6.6, 3H). 13C NMR (50 MHz, CDCl3) δ 166.91, 160.54, 157.24, 144.06, 136.47, 136.38, 136.09, 131.89, 128.89, 128.76, 128.61, 128.48, 128.44, 128.17, 128.12, 127.75, 127.30, 123.86, 120.06, 119.89, 109.67, 101.97, 93.47, 86.28, 70.80, 70.53, 67.30, 36.15, 32.12, 31.47, 29.81, 29.71, 29.55, 29.46, 22.91, 14.34. MS (APPI): m/z 665.2 [M+H+].
4.1.38. 2-(4-decylphenethyl)-4,6-dihydroxybenzoic acid (20)
Benzyl 2,4-bis(benzyloxy)-6-((4-decylphenyl)ethynyl)benzoate (0.28 g, 0.42 mmol) was dissolved in EtOAc (due to low solubility in MeOH) and added to 10 mol% Pd/C (10%) (45 mg, 42 nmol). The suspension was shaken with 3 atm H2-pressure in a Parr apparatus at 45 °C for 24 hours. Subsequently, the mixture was filtered through Celite. The solvent was evaporated under reduced pressure and the residue was purified by column chromatography with heptane/EtOAc 9:1 (v/v) as eluent, followed by EtOAc as eluent to obtain the desired product. Yield 72%. White solid. Rf = 0.40 (heptane:EtOAc 1:1 + acetic acid). The product was more then 95% pure as judged from TLC and NMR. 1H NMR (200 MHz, CD3OD) δ 7.12 – 7.02 (m, 4H), 6.18 (d, J=2.4 Hz, 1H); 6.16 (d, J= 2.4 Hz, 1H), 3.18 – 3.10 (m, 2H), 2.83 – 2.75 (m, 2H), 2.62 – 2.48 (m, 2H), 1.60 – 1.50 (m, 2H), 1.28 – 1.20 (m, 14H), 0.89 (t, J = 6.5, 3H). 13C NMR (50 MHz, CD3OD) δ 167.82, 164.57, 150.02, 142.23, 141.60, 130.15, 112.86, 106.44, 102.82, 41.15, 40.02, 37.41, 33.93, 33.67, 31.59, 31.47 (2C), 31.31, 31.19, 24.59, 15.30. HRMS: m/z [M–H]− calcd for C25H33O4 397.23843, found 397.2388.
4.1.39. 2-(benzo[d]oxazol-2-yl)phenol (26)
Salicylic acid (0.138 g, 1.0 mmol) was mixed with 2-aminophenol (0.11 g, 1.0 mmol) and 3 mL of polyphosphoric acid was added to the mixture. The mixture was heated in oil bath at 180 °C under N2 for 6.5 hours. The reaction mixture was cooled to room temperature and subsequently poured into 150 mL demi water. The mixture was extracted with ethylacetate (2 × 30 mL). The combined organic layers were washed with demiwater (2 × 40 mL), brine (1 × 40mL), dried with MgSO4, filtered and concentrated in vacuo. The product was obtained after crystallization or purification using column chromatography with heptane/EtOAc 50:1 (v/v) as eluent. Yield 47%. Colorless to white crystals. Rf = 0.69 (heptane:EtOAc 1:1). The product was more than 95% pure as judged from TLC and NMR. 1H NMR (200 MHz, DMSO) δ 8.04 (dd, J = 7.9, 1.7, 1H), 7.83–7.89 (m, 2H), 7.53–7.58 (m, 1H), 7.45–7.50 (m, 2H), 7.06–7.16 (m, 2H). 13C NMR (50 MHz, DMSO) 158.17, 151.40, 149.24, 139.90, 134.41, 128.04, 126.31, 125.77, 120.43, 119.64, 117.63, 111.50, 110.84. HRMS: m/z [M+H]+ calcd for C13H10O2N1 212.07061, found 212.07051.
4.2. Enzyme activity studies
4.2.1. Human 5-LOX inhibition screening UV Assay
Enzyme inhibition was measured by the residual enzyme activity after 10 minutes incubation with the inhibitor at room temperature. The enzyme activity was determined by conversion of lipoxygenase substrate linoleic acid into hydroperoxy-octadecadienoate (HPOD). The conversion rate was followed by UV absorbance of the conjugated diene at 234 nm (ε = 25000 M−1cm−1) over a period of 20 minutes. The UV absorbance increase over time was used to determine the enzyme activity.
