Abstract
Background
One-tenth of all infectious diseases are attributable to emerging organisms. As emerging organisms sporadically affect a relatively small percentage of population they are not studied at large. This study was aimed at studying the characteristics of emerging organisms encountered from various clinical samples in an apex tertiary care multispeciality teaching and research hospital.
Methods
16,918 positive isolates obtained from 66,323 culture samples processed in the clinical microbiology lab of an apex multispeciality hospital during 2011–2012 were included after a pilot study. Both manual and automated systems were used for identification and antimicrobial susceptibility. The frequency of isolation, sources, referring centers, resistance and susceptibility profiles, phenotypic characteristics and number of reports in PubMed were studied.
Results
Out of 16,918 isolates, 13,498 (79.78%) were Gram negative bacteria, 3254 (19.23%) were Gram positive bacteria and 166 (0.98%) were yeasts. A total of 483 (2.85%, 95% CI 2.6%–3.1%) emerging organisms including 116 (0.69%, 95% CI 0.57%–0.81%) emerging species were identified comprising 54 genera.
Conclusion
Emerging organisms are likely to evade routine identification or be disregarded as non-contributory. Astute efforts directed at identification of emerging isolates, decisions by clinical microbiologists and treating physicians and containment of infection are required.
Keywords: Emerging organisms, Microbial identification, Antimicrobial resistance, Automated microbiology systems
Introduction
The resurgence of infectious diseases in 1980s parallel to the emergence of Human Immunodeficiency Virus-Acquired Immunodeficiency Syndrome (HIV-AIDS) pandemic, resulted in 1.5 fold increase in death rate from infectious diseases between 1980 and 1992.1 Presently, a quarter of physician visits are attributed to infectious diseases, of which one-tenth are attributable to emerging organisms, of which bacteria constitute more than half and fungi one-tenth.2 Emerging organisms are organisms that have newly appeared in a cohort/population or have existed but are rapidly increasing in incidence, geographic or host range. Recently discovered etiological agents of known diseases are also considered as emerging organisms. However, operationally defining an organism as emerging is a subjective endeavor.3 The evolution of microbes has increased their virulence, infectivity, pathogenicity, resistance and paved the way for emergence of new infectious organisms.3–5 Established non-pathogens and commensals are now increasingly being encountered as opportunistic pathogens in patients with special conditions such as organ transplantation, immunocompromised states, cancer chemotherapy and radiotherapy, altered metabolic states, extensive burns, prematurity, old age and terminal illness. As emerging organisms sporadically affect a relatively small percentage of population they are not studied at large. As the burden of treating compromised patients falls under tertiary healthcare, a high index of suspicion is required in the diagnosis and management of infectious diseases caused by emerging organisms. This study was aimed at studying the frequency, sources, resistance and susceptibility profiles, and phenotypic characteristics of emerging organisms encountered from various clinical samples in an apex tertiary care teaching and research hospital.
Materials and methods
16,918 positive isolates obtained from 66,323 culture samples processed in the clinical microbiology lab of an apex multispeciality hospital during Jan 2011–Dec 2012 were included in the retrospective study after inferences from a pilot study conducted for the period covering Jul 2010–Dec 2010 and due approval from the Hospital Ethics Committee. The pilot study was conducted to improve upon the isolation modalities of the laboratory when adequate species level identification of many isolates could not be attempted satisfactorily utilizing standard manual identification methods. Out of 2040 positive isolates obtained from 12,885 samples, 1500 (73.53%, 95% Confidence Interval 71.62%–75.44%) were Gram negative bacteria, 380 (18.63%, 95% CI 16.94%–20.32%) were Gram positive bacteria and 16 (0.78%, 95% CI 0.4%–1.16%) were yeasts. The isolates hitherto unidentifiable manually were subjected to species level identification by automated microbiology system, MicroScan WalkAway 40 SI (Siemens Healthcare Diagnostics, Inc., West Sacramento, CA 95691, USA). The pilot study revealed the isolation of rarer organisms including nonfermenters such as Providencia rettgeri (4), Stenotrophomonas maltophilia (3), and increase in frequency of isolation of Acinetobacter baumannii and Burkholderia cepacia was noted.
