Abstract
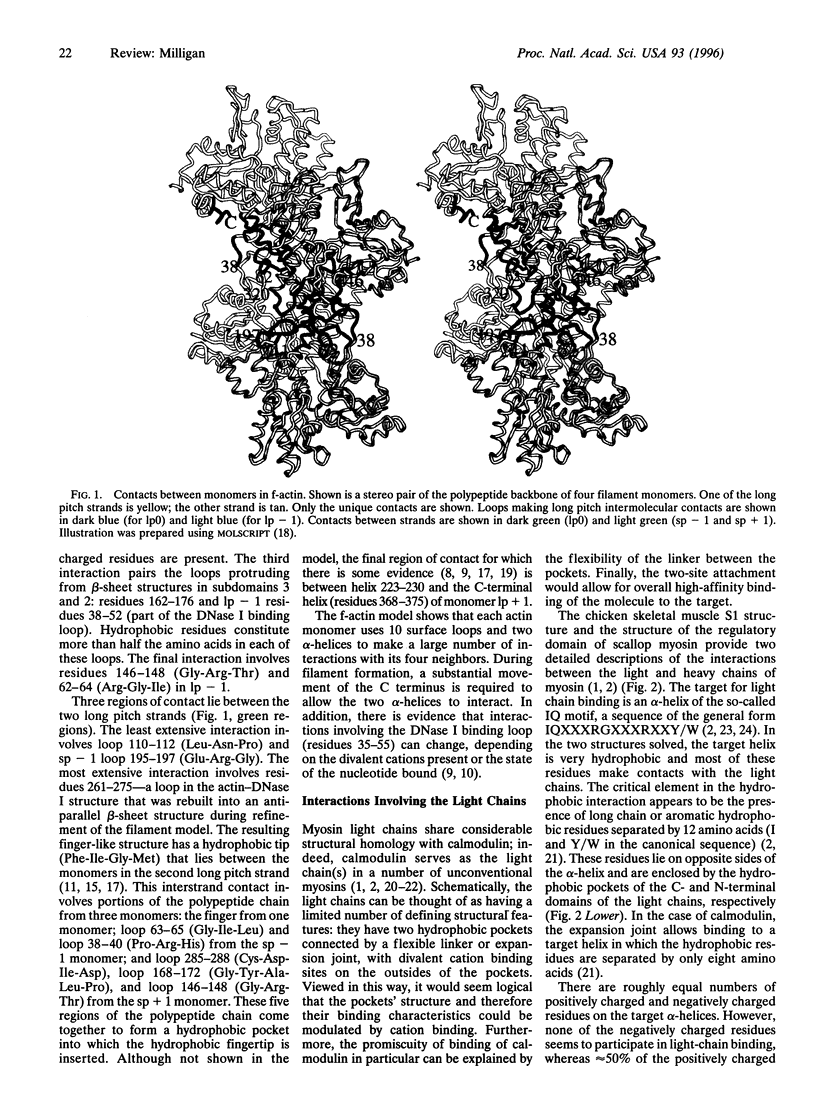
Since it has not been possible to crystallize the actomyosin complex, the x-ray structures of the individual proteins together with data obtained by fiber diffraction and electron microscopy have been used to build detailed models of filamentous actin (f-actin) and the actomyosin rigor complex. In the f-actin model, a single monomer uses 10 surface loops and two alpha-helices to make sometimes complicated interactions with its four neighbors. In the myosin molecule, both the essential and regulatory light chains show considerable structural homology to calmodulin. General principles are evident in their mode of attachment to the target alpha-helix of the myosin heavy chain. The essential light chain also makes contacts with other parts of the heavy chain and with the regulatory light chain. The actomyosin rigor interface is extensive, involving interaction of a single myosin head with regions on two adjacent actin monomers. A number of hydrophobic residues on the apposing faces of actin and myosin contribute to the main binding site. This site is flanked on three sides by charged myosin surface loops that form predominantly ionic interactions with adjacent regions of actin. Hydrogen bonding is likely to play a significant role in actin-actin and actin-myosin interactions since many of the contacts involve loops. The model building approach used with actomyosin is applicable to other multicomponent assemblies of biological interest and is a powerful method for revealing molecular interactions and providing insights into the mode of action of the assemblies.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bremer A., Henn C., Goldie K. N., Engel A., Smith P. R., Aebi U. Towards atomic interpretation of F-actin filament three-dimensional reconstructions. J Mol Biol. 1994 Oct 7;242(5):683–700. doi: 10.1006/jmbi.1994.1617. [DOI] [PubMed] [Google Scholar]
- Cheney R. E., Mooseker M. S. Unconventional myosins. Curr Opin Cell Biol. 1992 Feb;4(1):27–35. doi: 10.1016/0955-0674(92)90055-h. [DOI] [PubMed] [Google Scholar]
- Cuda G., Fananapazir L., Zhu W. S., Sellers J. R., Epstein N. D. Skeletal muscle expression and abnormal function of beta-myosin in hypertrophic cardiomyopathy. J Clin Invest. 1993 Jun;91(6):2861–2865. doi: 10.1172/JCI116530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher A. J., Smith C. A., Thoden J., Smith R., Sutoh K., Holden H. M., Rayment I. Structural studies of myosin:nucleotide complexes: a revised model for the molecular basis of muscle contraction. Biophys J. 1995 Apr;68(4 Suppl):19S–28S. [PMC free article] [PubMed] [Google Scholar]
- Geisterfer-Lowrance A. A., Kass S., Tanigawa G., Vosberg H. P., McKenna W., Seidman C. E., Seidman J. G. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990 Sep 7;62(5):999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- Holmes K. C., Popp D., Gebhard W., Kabsch W. Atomic model of the actin filament. Nature. 1990 Sep 6;347(6288):44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- Johnson J. E. Functional implications of protein-protein interactions in icosahedral viruses. Proc Natl Acad Sci U S A. 1996 Jan 9;93(1):27–33. doi: 10.1073/pnas.93.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W., Mannherz H. G., Suck D., Pai E. F., Holmes K. C. Atomic structure of the actin:DNase I complex. Nature. 1990 Sep 6;347(6288):37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- Lorenz M., Popp D., Holmes K. C. Refinement of the F-actin model against X-ray fiber diffraction data by the use of a directed mutation algorithm. J Mol Biol. 1993 Dec 5;234(3):826–836. doi: 10.1006/jmbi.1993.1628. [DOI] [PubMed] [Google Scholar]
- McLaughlin P. J., Gooch J. T., Mannherz H. G., Weeds A. G. Structure of gelsolin segment 1-actin complex and the mechanism of filament severing. Nature. 1993 Aug 19;364(6439):685–692. doi: 10.1038/364685a0. [DOI] [PubMed] [Google Scholar]