Tris buffer (50 mM) pH 7.5 containing 2 mM EDTA and 2 mM CaCl2 was used as an assay buffer for human 5-LOX experiments. The human 5-LOX enzyme was diluted 1:4000 with the assay buffer. The inhibitor (100 mM in DMSO) was diluted with the assay buffer to 1 mM. The substrate, linoleic acid was diluted with EtOH to 20 mM. Subsequently, 1 mL of enzyme solution (1:4000) was mixed with 100 μL ATP (2 mM), 100 μL inhibitor (1 mM) and 790 μL Tris buffer and incubated for 10 minutes. After that the linoleic acid solution (10 μL, 20 mM) was added to a mixture and the conversion rate of the substrate was measured after 10 second mixing of the enzyme with the substrate. The reaction rate in the absence of the inhibitor was used as positive control. In the positive control experiment, the assay buffer was supplemented with a small amount of DMSO (3 μL of DMSO in 1.5 mL) in order to replace the DMSO inhibitor solution, which was also pre-incubated for 10 minutes.
4.2.2. COX-2 inhibition screening assay
The COX-2 activity was measured spectrophotometrically by measuring the formation of oxidized N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD). Tris buffer (0.1M) pH 8.0 was used as an assay buffer for COX-2 experiments. The COX-2 enzyme was diluted 1:1000 with the assay buffer. Hematin (7.5 mM in DMSO) was diluted with assay buffer to 330 μM. The inhibitor was diluted to 1.1 mM in DMSO. The substrate, arachidonic acid was diluted by adding 50 μL KOH 0.1 M and 800 μL H2O into 150 μL of a 22 mM arachidonic acid solution in EtOH. The colorimetric substrate TMPD (2.64 mM in H2O) was prepared freshly prior to the experiment. The substrate, linoleic acid was diluted with EtOH to 20 mM. Subsequently, in a 96 wells plate, 150 μL assay buffer, 10 μL hematin 330 μM, 10 μL COX-2 enzyme (1:1000) was added. 10 μL inhibitor solution (1.1 mM in DMSO) was added to the inhibitor well and 10 μL of DMSO was added to the positive control wells. The plate was incubated for 5 minutes at room temperature. 20 μL of freshly prepared TMPD was added followed by the addition of 20 μL arachidonic acid. Arachidonic acid addition was excluded from the negative control wells. The plate was incubated for another 5 mintes at room temperature. The absorbance was measured at 550 nm, and the calculations were performed with Excel 2010. The experiments were performed in triplicate, and the presented results were the average of three measurements with the standard deviation.
4.2.3. Michaelis Menten Enzyme Kinetics
The enzyme kinetics of human 5-LOX were also studied by the formation of the conjugated diene product at 234 nm (ε = 25000 M−1cm−1) using the same experimental setup as for the concentration dependent and the inhibitor screening. Enzyme kinetics were performed against linoleic acid and ATP to get a complete indication about the regulation of human 5-LOX enzyme activity.
In the kinetic study of the human 5-LOX against ATP, the ATP concentration was varied between 12.5–200 μM while the concentration of linoleic acid was fixed (100 μM). In the kinetic study of the human 5-LOX against the linoleic acid, substrate concentration was varied between 37.5 – 300 μM. The enzyme activity was measured in the absence or presence of fixed concentrations of activator 23a (0 μM, 12.5 μM, and 25 μM) or inhibitor 23d (0 μM, 25 μM, and 50 μM). The reaction velocities (v), which are the concentration changes over time, were plotted against the substrate concentrations in Michaelis-Menten plots and the Km and Vmax and the apparent values (Kmapp and Vmaxapp) in the presence of the activator were derived. The results from the non-linear curve fitting were in line with the results from Lineweaver-Burk plot.