For the present study, various samples were plated either directly on solid agar or after positive culture screen from BACTEC™ 9120 (BD Diagnostics, 1 Becton Drive, Franklin Lakes, NJ, USA 07417) and BacT/ALERT® 3D (bioMérieux SA, F-69280 Marcy l'Etoile, France) blood culture systems and incubated in O2 at 37 °C for 18–120 h. Blood samples included aerobic, anaerobic blood cultures and central line tip; urine samples included urine and urinary catheter tips; respiratory samples included sputum, tracheal aspirate, bronchoalveolar lavage, throat swab and nasal swab; body fluid samples included pleural fluid, ascitic fluid and cerebrospinal fluid; pus included pus from various sites; miscellaneous samples included semen, high vaginal swab, stool, tissue and drain fluid; and isolation of organisms from multiple samples was considered. Both manual and automated systems were used for identification and antimicrobial susceptibility. The organisms were identified manually by Gram staining, tests for motility, carbon source utilization, enzymatic activity and special characteristics, and antibiograms were obtained by Kirby–Bauer disc diffusion method on Mueller Hinton agar. MicroScan WalkAway 40 SI (Siemens Healthcare Diagnostics, Inc., West Sacramento, CA 95691, USA) and Vitek 2 compact (bioMérieux SA, F-69280 Marcy l'Etoile, France) automated systems were used in parallel to manual methods for identification and antimicrobial susceptibility, especially for uncommon isolates wherein manual identification was difficult. Inbuilt standards for identification comparison were utilized. Identification percentage >85% for both systems were taken as cutoffs for final validation.6,7 Non-repeat positive cultures with respective antibiograms were taken into account for profiling of isolates and antimicrobial susceptibility. All identified isolates were interpreted in conjunction with colony characteristics, cellular morphology after staining, motility testing, reactions on various isolation media, results of presumptive biochemical reactions, disc diffusion antimicrobial susceptibility patterns and clinical correlates. Isolates, sources of isolates, referring centers and drug resistance from lab reports were noted. A literature search was done to identify reports on human pathogenicity. An advanced PubMed search for human pathogen records mentioning the organism in main title was done (name of the organism[title], no space between organism and [title], searched at http://www.ncbi.nlm.nih.gov/pubmed/advanced). Surveillance studies of various centers along with temporal pattern were correlated with diagnosis of the patient. Descriptive statistics including frequency, percentages, 95% Confidence Intervals (95% CI) were worked out.
Results
Out of 16,918 isolates, 13,498 (79.78%, 95% CI 79.17%–80.39%) were Gram negative bacteria, 3254 (19.23%, 95% CI 18.64%–19.83%) were Gram positive bacteria and 166 (0.98%, 95% CI 0.83%–1.13%) were yeasts. A total of 483 (2.85%, 95% CI 2.6%–3.1%) emerging organisms including 116 (0.69%, 95% CI 0.57%–0.81%) emerging species were identified comprising 54 genera amongst 16,918 isolates (Fig. 1). A few lactose fermenting isolates initially suspected to be Escherichia coli were identified as other Enterobacteriaceae (Table 1). Many unidentifiable nonfermenters by standard methods were identified to be organisms from various families (Table 2). Similarly, a host of emerging organisms and newer species of Staphylococci and Streptococci were isolated (Table 3). Many new species of Candida were also isolated (Table 4). Twelve isolates of Prototheca, an extremely rare alga, were also identified by automated system (Table 4). Most common emerging genera were Serratia and Citrobacter amongst Enterobacteriaceae; Sphingomonas and Pseudomonas amongst nonfermenters; newer species of Staphylococci and Streptococci amongst Gram positive cocci; Candida and Prototheca amongst yeasts and algae. Most common source of isolation for nonfermenters, Gram positive cocci and yeasts was blood while it was urine for coliforms. These emerging organisms were identified from samples sent from various centers. Variable susceptibility and resistance patterns were encountered. PubMed search for human pathogens revealed Kingella, Achromobacter xylosoxidans, Rhodococcus equi, Lactococcus garvieae, Staphylococcus lugdunensis, Streptococcus mitis, Streptococcus pasteurianus and Malassezia furfur with maximum records mentioning them in titles. The frequency of isolation, sources, referring centers, resistance and susceptibility profiles, phenotypic characteristics and number of reports in PubMed are listed in Tables 1–4.
