Abstract
We used NMR directly in live human cells to describe the complete post-translational maturation process of human superoxide dismutase 1 (SOD1). We could follow, at atomic resolution, zinc binding, homodimer formation and copper uptake, and discover that copper chaperone for SOD1 (CCS) oxidation of the SOD1 intrasubunit disulfide bond occurs through both copper-dependent and independent mechanisms. Our approach represents a new strategy for structural investigation of endogeneously expressed proteins within a physiological (cellular) environment.
Functional understanding of cellular processes requires a detailed characterization of molecular players, their structural and dynamic properties, and their networks of interactions. Biomolecules should ideally be characterized within their cellular milieu, to match the physiological environment including pH, redox potential, viscosity, and the presence of all relevant interaction partners. A new approach to structural biology is therefore needed to explore the cellular context with atomic resolution techniques. In principle, in-cell NMR1-5 represents an ideal method to monitor protein structure during functional processes, in “close to physiological” conditions. However, in practical terms, one must overcome a number of technological limitations in order to: 1) express (or co-express, where appropriate) isotopically labelled proteins within cells derived from a suitable organism: human proteins, for example, should be endogenously synthesised and studied within human cells; 2) establish experimental conditions to maintain cellular viability inside the NMR tube and to reduce data acquisition time, while increasing measurement sensitivity. We sought to address these challenges and attempt to directly monitor the steps involved in post-translational modifications of a model protein, human superoxide dismutase 1 (SOD1), within live human cells. Furthermore, we aimed to define the role of the copper chaperone for SOD1 (CCS), which was proposed to make a major contribution to SOD1 maturation6. A correct understanding of this process is important considering the fundamental role played by SOD1 in the cellular defence against oxidative stress7. Impaired SOD1 maturation has been linked to disease states, including the onset of amyotrophic lateral sclerosis8,9.
We expressed human SOD1 transiently in HEK293T cells10 and modulated its expression levels by varying the amount of transfected cDNA (see Online Methods). The endogenous concentration of SOD1 in HEK293T cells, under our culture conditions, was 10±2 μM (Supplementary Results, Supplementary Fig. 1). Levels up to 40 μM SOD1 have been previously reported in the cytoplasm of mammalian cells11. Following recombinant expression, the maximal intracellular SOD1 monomer concentration obtained was 360±30 μM. However, we could still detect SOD1 by in-cell NMR at closer to physiological intracellular concentrations of 45±10 μM (Supplementary Fig. 2). Cellular distribution of SOD1 was assessed in isolated nuclear, cytoplasmic and mitochondrial fractions. Irrespectively of the total protein level, the majority of SOD1 was present in the cytoplasm and less than 1% was localized in the mitochondrial fraction (Supplementary Fig. 3), a value somewhat lower than previously reported12.