The slopes and y-intercepts from the Lineweaver-Burk plot of h-5-LOX activation was derived and then re-plotted as 1/Δslope or 1/Δy-intercept versus 1/[activator]. From these plots α, β and KA values were derived as described by Leskovac [44] for non-essential activation. According to this method in the re-plot of 1/Δy-intercept versus 1/[activator] the y intercept corresponds to β (Vmax/(β-1), whereas in the re-plot of 1/Δslope versus 1/[activator] the y intercept correspond to β. Vmax/Km(β-α). The x intersection point of two lines from the plot of 1/Δy-intercept and 1/Δslope is –β/αKA, which can be employed to derive the KA value if α and β are known. All the experiments were performed in triplicate and the average triplicate values and their standard deviations are plotted. Calculations were performed with Excel 2010.
4.3. Docking Studies
4.3.1. Protein and ligand structures
Structures of compounds 23a, 23b, 23c, and 23d were built using Omega2 from OpenEye [53] We fixed the rotatable bonds between the benzyl ring and the carboxylic acid group on all molecules using AutoDockTools to dock structures that kept a planar geometry for the benzyl ring and carboxylic acid [54].
4.3.2. Docking and scoring
The receptor structure was prepared for docking using the prepare_receptor4.py script from AutoDockTools. Compounds were docked using smina55 with the default vina scoring function and the argument ‘--num_modes=40’. The docked poses were rescored using the ‘--score_only’ command in smina and then reranked. The top ranked poses were used for further analysis.
Supplementary Material
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. RW, PAMK and NE synthesized the molecules and performed the biochemical analysis. MPB and CJC performed and analyzed the docking studies and were supported by grant R01GM097082-01 from the National Institutes of Health. HJH and FJD supervised the research. RW and FJD wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Martel-Pelletier J, Lajeunesse D, Reboul P, Pelletier J. Ann Rheum Dis. 2003;62:501. doi: 10.1136/ard.62.6.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gillmor SA, Villasenor A, Fletterick R, Sigal E, Browner MF. Nat Struct Biol. 1997;4:1003. doi: 10.1038/nsb1297-1003. [DOI] [PubMed] [Google Scholar]
- 3.Sigal E. Am J Physiol. 1991;260:L13. doi: 10.1152/ajplung.1991.260.2.L13. [DOI] [PubMed] [Google Scholar]
- 4.Epp N, Fürstenberger G, Müller K, de Juanes S, Leitges M, Hausser I, Thieme F, Liebisch G, Schmitz G, Krieg P. J Cell Biol. 2007;177:173. doi: 10.1083/jcb.200612116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ikei KN, Yeung J, Apopa PL, Ceja J, Vesci J, Holman TR, Holinstat M. J Lipid Res. 2012;53:2546. doi: 10.1194/jlr.M026385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter JA, Finkbeiner WE, Nadel JA, Goetzl EJ, Holtzman MJ. Proc Natl Acad Sci. 1985;82:4633. doi: 10.1073/pnas.82.14.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nadel JA, Conrad DJ, Ueki IF, Schuster A, Sigal E. J Clin Invest. 1991;87:1139. doi: 10.1172/JCI115110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brash AR, Boeglin WE, Chang MS. Proc Natl Acad Sci. 1997;94:6148. doi: 10.1073/pnas.94.12.6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown CD, Kilty I, Yeadon M, Jenkinson S. Inflamm Res. 2001;50:321. doi: 10.1007/PL00000251. [DOI] [PubMed] [Google Scholar]