Fig. 1.
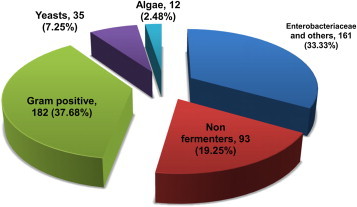
Distribution of emerging organisms by category.
Table 1.
Emerging Enterobacteriaceae, Vibrionaceae and Pasteurellaceae (161).
| S. no. | Organisms (37) | No | Source(s) | Referring center | Resistance | Susceptibility | Characteristics | PubMed records |
|---|---|---|---|---|---|---|---|---|
| 1. | Raoultella ornithinolytica | 11 | Urine | Multiple | Beta-lactams | Tigecycline | Nonmotile, IMViC − −++ | 4 |
| Raoultella planticola* | 1 | |||||||
| 2. | Cronobacter sakazakii | 4 | Multiple | OPD | Beta-lactams | Aminoglycosides, Fluoroquinolones | Motile, IMViC − −++ | 32 |
| C. dublinensis* | 1 | Urine | Int Medicine | Nil | ||||
| 3. | Kluyvera ascorbata | 4 | Blood | Gen Surgery OPD |
Beta-lactams, Fluoroquinolones | Aminoglycosides, Tetracyclines | Motile, IMViC − + −+ | 14 |
| K. intermedia* | 2 | Blood | Nil | |||||
| 4. | Lecleria adecarboxylata* | 2 | Body fluid | Gen Surgery | Multisensitive | Multisensitive | Motile, IMViC ++− − | Nil |
| 5. | Tatumella ptyseos* | 1 | Blood | Int Medicine | Multisensitive | Multisensitive | Motile, IMViC − − −+ | 4 |
| 6. | Cedecea lapagei* | 1 | Pus | Gen Surgery | 3 GC, Aminoglycosides | SXT | Motile, IMViC − −++ | 3 |
| 7. | Yokenella regensburgei* | 1 | Misc | OPD | Multisensitive | Multisensitive | Motile, IMViC − + −+ | 5 |
| 8. | Ewingella Americana* | 1 | Urine | OPD | Multiresistant | SXT | Motile, IMViC − +++ | 15 |
| 9. | Escherichia fergusonii | 3 | Urine | Multiple | Beta-lactams | Aminoglycosides, Carbapenems | Motile, IMViC ++− − | 11 |
| E. vulneris* | 1 | Blood | Int Medicine | 13 | ||||
| 10. | Citrobacter koseri | 15 | Urine, Pus | OPD | Beta-lactams | Aminoglycosides, Fluoroquinolones, Carbapenems, SXT | Motile, IMViC − −++ | 46 |
| C. amalonaticus | 7 | Urine | OPD | 3 | ||||
| C. sedlakii | 5 | Pus | Orthopedics | 4 | ||||
| C. youngae | 5 | Pus | Int Medicine | 2 | ||||
| 11. | Klebsiella ozaenae | 4 | Multiple | ICU | Pansensitive | Pansensitive | Motile, IMViC − − − − | 28 |
| K. rhinoscleromatis* | 1 | Pus | Gen surgery | Motile, IMViC − − − - | 21 | |||
| 12. | Enterobacter agglomerans | 4 | Multiple | Multiple | Beta-lactams, Aminoglycosides, Carbapenems | Piperacillin-Tazobactam | Motile, IMViC − −++ | 28 |
| E. durans* | 2 | Urine | Int Medicine | Nil | ||||
| E. cancerogenus* | 2 | Respiratory | Gen Surgery | Nil | ||||
| E. gergoviae* | 1 | Urine | OPD | 6 | ||||
| E. amnigenus* | 1 | Blood | ICU | 6 | ||||
| E. asburiae* | 1 | Blood | ICU | 5 | ||||
| 13. | Serratia fonticola | 28 | Urine | OPD | Beta-lactams | Aminoglycosides, Fluoroquinolones, Carbapenems, SXT, Piperacillin-Tazobactam | Motile, IMViC − + −+ | 4 |
| S. liquefaciens | 9 | Urine | Multiple | 28 | ||||
| S. odorifera | 7 | Urine | Multiple | 8 | ||||
| S. ficaria | 3 | Urine | ICU | 8 | ||||
| S. rubidaea | 3 | Blood | Oncology | 5 | ||||
| 14. | Providencia rettgeri | 13 | Multiple | Multiple | Multiresistant | Multiresistant | Motile, IMViC ++−+ | 13 |
| P. rustigianii | 4 | Multiple | Multiple | 1 | ||||
| 15. | Aeromonas salmonicida | 4 | Urine | Nephrology | Beta-lactams | Tetracyclines, Aminoglycosides, SXT | Nonmotile, IMViC + −++, Oxidase +, Catalase + | 18 |
| A. veronii | 2 | Urine | Hematology | 47 | ||||
| A. caviae* | 1 | Blood | Hematology | 52 | ||||
| A. sobria* | 1 | Blood | ICU | 62 | ||||
| 16. | Pasteurella pneumotropica | 3 | Urine | Nephrology | Multisensitive | Multisensitive | Nonmotile, IMViC + − − − Oxidase +, Catalase + | 19 |
| P. canis* | 1 | Blood | OPD |
Note: *All these isolates would require further surveillance for possible emerging infections.
Key: IMViC – Indole, Methyl Red, Voges–Proskauer, Citrate; 3GC – Third generation cephalosporins, SXT – Sulphamethoxazole-Trimethoprim, Int Medicine – Internal Medicine, Gen Surgery – General Surgery, Misc – Miscellaneous, ICU – Multidisciplinary Intensive Care Unit, OPD – Out Patient Department, Figures in brackets against table title represent the cumulative frequency, Figures in brackets against organisms represent sum of all species.
Table 2.
Emerging nonfermenters (93).