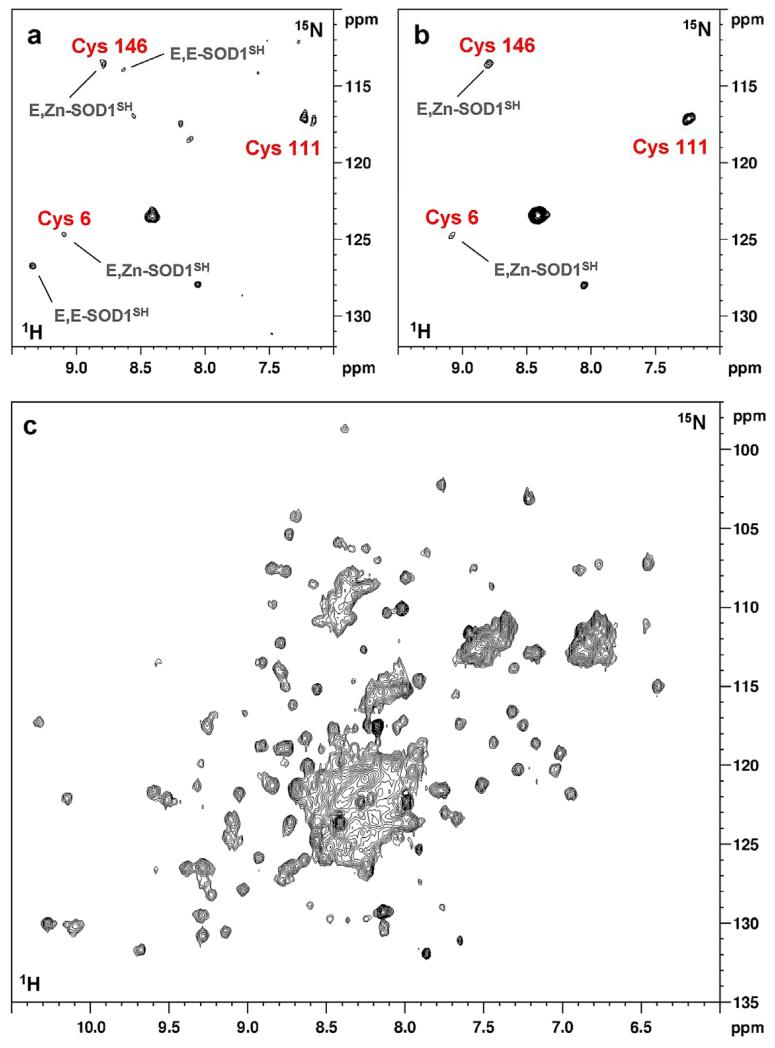
Two forms of SOD1 could be detected, in cells grown without supplements of zinc or copper ions, in similar amounts: the monomeric, metal-free species (apo-SOD1) and the dimeric species with one Zn2+ ion bound to each subunit (E,Zn-SOD1) (Supplementary Fig. 4a); the NMR data were analyzed by taking advantage of previous in vitro spectra and backbone assignments13,14. This mixture of species is likely due to residual zinc in the culture media (calculated ~4.5 μM in 10% foetal bovine serum (FBS)-supplemented medium and ~1 μM in 2% FBS-supplemented medium, see Online Methods). Cysteines 57 and 146, forming an intrasubunit disulfide bond in the mature enzyme, were reduced in both species (Fig. 1a). The presence of only these two species was further confirmed by NMR analysis of cell extracts (Supplementary Fig. 4b). The 1H-15N spectrum of the metal-free SOD1 species was consistent with its equivalent produced in E. coli cells without addition of metal ions15, only the crosspeaks of the less structured parts being detected (Supplementary Fig. 4a).
Figure 1. Zn(II) added to the culture medium promotes binding of one Zn2+ ion per apo-SOD1 subunit in the cytoplasm.
1H-15N SOFAST HMQC spectra were acquired on human cells expressing 15N-cysteine labelled SOD1: a, in absence of metals; b, with Zn(II) added to the culture medium. Assigned cysteine residues are indicated in red. When two species of SOD1 are present, labels indicate the species to which each crosspeak belongs (E,E-SOD1SH: reduced apo-SOD1; E,Zn-SOD1SH: reduced SOD1 containing one Zn2+ ion per subunit). In both species, cysteine 57 crosspeak is not detected. Unlabelled crosspeaks correspond to cellular background signals; c, 1H-15N SOFAST HMQC acquired on human cells expressing uniformly 15N-labelled SOD1 in Zn(II)-supplemented medium. The region between 8.0 and 8.5 (1H) ppm contains overlapped signals arising from non-specific labelling of the cells, which can be subtracted to obtain a cleaner spectrum (Supplementary Fig. 5). By comparing the chemical shift of the assigned crosspeaks in the in-cell spectrum with backbone assignments of SOD1 in different metallation and redox state in vitro13,14 it was possible to assess the species present in the cytoplasm (Supplementary Fig. 6).
Zn2+ addition to the culture medium eliminated apo-SOD1 species signals (Fig. 1b). The uniformly-15N labelled cell sample yielded a good quality spectrum of E,Zn-SOD1 (Fig. 1c), which could be improved by removing background signals arising from non-selective labelling of cellular components (Supplementary Fig. 5). Therefore, in culture, Zn2+ ions are efficiently uptaken by cells and bound in stoichiometric amounts, specifically to the native binding site of dimeric SOD1, as further confirmed by 1H-15N crosspeak analysis (Supplementary Fig. 6). On the contrary, when reduced apo-SOD1 is exposed to Zn2+ ions in vitro or in cell lysates, even at sub-stoichiometric concentrations, a mixture of apo-SOD1, E,Zn-SOD1 and Zn,Zn-SOD1 forms is generated15. Therefore, site-selectivity of Zn2+-binding can only be achieved within the physiological cellular context.