- 10.Chanez P, Bonnans C, Chavis C, Vachier I. Am J Respir Cell Mol Biol. 2002;27:655. doi: 10.1165/rcmb.f253. [DOI] [PubMed] [Google Scholar]
- 11.Chu HW, Balzar S, Westcott JY, Trudeau JB, Sun Y, Conrad DJ, Wenzel SE. Clin Exp Allergy. 2002;32:1558. doi: 10.1046/j.1365-2222.2002.01477.x. [DOI] [PubMed] [Google Scholar]
- 12.Shannon VR, Chanez P, Bousquet J, Holtzman MJ. Am Rev Respir Dis. 1993;147:1024. doi: 10.1164/ajrccm/147.4.1024. [DOI] [PubMed] [Google Scholar]
- 13.Piao Y, Du Y, Oshima H, Jin J, Nomura M, Yoshimoto T, Oshima M. Carcinogenesis. 2008;29:440. doi: 10.1093/carcin/bgm274. [DOI] [PubMed] [Google Scholar]
- 14.Rásó E, Döme B, Somlai B, Zacharek A, Hagmann W, Honn KV, Tímár J. Melanoma Res. 2004;14:245. doi: 10.1097/01.cmr.0000138479.00897.d8. [DOI] [PubMed] [Google Scholar]
- 15.Gupta S, Srivastava M, Ahmad N, Sakamoto K, Bostwick DG, Mukhtar H. Cancer. 2001;91:737. doi: 10.1002/1097-0142(20010215)91:4<737::aid-cncr1059>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 16.Melstrom LG, Bentrem DJ, Salabat MR, Kennedy TJ, Ding X, Strouch M, Rao SM, Witt RC, Ternent CA, Talamonti MS, Bell RH, Adrian TA. Clin Cancer Res. 2008;14:6525. doi: 10.1158/1078-0432.CCR-07-4631. [DOI] [PubMed] [Google Scholar]
- 17.Nathoo N, Prayson RA, Bondar J, Vargo L, Arrigain S, Mascha EJ, Suh JH, Barnett GH, Golubic M. Neurosurgery. 2006;58:347. doi: 10.1227/01.NEU.0000195096.43258.94. [DOI] [PubMed] [Google Scholar]
- 18.Wilborn J, Bailie M, Coffey M, Burdick M, Strieter R, Peters-Golden M. J Clin Invest. 1996;97:1827. doi: 10.1172/JCI118612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou LY. J Cancer Mol. 2006;2:227. [Google Scholar]
- 20.Serhan CN, Chiang N, Van Dyke TE. Nat Rev Immunol. 2008;8:349. doi: 10.1038/nri2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edenius C, Haeggström J, Lindgren JÅ. Biochem Biophys Res Commun. 1988;157:801. doi: 10.1016/s0006-291x(88)80320-6. [DOI] [PubMed] [Google Scholar]
- 22.Hashimoto A, Hayashi I, Murakami Y, Sato Y, Kitasato H, Matsushita R, Iizuka N, Urabe K, Itoman M, Hirohata S, Endo H. J Rheumatol. 2007;34:2144. [PubMed] [Google Scholar]
- 23.Sobrado M, Pereira MP, Ballesteros I, Hurtado O, Fernández-López D, Pradillo JM, Caso JR, Vivancos J, Nombela F, Serena J, Lizasoain I, Moro MA. J Neurosci. 2009;29:3875. doi: 10.1523/JNEUROSCI.5529-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matschinsky FM. Nat Rev Drug Discov. 2009;8(5):399. doi: 10.1038/nrd2850. [DOI] [PubMed] [Google Scholar]
- 25.Smith WL, DeWitt DL, Garavito RM. Annu Rev Biochem. 2000;69:145. doi: 10.1146/annurev.biochem.69.1.145. [DOI] [PubMed] [Google Scholar]
- 26.Anderson GD, Hauser SD, McGarity KL, Bremer ME, Isakson PC, Gregory S. J Clin Invest. 1996;97:2672. doi: 10.1172/JCI118717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reuter BK, Asfaha S, Buret A, Sharkey KA, Wallace JL. J Clin Invest. 1996;98:2076. doi: 10.1172/JCI119013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendel J, Nielsen OH. Am J Gastroenterol. 1997;92:1170. [PubMed] [Google Scholar]
- 29.Kargman SL, O’Neill GP, Vickers PJ, Evans JF, Mancini JA, Jothy S. Cancer Res. 1995;55:2556. [PubMed] [Google Scholar]
- 30.Cianchi F, Cortesini C, Bechi P, Fantappiè O, Messerini L, Vannacci A, Sardi I, Baroni G, Boddi V, Mazzanti R, Masini E. Gastroenterology. 2001;121:1339. doi: 10.1053/gast.2001.29691. [DOI] [PubMed] [Google Scholar]