| S. no. | Organisms (26) | No | Source(s) | Referring center | Resistance | Susceptibility | Characteristics | PubMed records |
|---|---|---|---|---|---|---|---|---|
| 1. | Sphingomonas paucimobilis | 21 | Blood | Multiple | Multiresistant | Multiresistant | Nonmotile, Ox +, Cat + | 30 |
| 2. | Pseudomonas luteola | 9 | Multiple | Multiple | Beta-lactams, Aminoglycosides | Fluoroquinolones, SXT, Carbapenems, Tetracycline, Piperacillin-Tazobactam | Motile, Ox +, Cat + | 8 |
| P. pseudoalcaligenes | 2 | Blood | ICU | 3 | ||||
| P. alcaligenes* | 1 | Pus | OPD | 8 | ||||
| P. oryzihabitans* | 1 | Misc | OPD | 1 | ||||
| 3. | Achromobacter xylosoxidans | 8 | Urine | Urology | Multiresistant | SXT | Motile, Ox +, Cat +, Urease + | 103 |
| A. denitrificans | 5 | Blood | ICU | 3 | ||||
| 4. | Ralstonia picketii | 3 | Urine | Nephrology | Multiresistant | Carbapenems, SXT, Piperacillin-Tazobactam | Motile, Ox +, Cat + | Nil |
| R. paucula | 3 | Blood | Nephrology | 4 | ||||
| R. mannitolytica* | 1 | Misc | OPD | 1 | ||||
| 5. | Chryseobacterium indologenes | 7 | Urine | Int Medicine | Multiresistant | SXT | Motile, Ox +, Cat + | 23 |
| 6. | Chromobacterium violaceum | 6 | Multiple | Multiple | Multiresistant | Carbapenems | Motile, Ox +, Cat + | 93 |
| 7. | Acinetobacter junii | 4 | Urine | OPD | Multisensitive | Multisensitive | Nonmotile, Ox −, Cat + | 13 |
| 8. | Cupriavidus pauculus | 3 | Blood | OPD | Beta-lactams | Carbapenems, SXT | Motile, Ox +, Cat + | 5 |
| 9. | Yersinia aldovae* | 2 | Multiple | Multiple | Multisensitive | Multisensitive | Motile, Ox −, Cat + | Nil |
| Y. ruckeri* | 1 | Blood | Int Medicine | 1 | ||||
| 10. | Comamonas testosterone* | 2 | Urine, Pus | Gynecology | Beta-lactams | SXT | Motile, Ox +, Cat + | 9 |
| 11. | Empedobacter brevis* | 2 | Body fluid | Burn center | Multiresistant | Carbapenems | Nonmotile, Ox -, Cat - | 1 |
| 12. | Rhizobium radiobacter* | 2 | Blood | Int Medicine | Multisensitive | Multisensitive | Motile, Ox −, Cat + | 16 |
| 13. | Myroides species* | 2 | Urine | Burn center | Beta-lactams | Carbapenems | Nonmotile, Ox +, Cat + | 7 |
| 14. | Kingella species* | 2 | Urine | OPD | Multisensitive | Multisensitive | Nonmotile, Ox +, Cat - | 192 |
| 15. | Sphingobacterium spiritovorum* | 1 | Blood | Int Medicine | Multiresistant | Levofloxacin, SXT | Nonmotile, Ox +, Cat + | 2 |
| 16. | Brevundimonas diminuta* | 1 | Pus | Int Medicine | Multiresistant | Piperacillin-Tazobactam | Motile, Ox +, Cat + | 4 |
| 17. | Burkholderia multivorans* | 1 | Blood | Burn center | Multiresistant | Multiresistant | Motile, Ox +, Cat + | 24 |
| 18. | Elizabetkingia meningoseptica* | 1 | Body fluid | OPD | Multiresistant | Multiresistant, SXT | Nonmotile, Ox +, Cat + | 11 |
| 19. | Oligella ureolytica* | 1 | Blood | OPD | Multisensitive | Multisensitive | Motile, Ox +, Cat + | 2 |
| 20. | Alkaligenes faecalis* | 1 | Urine | OPD | Multisensitive | Multisensitive | Motile, Ox +, Cat + | 1 |
Note: *All these isolates would require further surveillance for possible emerging infections.
Key: Ox – Oxidase, Cat – Catalase, SXT – Sulphamethoxazole-Trimethoprim, Int Medicine – Internal Medicine, ICU – Multidisciplinary Intensive Care Unit, OPD – Out Patient Department, Misc – Miscellaneous, Figures in brackets against table title represent the cumulative frequency, Figures in brackets against organisms represent sum of all species.
Table 3.
Emerging Gram positive bacteria (182).
| S. no. | Organisms (40) | No | Source(s) | Referring center | Resistance | Susceptibility | Characteristics | PubMed records |
|---|---|---|---|---|---|---|---|---|
| 1. | Kocuria kristinae | 19 | Blood, Pus | Multiple | Multisensitive | Multisensitive | Cat +, Coagulase −, Ox − | 8 |
| K. varians | 4 | Blood | ICU | 3 | ||||
| K. rosea* | 2 | Blood | Gynecology | 2 | ||||
| 2. | Micrococcus luteus | 7 | Blood | Multiple | Multisensitive | Multisensitive | Cat +, Coagulase −, Ox + | 26 |
| 3. | Rothia mucilaginosa* | 1 | Respiratory | OPD | Multisensitive | Multisensitive | Cat +, Coagulase −, Ox − | 7 |
| 4. | Lactococcus garvieae | 7 | Urine | Multiple | Beta-lactams, Macrolides | Vancomycin | Cat −, Coagulase -, Ox - | 143 |
| L. lactis* | 2 | Urine | Gen Surgery | 29 | ||||
| 5. | Kytococcus sedentarius* | 1 | Urine | OPD | Multisensitive | Multisensitive | Cat +, Coagulase −, Ox − | 5 |
| 6. | Gemella sanguinis | 4 | Respiratory | OPD | Multisensitive | Multisensitive | Cat -, Coagulase -, Ox - | 4 |
| G. morbillorum* | 2 | Respiratory | Neurology | 82 | ||||
