Abstract
Recent studies suggested that induced pluripotent stem cells (iPSCs) retain a residual donor cell gene expression which may impact their capacity to differentiate into cell of origin. Here we addressed a contribution of a lineage stage-specific donor cell memory in modulating the functional properties of iPSCs. iPSCs were generated from hepatic lineage cells at an early (hepatoblast-derived, HB-iPSCs) and end stage (adult hepatocyte, AH-iPSCs) of hepatocyte differentiation as well as from mouse fetal fibroblasts (MEF-iPSCs) using a lentiviral vector encoding four pluripotency-inducing factors Oct4, Sox2, Klf4, and c-Myc. All resulting iPS cell lines acquired iPSCs phenotype as judged by the accepted criteria including morphology, expression of pluripotency markers, silencing of transducing factors, capacity of multilineage differentiation in teratoma assay and normal diploid karyotype. However, HB-iPSCs were more efficient in directed differentiation towards hepatocytic lineage as compared to AH-iPSCs, MEF-iPSCs or mESCs. Extensive comparative transcriptome analyses of the early passage iPSCs, donor cells and mESCs revealed that despite global similarities in gene expression patterns between generated iPSCs and mESCs, HB-iPSCs retained a transcriptional memory (7 up- and 17 down-regulated genes) typical of the original cells. Continuous passaging of HB-iPSCs erased most of these differences including a superior capacity for hepatic re-differentiation. These results suggest that retention of lineage stage-specific donor memory in iPSCs may facilitate differentiation into donor cell type. The identified gene set help to improve hepatic differentiation for therapeutic applications and contribute to the better understanding of liver development.
Keywords: induced pluripotent stem cells, donor memory, hepatocyte lineage cells, hepatic differentiation
Introduction
Induced pluripotent stem cells (iPSCs) are derived by introducing a combination of four transcription factors (KLF4, Oct4, Sox2 and Myc) into somatic cells. iPSCs have been shown to be similar to embryonic stem cells (ESCs) in many aspects, including morphology, self-renewal and pluripotency, and the capacity to generate any lineage-specific cell types including hepatocytes [1–4]. The generation of iPSCs has significantly changed our perspective on regenerative medicine including cell replacement therapy, and disease modeling in vitro by generating patient-specific iPSC [5–8]. Although iPSCs share most characteristics of ES cells, recent evidence reveals important differences between iPSCs and ESCs at the level of DNA methylation, global gene expression [9–11], genome stability [12,13], and differentiation potential [14,15]. The unique properties of iPSCs are thought to be defined by a donor memory reflecting the epigenetic and transcriptional characteristics inherited from the cell of origin during the reprogramming process [16–18]. Although the molecular mechanisms of transcriptional donor memory retention in iPSCs are not fully understood, evidence is growing that it is due to the incomplete DNA methylation during reprogramming of parental cells into iPSC. Among the identified transcriptional memory genes is C9orf64 implicated in regulating the efficiency of iPSCs generation from human foreskin fibroblasts [18].
Functionally, the retention of donor memory in iPSCs has been shown to facilitate the re-differentiation capacity of iPSCs into the donor cells rather than into other cell types [16,17,19]. Recent work demonstrated that a unique pattern of methylation and/or gene expression characteristic for low passage iPSCs is not stable but gradually reverses to the ESC-type gene expression profile upon extended iPSC culture [9–11,16–18]. The latter suggests that donor memory can be one of the potential factors to selectively regulate the re-differentiation potency of iPSCs. However, it remains unknown whether a lineage stage-specific donor memory can modulate iPSCs functional properties.
In this study, to address the lineage stage specific donor memory we generated iPSC from the early and late stages of hepatocytic differentiation, including E16.5 hepatoblasts (HB-iPSC) and adult hepatocytes (AH-iPSC). We chose the hepatocyte lineage as a reliable experimental system with well described structural and molecular changes along the hepatocytic differentiation, and availability of cell isolation methods [20–22]. Furthermore, in the past, hepatocytes have been successfully generated from both iPSC and ESC [23–25], and are considered to be an attractive tool for treating end-stage liver diseases hampered by a shortage of donor organs for transplantation and the difficulties in cryopreservation and long term culture of mature hepatocytes [26,27]. Using a comparative analysis of transcriptional profiles of the resulting iPSC and mouse embryonic stem cells (mESCs), we identified the hepatoblast lineage stage-specific donor memory genes which may be functionally relevant during induced re-differentiation towards hepatocytes.
MATERIALS AND METHODS
Isolation of Hepatoblasts, Hepatocytes and Mouse Embryonic Fibroblasts (MEF)
Hepatoblasts (HB), adult hepatocytes (AH) and mouse embryonic fibroblasts (MEF) were isolated from C57BL/6 mice (Jackson Laboratories, Rochester, New York). HB isolated from a pool of fetal (E16.5) livers were purified using MACS system and hepatoblast-specific E-cadherin antibody (Miltenyi, Auburn, California) as described previously [22]. In brief, cells were first blocked with 5% normal goat serum, incubated with a rat anti-mouse ECCD-2 antibody (Clontech) for 15 minutes at 4°C in PBS with 1% bovine serum albumin (BSA) followed by washing and incubation with goat anti-rat IgG microbeads (Miltenyi, Auburn, California). Hepatoblasts were plated on collagen I coated plates (BD) in DMEM supplemented with 10% FBS. After 4 h attachment, a medium was replaced with a fresh DMEM containing 50% DMEM conditioned by fetal liver cells cultured without the separation of hepatocytes and nonparenchymal cells (1:1). Adult hepatocytes were isolated from 3-month-old male mice by a two step collagenase perfusion method as reported previously [28] and seeded at 1.5×103 cells/cm2 in hepatocyte growth medium supplemented with 10% FBS. Mouse embryonic fibroblasts were isolated by a standard protocol as reported [29].