- 31.Wisastra R, Ghizzoni M, Boltjes A, Haisma HJ, Dekker FJ. Bioorg Med Chem. 2012;20:5027. doi: 10.1016/j.bmc.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 32.Brash AR. J Biol Chem. 1999;274:23679. doi: 10.1074/jbc.274.34.23679. [DOI] [PubMed] [Google Scholar]
- 33.Ghizzoni M, Boltjes A, Graaf Cd, Haisma HJ, Dekker FJ. Bioorg Med Chem. 2010;18:5826. doi: 10.1016/j.bmc.2010.06.089. [DOI] [PubMed] [Google Scholar]
- 34.Ghizzoni M, Wu J, Gao T, Haisma HJ, Dekker FJ, George Zheng Y. Eur J Med Chem. 2012;47:337. doi: 10.1016/j.ejmech.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uchiyama M, Ozawa H, Takuma K, Matsumoto Y, Yonehara M, Hiroya K, Sakamoto T. Org Lett. 2006;8:5517. doi: 10.1021/ol062190+. [DOI] [PubMed] [Google Scholar]
- 36.Harrowven DC, Woodcock T, Howes PD. Angew Chem Int Ed Engl. 2005;44:3899. doi: 10.1002/anie.200500466. [DOI] [PubMed] [Google Scholar]
- 37.Sista P, Nguyen H, Murphy JW, Hao J, Dei DK, Palaniappan K, Servello J, Kularatne RS, Gnade BE, Xue B, Dastoor PC, Biewer MC, Stefan MC. Macromolecules. 2010;43:8063. [Google Scholar]
- 38.Tranchimand S, Tron T, Gaudin C, Iacazio G. Synth Comm. 2006;36:587. [Google Scholar]
- 39.Back DF, Manzoni de Oliveira G, Ballin MA, Corbellini VA. Inorg Chim Acta. 2010;363:807. [Google Scholar]
- 40.Gibian MJ, Galaway RA. Biochemistry. 1976;15:4209. doi: 10.1021/bi00664a012. [DOI] [PubMed] [Google Scholar]
- 41.Ha TJ, Kubo I. J Agric Food Chem. 2005;53:4350. doi: 10.1021/jf048184e. [DOI] [PubMed] [Google Scholar]
- 42.Copeland RA, Williams JM, Giannaras J, Nurnberg S, Covington M, Pinto D, Pick S, Trzaskos JM. Proc Natl Acad Sci. 1994;91:11202. doi: 10.1073/pnas.91.23.11202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verhagen J, Vliegenthart JF, Boldingh J. Chem Phys Lipids. 1978;22:255. doi: 10.1016/0009-3084(78)90014-2. [DOI] [PubMed] [Google Scholar]
- 44.Leskovac V. Comprehensive Enzyme Kinetics. 2004:111. [Google Scholar]
- 45.Gilbert NC, Bartlett SG, Waight MT, Neau DB, Boeglin WE, Brash AR, Newcomer ME. Science. 2011;331:217–219. doi: 10.1126/science.1197203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilbert NC, Rui Z, Neau DB, Waight MT, Bartlett SG, Boeglin WE, Brash AR, Newcomer ME. FASEB J. 2012;26:3222–3229. doi: 10.1096/fj.12-205286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The PyMOL Molecular Graphics System, Version 1.5.0.4. Schödinger, LLC; [Google Scholar]
- 48.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 49.Lee KS, Kim SR, Park HS, Park SJ, Min KH, Lee KY, Jin SM, Lee YC. J Allergy Clin Immunol. 2007;119:141–149. doi: 10.1016/j.jaci.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 50.Okamoto F, Saeki K, Sumimoto H, Yamasaki S, Yokomizo T. J Biol Chem. 2010;285:41113–41121. doi: 10.1074/jbc.M110.175497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson C, Cloutier A, Bossé Y, Poisson C, Larivée P, McDonald PP, Stankova J, Rola– Z, Pleszczynski M. J Biol Chem. 2008;283:1974–1984. doi: 10.1074/jbc.M608197200. [DOI] [PubMed] [Google Scholar]
- 52.Deb A, Haque SJ, Mogensen T, Silverman RH, Williams BRG. J Immunol. 2001;166:6170–6180. doi: 10.4049/jimmunol.166.10.6170. [DOI] [PubMed] [Google Scholar]
- 53.Boström J, Greenwood JR, Gottfries J. J Mol Graph Model. 2003;21:449–462. doi: 10.1016/s1093-3263(02)00204-8. [DOI] [PubMed] [Google Scholar]
- 54.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koes DR, Baumgartner MP, Camacho CJ. J Chem Inf Model. 2013;53:1893–1904. doi: 10.1021/ci300604z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.