- Meador W. E., Means A. R., Quiocho F. A. Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science. 1993 Dec 10;262(5140):1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- Meador W. E., Means A. R., Quiocho F. A. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science. 1992 Aug 28;257(5074):1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- Mendelson R. A., Morris E. The structure of F-actin. Results of global searches using data from electron microscopy and X-ray crystallography. J Mol Biol. 1994 Jul 8;240(2):138–154. doi: 10.1006/jmbi.1994.1428. [DOI] [PubMed] [Google Scholar]
- Mercer J. A., Seperack P. K., Strobel M. C., Copeland N. G., Jenkins N. A. Novel myosin heavy chain encoded by murine dilute coat colour locus. Nature. 1991 Feb 21;349(6311):709–713. doi: 10.1038/349709a0. [DOI] [PubMed] [Google Scholar]
- Milligan R. A., Flicker P. F. Structural relationships of actin, myosin, and tropomyosin revealed by cryo-electron microscopy. J Cell Biol. 1987 Jul;105(1):29–39. doi: 10.1083/jcb.105.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan R. A., Whittaker M., Safer D. Molecular structure of F-actin and location of surface binding sites. Nature. 1990 Nov 15;348(6298):217–221. doi: 10.1038/348217a0. [DOI] [PubMed] [Google Scholar]
- Mornet D., Pantel P., Audemard E., Kassab R. The limited tryptic cleavage of chymotryptic S-1: an approach to the characterization of the actin site in myosin heads. Biochem Biophys Res Commun. 1979 Aug 13;89(3):925–932. doi: 10.1016/0006-291x(79)91867-9. [DOI] [PubMed] [Google Scholar]
- Orlova A., Egelman E. H. Structural dynamics of F-actin: I. Changes in the C terminus. J Mol Biol. 1995 Feb 3;245(5):582–597. doi: 10.1006/jmbi.1994.0048. [DOI] [PubMed] [Google Scholar]
- Orlova A., Prochniewicz E., Egelman E. H. Structural dynamics of F-actin: II. Cooperativity in structural transitions. J Mol Biol. 1995 Feb 3;245(5):598–607. doi: 10.1006/jmbi.1994.0049. [DOI] [PubMed] [Google Scholar]
- Owen C., DeRosier D. A 13-A map of the actin-scruin filament from the limulus acrosomal process. J Cell Biol. 1993 Oct;123(2):337–344. doi: 10.1083/jcb.123.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment I., Holden H. M., Whittaker M., Yohn C. B., Lorenz M., Holmes K. C., Milligan R. A. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 1993 Jul 2;261(5117):58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- Schröder R. R., Manstein D. J., Jahn W., Holden H., Rayment I., Holmes K. C., Spudich J. A. Three-dimensional atomic model of F-actin decorated with Dictyostelium myosin S1. Nature. 1993 Jul 8;364(6433):171–174. doi: 10.1038/364171a0. [DOI] [PubMed] [Google Scholar]
- Schutt C. E., Myslik J. C., Rozycki M. D., Goonesekere N. C., Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993 Oct 28;365(6449):810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- Sweeney H. L. Function of the N terminus of the myosin essential light chain of vertebrate striated muscle. Biophys J. 1995 Apr;68(4 Suppl):112S–119S. [PMC free article] [PubMed] [Google Scholar]
- Tirion M. M., ben-Avraham D., Lorenz M., Holmes K. C. Normal modes as refinement parameters for the F-actin model. Biophys J. 1995 Jan;68(1):5–12. doi: 10.1016/S0006-3495(95)80156-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus M. A. Myosins. Curr Opin Cell Biol. 1993 Feb;5(1):77–81. doi: 10.1016/s0955-0674(05)80011-0. [DOI] [PubMed] [Google Scholar]
- Uyeda T. Q., Ruppel K. M., Spudich J. A. Enzymatic activities correlate with chimaeric substitutions at the actin-binding face of myosin. Nature. 1994 Apr 7;368(6471):567–569. doi: 10.1038/368567a0. [DOI] [PubMed] [Google Scholar]
- Xie X., Harrison D. H., Schlichting I., Sweet R. M., Kalabokis V. N., Szent-Györgyi A. G., Cohen C. Structure of the regulatory domain of scallop myosin at 2.8 A resolution. Nature. 1994 Mar 24;368(6469):306–312. doi: 10.1038/368306a0. [DOI] [PubMed] [Google Scholar]