| 7. | Granulicatella adiacens | 5 | Multiple | OPD | Beta-lactams | Vancomycin | Cat −, Coagulase −, Ox − | 16 |
| 8. | Enterococcus gallinarum* | 2 | Urine | Nephrology | Multisensitive | Multisensitive | Cat −, Coagulase −, Ox - | 32 |
| E. avium* | 2 | Urine | OPD | 24 | ||||
| 9. | Leuconostoc mesenteroides | 3 | Multiple | Multiple | Beta-lactams | Aminoglycosides | Cat −, Coagulase −, Ox − | 17 |
| 10. | Aerococcus viridians* | 1 | Pus | Burn center | Beta-lactams | 3 GC, Aminoglycosides | Cat +, Coagulase -, Ox - | 27 |
| 11. | Dermacoccus nishinomiyaensis* | 1 | Misc | Int Medicine | Multisensitive | Multisensitive | Cat +, Coagulase −, Ox + | Nil |
| 12. | Rhodococcus equi* | 1 | Pus | OPD | Multisensitive | Multisensitive | Cat +, Coagulase -, Ox - | 304 |
| 13. | Staphylococcus sciuri | 61 | Multiple | Multiple | Multiresistant | Vancomycin, Linezolid | Cat +, Coagulase +, Ox+ | 31 |
| S. cohnii | 5 | Blood | Multiple | Multiresistant | Multiresistant | Cat +, Coagulase −, Ox - | 24 | |
| S. xylosus | 5 | Blood | Multiple | Multiresistant | Multiresistant | Cat +, Coagulase −, Ox − | 12 | |
| S. lentus | 4 | Multiple | Multiple | Multiresistant | Multiresistant | Cat +, Coagulase −, Ox − | 2 | |
| S. warneri | 4 | Multiple | Multiple | Multiresistant | Multiresistant | Cat +, Coagulase −, Ox − | 24 | |
| S. lugdunensis | 3 | Multiple | Multiple | Multiresistant | Multiresistant | Cat +, Coagulase −, Ox − | 161 | |
| S. capitis | 3 | Blood | NICU | Multiresistant | Multiresistant | Cat −, Coagulase −, Ox − | 14 | |
| S. hyicus* | 2 | Multiple | Multiple | Multiresistant | Tetracycline, Rifampin | Cat +, Coagulase +, Ox − | 31 | |
| S. simulans* | 2 | Multiple | Multiple | Multiresistant Multiresistant | Quinolones, Vancomycin | Cat +, Coagulase −, Ox − | 13 | |
| S. caprae* | 1 | Body fluid | Orthopedics | Multiresistant | Multiresistant | Cat +, Coagulase −, Ox - | 13 | |
| S. chromogenes* | 1 | Pus | NICU | Multiresistant | Cat +, Coagulase −, Ox - | 1 | ||
| 14. | Streptococcus mitis | 7 | Multiple | Multiple | Multisensitive | Multisensitive | Cat −, Coagulase −, Ox - | 124 |
| S. thoraltensis | 4 | Multiple | Multiple | Nil | ||||
| S. pasteurianus | 3 | Multiple | Multiple | 310 | ||||
| S. ciferrii | 3 | Blood | ICU | 35 | ||||
| S. ovis* | 2 | Misc | OPD | 1 | ||||
| S. alactolyticus* | 2 | Misc | Int Medicine | Nil | ||||
| S. bovis* | 1 | Multiple | Multiple | Nil | ||||
| S. sanguinis* | 1 | Multiple | Multiple | 29 | ||||
| S. parasanguinis* | 1 | Pus | Int Medicine | 6 | ||||
| S. pluranimalium* | 1 | Misc | Int Medicine | Nil | ||||
| S. porcinus* | 1 | Pus | Oncology | 4 | ||||
| S. equisimilis* | 1 | Blood | OPD | Nil |
Note: *All these isolates would require further surveillance for possible emerging infections.
Key: Cat – Catalase, Ox – Oxidase, 3GC – Third generation cephalosporins, SXT – Sulphamethoxazole-Trimethoprim, Int Medicine – Internal Medicine, Misc – Miscellaneous, ICU – Multidisciplinary Intensive Care Unit, NICU – Neonatal Intensive Care Unit, OPD – Out Patient Department, Figures in brackets against table title represent the cumulative frequency, Figures in brackets against organisms represent sum of all species.
Table 4.
Emerging yeasts and algae (47).
| S. no. | Organisms (13) | No | Source(s) | Referring center | Resistance | Susceptibility | Characteristics | PubMed records |
|---|---|---|---|---|---|---|---|---|
| Yeasts (35) | ||||||||
| 1. | Cryptococcus laurentii | 5 | Pus, Blood | ICU | Azoles | Amphotericin B | Urease −, Germ tube − | 21 |
| 2. | Candida haemulonii | 10 | Blood | ICU | Azoles, Amphotericin B | Echinocandins | Urease −, Germ tube − | 10 |
| C. famata | 9 | Blood | Multiple | 17 | ||||
| C. rugosa* | 2 | Blood | Int Medicine | 19 | ||||
| C. guilliermondii* | 1 | Blood | BMT | 40 | ||||
| C. lusitaniae* | 1 | Body fluid | Paediatrics | 60 | ||||
| C. utilis* | 1 | Blood | ICU | 10 | ||||
| C. zeylanoides* | 1 | Blood | ICU | 6 | ||||
| C. sphaerica* | 1 | Blood | ICU | Nil | ||||
| 3. | Malassezia furfur* | 2 | Misc | ICU | Multisensitive | Multisensitive | Urease −, Germ tube − | 166 |
| 4. | Trichosporon asahii* | 2 | Urine | ICU | Azoles | Amphotericin B | Urease +, Germ tube − | 83 |
| Algae (12) | ||||||||
| 1. | Prototheca species | 11 | Blood | Oncology | Multisensitive | Multisensitive | Urease +, Propanol − | 56 |
| P. wickerhamii* | 1 | Blood | Oncology | 23 | ||||
Note: *All these isolates would require further surveillance for possible emerging infections.