Prokaryotes have simple mechanisms for copper uptake and excretion, while they are tightly regulated in eukaryotic cells16. Accordingly, copper added as Cu(II) salt to E. coli cells culture medium was reduced to Cu(I) and readily and stoichiometrically bound only in this redox state to recombinantly expressed SOD1, forming Cu(I),Zn-SOD1 (Supplementary Fig. 7a,b). Importantly, Cu(I) added to E. coli cells either as an acetonitrile or glutathione complex did not become available to E,Zn-SOD1 (Supplementary Fig. 7c). In-cell NMR spectra also showed that in E.coli the SOD1 intrasubunit disulfide bridge is oxidized in a sizable fraction (around 50%, Supplementary Fig. 7d). Unlike bacteria, copper entrance in eukaryotic cells and its delivery to copper-binding proteins require a number of steps, involving specific chaperones responsible of its intracellular trafficking17,18. Accordingly, only around 25% of the recombinant SOD1 protein incorporated copper, again in the Cu(I) state, when human cells were cultured in the presence of Cu(II) (Fig. 2a). The remaining SOD1 fraction contained only one zinc ion per subunit, as observed from the 1H histidine signals in the 1D 1H NMR spectrum (Fig. 2a-c). Additionally, the spectra of the 15N-Cys selectively labelled protein showed only ~20% SOD1 intrasubunit disulfide bond formation (Supplementary Fig. 8). Copper incorporation of SOD1 in eukaryotes is known to be dependent on the CCS protein19,20. Although a basal expression-level of the hCCS gene does occur during normal cell growth, it is likely that the amount of SOD1 produced in our experimental setup was too high to allow for complete copper insertion via the CCS-dependent pathway. However, a CCS-independent copper insertion pathway has also been reported for human SOD121,22 and might have contributed to the partial formation of Cu(I),Zn-SOD1 observed (Fig 2a).
Figure 2. Cu(II) addition to the culture medium induces Cu(I) binding to a fraction of cytoplasmic SOD1.
Histidine region of 1H NMR spectra were acquired on human cells expressing unlabelled SOD1: a, in Zn(II)-supplemented medium, after incubation with Cu(II); b, in Zn(II)-supplemented medium without incubation with Cu(II); c, in medium without added metals. d, 1H NMR spectrum of human cells co-expressing SOD1 and CCS in Zn(II)-supplemented medium, after incubation with Cu(II). Histidine protons unambiguously assigned to Cu(I),Zn-SOD1 species are indicated.
Simultaneous overexpression and isotopic enrichment of both SOD1 and CCS caused a reduction in the overall SOD1 expression levels (Supplementary Fig. 2). Nevertheless, SOD1 signals were still easily identified (Fig. 3a and 3b), with minimum interference from CCS due to the larger molecular mass of the latter. Intracellular protein concentration in these experiments ranged between 70±10 μM and 45±10 μM SOD1, and between 50±10 μM and 15±3 μM CCS (Supplementary Fig. 2). Co-expression of SOD1 and CCS in zinc supplemented medium resulted in dimeric, zinc-containing, SOD1 species. No difference in SOD1 metal content was found with respect to the cell sample with basal CCS level (Supplementary Fig. 9). However, spectra recorded from cells co-expressing the two proteins revealed a partial oxidation (around 50%) of the SOD1 intrasubunit disulfide bond (Fig. 3a). This result is consistent with a mechanism of CCS-mediated SOD1 disulfide oxidation which, unexpectedly, does not require copper insertion into SOD1.
Figure 3. The redox state of SOD1 is influenced by both copper binding and the presence of CCS.