Key: Int Medicine – Internal Medicine, BMT – Bone Marrow Transplant Center, Misc – Miscellaneous, ICU – Multidisciplinary Intensive Care Unit, Figures in brackets against Yeasts and Algae represent the cumulative frequency, Figures in brackets against organisms represent sum of all species.
Discussion
The study reveals that most of the emerging organisms have been isolated from samples received from high workload centers like OPD, ICU, Internal Medicine and General Surgery. Surveillance studies of various centers revealed bacteria from air, floor and bed rails but the individual organisms were not frequent enough to make a correlation between clinical isolates and resident flora. The isolation of azole resistant Candida from ICU is in consonance with opportunistic infections in patients receiving long-term parenteral antibacterial drugs. It is debated that individual emergence of such organisms is impossible to predict.8 The isolation of a plethora of organisms in just a two-year period is likely to represent the tip of an iceberg as many organisms elude the clinical and laboratory set ups. The processes involved in microbial invasion, colonization, infection, clinical presentation, laboratory diagnosis, interpretation and treatment are dynamic, complex and cannot be standardized preventing conclusive studies. Out of 116 emerging species described in the study, only 8 have been reported more than a 100 times in PubMed and 14 have never been reported as pathogens in disease process. Most of them have been reported to be multidrug resistant in the available literature, which is in consonance with this study (Tables 1–4). As these organisms are present in the hospital environment, they are likely to be opportunistic multidrug resistant organisms targeting compromised hosts. This study is intended to be presented in an observational fashion. Comparative analysis between the pilot study and present study is limited by little data in pilot study. Retrospective analysis, small number of isolates of individual organisms, chances of misidentification by automated systems and lack of control set up may limit conclusive inference necessitating large parallel multicentric studies and laid down standard operating procedures for microbiological laboratories especially with regard to quality control and dealing with scanty isolates of unusual organisms.
Identification and susceptibility of emerging organisms
Coliforms, comprising over 100 species in 27 genera, constitute half of all clinically significant bacterial isolates and are causative in 50% septicemia and 70% urinary infections. Nonfermenters, comprising 15 heterogenous families, constitute 10–15% clinically significant isolates. Many emerging organisms go unreported, under-reported or uncharacterized either due to limited isolation techniques or isolates being labeled as “commensal” or “contaminant” by clinical microbiologists and/or treating clinicians in view of unknown or uncertain pathogenicity. It is a common practice in many resource limited labs to provide antibiograms without species/genus level identification to facilitate early treatment of patients. Problems in identification include inadequate sample processing, no or scanty growth on routine isolation media, unknown patterns of substrate utilization, unavailability of specialized tests, understaffed/under-skilled manpower and unusual antibiograms. Many unsuspected fastidious organisms fail to grow on routine isolation media or require prolonged incubation, which may not be attempted in routine diagnostic laboratories. Improper isolation, single colony on the entire plate and mixed growth are often labeled as “culture negative”, “insignificant growth” or “contaminants grown”. Reports from certain labs may be restricted to reference to diverse groups such as “non-fermenter”. In the absence of guidelines for antimicrobial susceptibility testing of emerging organisms, interpretation is difficult and treatment jeopardized. Certain organisms may have elevated minimal inhibitory concentrations (MICs), though remains within the susceptible range leading to disparity in breakpoint concentrations in vivo, making susceptibility patterns difficult to characterize.9
The problem of limited isolation has largely been addressed by the advent of automated phenotypic microbial identification systems and molecular microbiology both of which can be used for organism identification, antimicrobial susceptibility, characterization of resistance mechanisms and possibly epidemiological typing. These advancements are increasingly being utilized in progressive labs though accessibility is restricted in resource deficient settings owing to limitations in acquisition, maintenance and output capacities. While molecular microbiology is rapidly emerging as the new gold standard, it is limited by availability or designing capacity for organism-specific sequences, requirement of expertise, standardization, quality assurance and cost effectiveness.4 The performance evaluation of automated systems has long been established.10,11 The advanced microbial database of automated systems provides reasonable ease of operation, reliability, reproducibility and standardization in characterization of emerging organisms and resistance patterns.3,7,12 These systems have facilitated compaction, shelf life, convenience, time management, visibility of reactions, standardization and quality control under one umbrella. They can be entrusted for routine identification and susceptibility, though results need to be interpreted in conjunction with microbial morphology, biochemical tests and clinical correlates as misidentification has also been reported.13,14
Emerging organisms
All isolated organisms are ubiquitous in animate and inanimate environments, thereby increasing the ease of transmission, colonization and development of resistance. Societal, technological, environmental and biological factors contribute to the emergence of pathogens and drug resistance. Ecological disturbance due to rapid urbanization, industrialization, alteration of land, forest and water resources, and climate change may lead to increased exposure to pathogen reservoirs or vectors such as insects, animals, plants or other environmental sources. Globalization, large-scale human migration from rural and war-stricken areas, travel and lifestyle changes have lead to geographical expansion and increased exposure to hitherto geographically sequestered pathogens.4 Developing tropical countries may form the cradle of emerging pathogens owing to overwhelming patient population, limited access to healthcare facilities and unmonitored antimicrobial stewardship.15 In addition, overcrowding, substandard socioeconomic conditions, inadequate nutrition, improper hygiene and sanitation, limited education and health awareness, inadequate public health infrastructure, restricted national health budgets and human resource attrition indirectly contribute to increased host susceptibility and prevent effective screening, quarantine and treatment. Favorable environmental temperature and humidity contribute to increased vector population and increased survival of pathogens thereby enhancing transmissibility.16 Unjust organizational practices to enhance farm yield in agriculture, animal husbandry and fisheries as well as individual practices such as self medication, antimicrobial abuse and uncompleted regimens contribute to the development of widespread antimicrobial resistance in the community even in hitherto nonpathogenic organisms.17 These multidrug resistant organisms opportunistically infect compromised hosts. Community acquired multidrug resistant infections are on the rise which in turn reach hospital environments.18 The selection pressure under higher generation antimicrobials further enhances resistance. This resistance spreads far and wide through bacterial conjugation, cross infections and patient movement. Undetected novel multidrug resistant organisms in patients and carriers serve as reservoirs of infection and may cause community or nosocomial outbreaks. Their obscurity is compounded by silent colonization, prolonged incubation period, opportunistic infections, uncertain pathogenicity and inadequate isolation.
Attributing pathogenicity
Attributing pathogenicity to emerging organisms is difficult.3,5 Positive isolates may be due to colonization of drug resistant hospital flora in the absence of infectious disease. Skin of patients and healthcare providers, personal protective equipment, medical equipment and hospital environment may harbor unusual organisms and get transferred to patients. These organisms may form biofilms on invasive devices causing a low-level infection, evading identification and antimicrobial treatment. Clinical samples, laboratory equipment, stains and media may get contaminated and lead to pseudo-outbreaks.19 One-time isolation of hitherto zoonotic pathogens may remain inconclusive. Positive isolates may not be associated with clinically manifest infection owing to muted immune response in premature neonates, elderly and immunocompromised patients. Also, clinical inflammatory response may not always be related to microbial infection. Single isolate may not seem to fit in the overall presentation, management and prognosis of patient. Polymicrobial isolates may emanate misunderstanding about dominant pathogen.20
The clinical presentation may get altered by the time microbiological identification and susceptibility reports are ready as most seriously ill patients are put on empirical antimicrobials. The other limitation in attributing pathogenicity to unusual isolates is the problem of patients being lost to follow-up. Clinical improvement of patients due to empirical antimicrobials or management of primary condition may lead to patient being transferred from intensive care to low dependency units and discharge from the hospital. Getting such patient to come back for further testing is looked upon as unimportant both by treating physicians and patients themselves. However, emerging organisms are increasingly being reported as either causative agents in infectious disease or contributory factors in exacerbation of other comorbidities. Antimicrobials in such cases have to be changed, escalated or de-escalated after susceptibility reports. Ignoring any isolate may be risky and decisions can adversely affect the patients, hospital and community environment and development of antimicrobial resistance. Clinical decisions remain a challenge as to which isolate should be ignored and which one should be considered significant.
Bioweaponization
Emerging multidrug resistant organisms can be exploited as bioweapons. They can be further modified to increase their infectivity, virulence, resistance, transmission and stability in the environment. Given the ease of maintenance, genetic modification, dispersion and person-to-person transmission in the present day, they can be clandestinely deployed against the human race. Their detection and control can be challenging in the absence of requisite knowledge, diagnostic and management experience. Uncontrolled infection and ongoing transmission can lead to outbreaks and epidemics, which may have long-term effects on furtherance of pathogenicity and drug resistance. Inadvertent or intended release of modified organisms from diagnostic and research labs is a possible after effect of bioterrorism.21
Prevention and future
The emergence of new organisms is perpetual and requires ongoing surveillance.22 Astute research incorporating parallel phenotypic and molecular identification including typing and extended surveillance can augment knowledge and experience on emerging organisms, newer strains and acquisition of antimicrobial resistance. Biomedical and social interventions on a mass scale are required to subdue emerging organisms.15 CDC has strategized target areas, which can form broad guidelines for public health systems worldwide.14 Core competencies for rapid detection and management of emerging organisms by enhancing lab capacity through resource allocation for infrastructure, expertise, automation and knowledge sharing are necessitated to overcome existing deficiencies, in line with International Health Regulations.15 Antimicrobial policy regulations and infection control measures should be strengthened. Judicious decision making aimed at “antimicrobial austerity” at the end of industrialists, healthcare providers and patients is crucial. Faster genotypic automated systems along with better understanding of pathogenicity will help in detecting clinically relevant emerging organisms.
Conclusion
Emerging organisms have the potential to infect compromised hosts posing difficulty in management due to multidrug resistance. They are likely to evade routine identification or be disregarded as insignificant contaminants. A greater engagement between clinicians and microbiologists, effective hospital infection control practices and faster microbial identification technology are required to identify emerging organisms and contain them effectively.
Conflicts of interest
All authors have none to declare.