1H-15N SOFAST HMQC spectra were acquired on human cells: a, co-expressing 15N-cysteine labelled SOD1 and CCS in Zn(II)-supplemented medium; b, co-expressing 15N-cysteine labelled SOD1 and CCS in Zn(II)-supplemented medium, after incubation with Cu(II). Assigned cysteine residues are indicated in red. When two species of SOD1 are present, labels indicate the disulfide redox state of each species. Unlabelled crosspeaks are cellular background signals. c, drawing summarizing SOD1 maturation steps. Left cell: SOD1 expressed in cells with no addition of metals is present mainly it the apo form, which is monomeric and partially unfolded. A fraction of SOD1 binds the zinc present in the expression medium. Central cell: when Zn(II) (cyan) is added to the expression medium SOD1 quantitatively binds one zinc ion per monomer and dimerizes; the intrasubunit disulfide bridge is completely reduced. Right cell: when both Zn(II) and Cu(II) (blue) are added to the expression medium, and CCS is co-expressed, a fraction of SOD1 binds Cu(I) (orange); the disulfide bridge is completely oxidized (yellow circles).
When Cu(II) was added to cells co-expressing SOD1 and CCS, a higher ratio of Cu(I),Zn-SOD1 to E,Zn-SOD1 was obtained (~1:1, Fig. 2c) compared with cell samples with basal CCS level (Fig. 2a), indicating that CCS promotes copper incorporation in SOD1. The redox state of SOD1 cysteines 57 and 146 is affected by both CCS overexpression and copper presence. Indeed, when cells overexpressing both SOD1 and CCS were incubated with Cu(II), the intrasubunit disulfide bridge of SOD1 was completely oxidized (Fig. 3b).
We attempted to explore the above mechanism at lower concentrations of SOD1 and CSS, as close as possible to the reported physiological levels11. At 45±10 μM SOD1 and 15±3 μM CCS, complete disulfide formation in E,Zn-SOD1 was observed (Supplementary Fig. 10a,b), whereas additional incubation with copper resulted in the complete formation of oxidized Cu(I),Zn-SOD1 (Supplementary Fig. 10c,d). These results suggest that only the relative amounts of species are affected by recombinant protein expression levels, but not the sequence of maturation events. We speculate that even lower levels of SOD1, undetectable by NMR, would result in complete formation of mature SOD1, even at endogenous levels of CCS and copper. In such conditions, however, no information on intermediate maturation steps would be obtained. By overexpressing only SOD1, components of its maturation pathway are not sufficiently abundant to complete the process, and therefore intermediate SOD1 maturation states are detected. Furthermore, by selectively increasing single components (e.g. CCS or copper), specific steps of SOD1 maturation can be recovered, and different SOD1 states detected. With this “knock-in” approach, combined with some a priori knowledge of the components involved, one can understand which of them are necessary for each step of the process.
The key steps of SOD1 maturation observed by in-cell NMR are schematically summarized in Fig. 3c. Specifically, we found that: apo-SOD1 is largely unfolded and monomeric in the HEK293T cytoplasm; zinc uptake occurs without the need of a chaperone; copper uptake and oxidation of the Cys57-Cys146 disulfide bond occur partially in cells exposed to Cu(II) (which is reduced to Cu(I)); copper loading and complete oxidation of the above mentioned cysteines is achieved in cells co-expressing SOD1 and CCS. Importantly, our experiments also reveal that, within a physiological context, CCS is able to oxidize the intramolecular SOD1 disulfide bond in the absence of copper binding to SOD1. This finding, while consistent with the previously reported effect of CCS in promoting SOD1 disulfide bond formation23, demonstrates that the cysteine oxidation step can occur in vivo independently of copper transfer, thus differing from the mechanism observed in vitro24.