References
- 1.Armstrong G.L., Conn L.A., Pinner R.W. Trends in infectious disease mortality in the United States during the 20th century. JAMA. 1999;281:61–66. doi: 10.1001/jama.281.1.61. [DOI] [PubMed] [Google Scholar]
- 2.Taylor L.H., Latham S.M., Woolhouse M.E. Risk factors for human disease emergence. Philosophical Trans R Soc Lond B Biol Sci. 2001;356(1411):983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hazen K.C. New and emerging yeast pathogens. Clin Micobiol Rev. 1995;8(4):462–478. doi: 10.1128/cmr.8.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong J., Olano J.P., McBride J.W., Walker D.H. Emerging pathogens: Challenges and Successes of molecular diagnostics. J Mol Diagn. 2008;10(3):185–197. doi: 10.2353/jmoldx.2008.070063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miceli M.H., Diaz J.A., Lee S.A. Emerging opportunistic yeast infections. Lancet Infect Dis. 2011;11:142–151. doi: 10.1016/S1473-3099(10)70218-8. [DOI] [PubMed] [Google Scholar]
- 6.Pincus DH. Microbial identification using the Biomerieux Vitek2 system. Encyclopedia rapid microbiol methods. http://www.pda.org/bookstore. Accessed 30.12.12.
- 7.Rhoads S., Marnielli L., Imperatrice C.A., Nachamkin I. Comparison of MicroScan WalkAway system and Vitek system for identification of gram-negative bacteria. J Clin Microbiol. 1995;33(11):3044–3046. doi: 10.1128/jcm.33.11.3044-3046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centre for Disease Control and Prevention Preventing Emerging Infectious Diseases: a strategy for the 21st Century. Overview of updated CDC plan. MMWR. 1998;47(RR-15):1–14. [PubMed] [Google Scholar]
- 9.McGettigan S.E., Andreacchio K., Edelstein P.H. Specificity of Ertapenem susceptibility screening for detection of Klebsiella pneumoniae Carbapenemases. J Clin Microbiol. 2009;47(3):785–786. doi: 10.1128/JCM.02143-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatzigeorgiou K.S., Sergentanis T.N., Tsiodras S., Hamodrakas S.J., Bagos P.G. Phoenix 100 versus Vitek 2 in the identification of gram-positive and gram-negative bacteria: a comprehensive meta-analysis. J Clin Microbiol. 2011;49(9):3284–3291. doi: 10.1128/JCM.00182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin W.Y., Jang S.J., Lee M.J. Evaluation of VITEK 2, MicroScan, and Phoenix for identification of clinical isolates and reference strains. Diag Microbiol Inf Dis. 2011;70(4):442–447. doi: 10.1016/j.diagmicrobio.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 12.Jethwani U.M., Mulla S.A., Shah L.N. Detection of inducible clindamycin resistance by an automated system in a tertiary care hospital. African J Microbiol Res. 2011;5(18):2870–2872. [Google Scholar]
- 13.NNIS System. National Nosocomial Infections Surveillance (NNIS) System Report. Centers for Disease Control and Prevention, Department of Health and Human Services; Atlanta, GA: 2004. http://www.cdc.gov Data summary from January 1992 through June 2004, issued October 2004. Accessed 30.12.12. [Google Scholar]
- 14.Centre for Disease Control and Prevention. National Center for Emerging and Zoonotic Infectious Diseases (NCEZID) Division of Preparedness and Emerging Infections (DPEI) CDC; Oct 2011. Emerging Infections Programs (EIP)http://www.cdc.gov/ncezid/dpei/eip Accessed 30.12.12. [Google Scholar]
- 15.Narain J.P., Bhatia R. The challenge of communicable diseases in the WHO South-East Asia Region. Bull World Health Organ. 2010 March;88(3):162. doi: 10.2471/BLT.09.065169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morse S.S. Factors in the emergence of infectious diseases. Emerging Infect Dis. 1995;1:7–15. doi: 10.3201/eid0101.950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhatia R., Narain J.P. The growing challenge of antimicrobial resistance in South East Asia region – Are we losing the battle? Ind J Med Res. 2010;132(5):482–486. doi: 10.4103/0971-5916.73313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hidron A.I., Edwards J.R., Patel J. NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006–2007. Infect Control Hosp Epidemiol. 2008;29(11):996–1011. doi: 10.1086/591861. [DOI] [PubMed] [Google Scholar]
- 19.Thiolet J.M., Jourdan-Da Silva N., Reggiani A., De Valk H., Coignard B., Weill F.X. Nationwide pseudo-outbreak of Salmonella enterica ssp. diarizonae, France. Clin Microbiol Infect. 2011;17(6):915–918. doi: 10.1111/j.1469-0691.2010.03343.x. [DOI] [PubMed] [Google Scholar]
- 20.Gupta P., Kumhar G.D., Kaur G., Ramachandran V.G. Clinical significance of polymicrobial bacteremia in newborns. J Paediatr Child Health. 2005;41(7):365–368. doi: 10.1111/j.1440-1754.2005.00633.x. [DOI] [PubMed] [Google Scholar]
- 21.Centers for Disease Control and Prevention . Centers for Disease Control and Prevention; 2008-02-12. Bioterrorism Overview.http://www.cdc.gov Accessed 30.12.12. [Google Scholar]
- 22.Fauci A.S. Emerging and reemerging infectious diseases: the perpetual challenge. Acad Med. 2005;80(12):1079–1085. doi: 10.1097/00001888-200512000-00002. [DOI] [PubMed] [Google Scholar]