We sought to establish whether the approach described here is applicable to proteins beyond hSOD1 and selected four other proteins [Mia40, Atox1, glutaredoxin-1 (Grx1) and thioredoxin (Trx)] with different properties including protein fold, binding of metal ions and redox potential of cysteine residues. Following the protocol we established for SOD1 and CCS, all these targets were highly expressed and visible in the 1H NMR spectra above the cellular background (Supplementary Fig. 11). Mia40 and Atox1 could be detected on 1H-15N spectra on uniformly 15N labelled cell samples (Supplementary Fig. 12a,b). Grx1 and Trx only became visible upon cell lysis, suggesting that some interaction occurs in the cytoplasm which makes the protein tumbling slower on average, thus broadening the amide crosspeaks beyond detection (Supplementary Fig. 12c,d). Such molecules may be successfully characterized through different NMR techniques (such as solid-state MAS NMR for slow tumbling proteins)25.
Successful application of in-cell NMR to proteins expressed endogenously in mammalian cells relies on efficient cDNA transfection, relatively high protein expression levels, applicability of different labelling strategies and maintenance of cell integrity during measurements. All these aspects have been successfully addressed in this study, allowing us to follow the sequential, physiological order of the events in the SOD1 post-translational modification process, information that cannot be retrieved in vitro. To our knowledge, this is the first time a complete protein maturation process has been followed in a living cell, in atomic detail. Importantly, this strategy may be applicable for many other protein targets, and thus opens the way for a broad range of molecular level, in-cell structural studies of proteins.
Online Methods
Constructs
cDNAs encoding full-length human SOD1 (amino acids 1-154, GenBank accession number: NP_000445.1), CCS (amino acids 1-274, GenBank accession number: NP_005116.1), Mia40 (amino acids 1-142, GenBank accession number: NP_001091972.1), Atox1 (amino acids 1-68, GenBank accession number: NP_004036.1), glutaredoxin-1 (amino acids 1-106, GenBank accession number: NP_001112362.1) and thioredoxin (amino acids 1-105, GenBank accession number: NP_003320.2) were amplified by PCR and cloned into the pHLsec10 vector between EcoRI and XhoI restriction enzyme sites to generate the mammalian expression plasmids. All clones were verified by DNA sequencing.
Cell culture and transfection
HEK293T cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM high glucose, D6546, Sigma) supplemented with L-glutamine, antibiotics (penicillin/streptomycin) and 10% foetal bovine serum (FBS, Gibco) in uncoated 75 cm2 plastic flasks, and were incubated at 310 K, 5% CO2 in a humidified atmosphere. Cells were transiently transfected with the pHLsec plasmid containing the hSOD1 cDNA using polyethylenimine (PEI), as described elsewhere10. Different DNA:PEI ratios were tested for maximizing protein expression (PEI was kept constant at 50 μg/flask), and an optimal ratio of 1:2 was found (25 μg/flask DNA, 50 μg/flask PEI). For co-expression of SOD1 and CCS, cells were transfected with plasmids containing hSOD1 and hCCS constructs in different amounts and ratios. The highest expression of both proteins was obtained by transfecting in a 1:1:2 hSOD1:hCCS:PEI ratio, thus doubling the total DNA amount. Lower expression levels of SOD1 were obtained by transfecting cells with a 1:4 hSOD:PEI ratio. To decrease the expression levels of both proteins, cells were transfected with a 1:1:4 hSOD1:hCCS:PEI ratio. PEI was always kept constant at 50 μg/flask. Different times of SOD1 expression were tested (1, 2, 3, 6 days), and the highest amount of protein was reached after 2 days (48 hours) of expression. During protein expression, cells were incubated at 310 K in 75 cm2 flasks. Commercial DMEM media were used for unlabelled in-cell NMR samples: BioExpress6000 medium (CIL) was used for uniform 15N labelling, while for selective 15N-cysteine labelling a reconstituted medium was prepared following the DMEM (Sigma) reported composition, in which 15N-cysteine was added together with all the other unlabelled components. All expression media were supplemented with 2% FBS. Zn(II) was supplemented as ZnSO4, which was added to the expression media to a final concentration of 10 μM immediately after transfection. Basal zinc concentration was calculated by considering 45 μM zinc present in the FBS, as reported by Sigma-Aldrich Media Expert (http://www.sigmaaldrich.com/life-science/cell-culture/learning-center/media-expert.html) Cu(II) was supplemented as CuCl2, added to a final concentration of 100 μM after 48 hours of protein expression, and incubated for 24 hours. Protein expression levels were monitored by comparing the protein band intensities in the cell extracts with bands of in vitro samples of known concentration run on Coomassie-stained SDS-PAGE.
Human cell samples for in-cell NMR
Samples for in-cell NMR were prepared following a reported protocol3 with some variations: HEK293T cells from a 75 cm2 culture flask were detached with trypsin-EDTA 0,05% (Gibco) and resuspended in 20 mL DMEM containing 10% FBS to inactivate trypsin. Cells were gently centrifuged (800 g), resuspended in 10 mL PBS, washed once with PBS and finally resuspended in one cell pellet volume of DMEM medium supplemented with 90 mM glucose, 16 mM HEPES buffer, 20% D2O (for a final 10% D2O amount). The cell suspension was transferred to a 3 mm Shigemi NMR tube; the glass plunger was not used. Cells were allowed to settle at the bottom of the tube, thus filling up the active coil volume. The supernatant was kept during the NMR experiments to obtain good field homogeneity. After the experiments, cells were resuspended in the supernatant, removed from the NMR tube and spun down again to collect the medium for protein leakage check (Supplementary Fig. 13). Cells were lysed by freeze/thaw method after suspending them in one pellet volume of PBS buffer supplemented with 0.5 mM EDTA and AEBSF (4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride). The lysate was centrifuged at 16000 g, 30′, 4°C and the cleared cell extract was collected for NMR and SDS-PAGE analysis.
NMR experiments
NMR experiments were acquired at a 950 MHz Bruker Avance™ III spectrometer equipped with a CP TCI CryoProbe™. 1D 1H and 2D 1H,15N-SOFAST-HMQC26 spectra were acquired at 305K. The total acquisition time for each cell sample ranged from 1 to 2 h. The supernatant of each cell sample was checked in the same experimental conditions, in order to exclude the presence of any signal arising from the protein leaked out of the cells. In the above experimental conditions, very low protein signal was detected in the external medium (< 10% of the signal in cells). The same NMR spectra were also acquired on the cell extracts. Cell viability before and after NMR experiments was assessed by Trypan Blue staining27. Cell viability remained above 90%, as damaged cells ranged from 3% before the experiments to 8% after the experiments.
E. coli cell samples
Samples of E. coli cells expressing human SOD1 were prepared as previously described15. For copper incorporation experiments, after 4 h expression of SOD1 cells were incubated with either 100 μM Cu(II)SO4, Cu(I)-acetonitrile complex or Cu(I)-glutathione complex for 15′. Cells were then washed once with M9 buffer and collected for NMR sample preparation. NMR spectra on E. coli cell samples were acquired at 305K at an 800 MHz Bruker Biospin™ spectrometer equipped with a TXI CryoProbe™.
Supplementary Material
Acknowledgements
This work was supported by the Access to Research Infrastructures activities in the 7th Framework Programme of the EC (Bio-NMR - Contract 261863 and P-CUBE – Contract 227764), by the Italian MIUR-PRIN 2009 ‘Biologia strutturale meccanicistica: avanzamenti metodologici e biologici’ and by Instruct, part of the European Strategy Forum on Research Infrastructures (ESFRI) and supported by national member subscriptions. Specifically, we thank the EU ESFRI Instruct Core Centres CERM – Italy, and University of Oxford – UK. We would like to thank E.Y. Jones and D.I. Stuart for critically reading the manuscript and providing advice. A.R.A. was supported by an MRC Career Development Award fellowship.
Footnotes
Competing financial interests:
The authors declare no competing financial interests.
Additional information:
Supplementary information (fourteen figures) is available in the online version of the paper.
References
- 1.Reckel S, Hänsel R, Löhr F, Dötsch V. Prog. NMR Spectrosc. 2007;51:91–101. [Google Scholar]
- 2.Burz DS, Shekhtman A. Plos ONE. 2008;3:e2571. doi: 10.1371/journal.pone.0002571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Inomata K, et al. Nature. 2009;458:106–109. doi: 10.1038/nature07839. [DOI] [PubMed] [Google Scholar]
- 4.Ogino S, et al. J. Am. Chem. Soc. 2009;131:10834–10835. doi: 10.1021/ja904407w. [DOI] [PubMed] [Google Scholar]
- 5.Selenko P, et al. Nat. Struct. Mol. Biol. 2009;15:321–329. doi: 10.1038/nsmb.1395. [DOI] [PubMed] [Google Scholar]
- 6.Culotta VC, Yang M, O’Halloran TV. Biochim. Biophys. Acta. 2006;1763:747–758. doi: 10.1016/j.bbamcr.2006.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michiels C, Raes M, Toussaint O, Remacle J. Free Radic. Biol. Med. 1994;17:235–248. doi: 10.1016/0891-5849(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 8.Lindberg MJ, Tibell L, Oliveberg M. Proc. Natl. Acad. Sci. USA. 2002;99:16607–16612. doi: 10.1073/pnas.262527099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banci L, et al. Proc. Natl. Acad. Sci. USA. 2007;104:11263–11267. doi: 10.1073/pnas.0704307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aricescu AR, Lu W, Jones EY. Acta Crystallogr. D. Biol. Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 11.Chang LY, Slot JW, Geuza HJ, Crapo JD. J. Cell Biol. 1988;107:2169–2179. doi: 10.1083/jcb.107.6.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sturtz LA, Diekert K, Jensen LT, Lill R, Culotta VC. J. Biol. Chem. 2001;276:38084–38089. doi: 10.1074/jbc.M105296200. [DOI] [PubMed] [Google Scholar]
- 13.Banci L, Bertini I, Cramaro F, Del Conte R, Viezzoli MS. Biochemistry. 2003;42:9543–9553. doi: 10.1021/bi034324m. [DOI] [PubMed] [Google Scholar]
- 14.Banci L, Bertini I, Cantini F, D’Amelio N, Gaggelli E. J. Biol. Chem. 2006;281:2333–2337. doi: 10.1074/jbc.M506497200. [DOI] [PubMed] [Google Scholar]
- 15.Banci L, Barbieri L, Bertini I, Cantini F, Luchinat E. Plos ONE. 2011;6:e23561. doi: 10.1371/journal.pone.0023561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim BE, Nevitt T, Thiele DJ. Nat. Chem. Biol. 2008;4:176–185. doi: 10.1038/nchembio.72. [DOI] [PubMed] [Google Scholar]
- 17.Puig S, Thiele DJ. Curr. Opin. Chem. Biol. 2002;6:171–180. doi: 10.1016/s1367-5931(02)00298-3. [DOI] [PubMed] [Google Scholar]
- 18.Banci L, et al. Nature. 2010;465:645–648. doi: 10.1038/nature09018. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt PJ, et al. J. Biol. Chem. 1999;274:23719–23725. doi: 10.1074/jbc.274.34.23719. [DOI] [PubMed] [Google Scholar]
- 20.Furukawa Y, Torres AS, O’Halloran TV. EMBO J. 2004;23:2872–2881. doi: 10.1038/sj.emboj.7600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carroll MC, et al. Proc. Natl. Acad. Sci. USA. 2004;101:5964–5969. doi: 10.1073/pnas.0308298101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leitch JM, Yick PJ, Culotta VC. J. Biol. Chem. 2009;284:24679–24683. doi: 10.1074/jbc.R109.040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Proescher JB, Son M, Elliott JL, Culotta VC. Hum. Mol. Genet. 2008;17:1728–1737. doi: 10.1093/hmg/ddn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banci L, et al. Proc. Natl. Acad. Sci. USA. 2012;109:13555–13560. doi: 10.1073/pnas.1207493109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reckel S, Lopez JJ, Löhr F, Glaubitz C, Dötsch V. ChemBioChem. 2012;13:534–537. doi: 10.1002/cbic.201100721. [DOI] [PubMed] [Google Scholar]
- 26.Schanda P, Brutscher B. J. Am. Chem. Soc. 2005;127:8014–8015. doi: 10.1021/ja051306e. [DOI] [PubMed] [Google Scholar]
- 27.Freshney R. Culture of Animal Cells: A Manual of Basic Technique. Alan R. Liss, Inc.; New York: 1987. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.