KOSM Lentiviral Generation and Establishment of iPSC Cell Lines from Mouse Hepatic Cells and MEF
The 293T cells were cultured in 100-mm dish at 75% confluence and transfected with a mixture of DNA containing 4 µg of pLentG-KOSM, 3.5 µg of pCMV-VSVG, and 2 µg of psPAX2 (Addgene) by Fugene HD (Roche), according to the manufacturer’s instruction. Twenty-four hours after transfection, the supernatant of transfected cells was collected and filtered through a 0.45 µm pore-size filter. The filtered lentiviral particles were concentrated by ultracentrifugation. For virus infection, mouse embryonic fibroblast (MEF) and mouse hepatic lineage cells were seeded in a 6-well plate at 1 × 105 cells per well one day before transduction. The medium was replaced with virus-containing supernatant, incubated for 2–3 hr and then cultured up to 7days with fresh media. For iPSC induction, the infected cells were trypsinized at day 7, plated in 1:5 ratio into six well plates containing feeder cells and incubated until appearance of ES-like cells. The mESC medium was changed every day.
Culture of Mouse Embryonic Stem Cells (mESCs) and iPSC Cells on Feeder Cells
mES cells (ASE) and iPSC cells were maintained as undifferentiated cells on inactivated mouse embryonic fibroblast (MEF) feeder cells as described previously [25].
Immunofluorescence and Alkaline Phosphatase Staining
For immunofluorescence staining, cells were fixed in 4% paraformaldehyde in PBS for 30 min at room temperature. After washing with PBS, cells were incubated with PBS containing 1% BSA and 0.1% Triton X-100 for one hour at room temperature, followed by incubation with primary antibodies , including OCT3/4 (1:50, Santa Cruz), Sox2 (1:100, Cell signaling), Nanog (1:200, Cosmo Bio), SSEA-1 (1:50, Santa Cruz), HNF4alpha (1:50, Santa Cruz), AFP (1:500, Dako) and albumin (1:100, Bethyl Laboratories). Texas Red-conjugated goat anti-mouse IgG (1: 100, Invitrogen), FITC-conjugated goat anti-rabbit IgG (1:100, Invitrogen) or Alexa Fluor 647 donkey anti-goat IgG (1:500, Invitrogen) were used as secondary antibodies. Alkaline phosphatase staining was performed by using the alkaline phosphatase detection kit (Millipore) according to the manufacturer’s instruction.
RNA Preparation and Real time Quantitative PCR (RT-qPCR)
Total RNA was prepared from cells as indicated using RNase mini kit (Quagen). A total of 1 µg of RNA was reverse transcribed using SuperScript First-Strand cDNA synthesis kit (Invitrogen, Carlsbad, CA). Quantitative PCR was performed using the SYBR Green PCR Core Reagents kit (Applied Biosystems, Foster City, CA). All reactions were performed in triplicate. mRNA expression levels were normalized to endogenous GAPDH and expressed relative to control cells. Primers and optimal annealing temperatures are listed in Supporting Information Table S1.
Karyotyping of iPSC Cell Lines
Karyotyping of HB-iPSC (passage 4), AH-iPSC (passage 3), and MEF-iPSC (passage 4) was performed by Sandra S. Burkett, Cytogenetic Core Facility, MCGP, Center for Cancer Research, National Cancer Institute at Frederick, Frederick, Maryland, USA.
Teratoma Formation
The iPSC cells were collected by trypsinization, resuspended in PBS, and subcutaneously inoculated into nude mice (3×106). Tumors were dissected 4–5 weeks after transplantation, embedded in paraffin and stained with hematoxylin-eosin (HE) for routine histology.
Microarray Analysis
A total of 200 ng RNA from three to four independent biological replicates of MACS-sorted mESCs and iPSCs were linearly amplified according to manufactures’ specification (Ambion, Austin, Texas). For in vitro transcription, reactions were incubated for 16 h at 37°C. The efficiency of the single round amplification was measured by NanoDrop (ND1000, Thermo Scientific). Hybridization, washing, detection (Cy3-streptavidin, Amersham Biosciences, GE Healthcare), and scanning were performed on an illumina iScan system (Illumina) using reagents and following protocols supplied by the manufacturer. The biotinylated cRNA (750 ng per sample) was hybridized on Sentrix beadchips human Ref-8v3 for 18 h at 58°C while rocking (5 rpm). The beadchip covers ~ 24,000 RefSeq transcripts. Image analysis and data extraction were performed automatically using illumina GenomeScan Software. Gene expression values were adjusted by subtracting a background noise in each spot by GenomeStudio (illumina ®), and normalized by quantile normalization method across all samples. Signal intensity with a detection P > 0.05 was treated as a missing value, and only genes with sufficient representation across the samples were included in further data analysis (presence in ≥2 replicates/group). Differentially expressed genes were identified by the Bootstrap ANOVA and Contrast test with 10,000 repetitions. Genes with a Bootstrap P-value ≤ 0.05 were considered significantly different. Ingenuity Pathway Analysis tool (Ingenuity Systems Inc.) was used to explore the functional relationships among the differentially expressed genes. The significance of each network, function and pathway was determined by a scoring system provided by Ingenuity Pathway Analysis tool. All microarray data were submitted to Gene Expression Omnibus database with the accession number of GSE33110.
In vitro Differentiation of mESC and iPSC into Hepatocytes
The protocol for differentiation of mES and iPS cells into hepatocytes was as described by Gillian M. Morrison et al. [30] and Karim Si-Tayeb et al. [24] with minor modifications. Briefly, 1×104 cells were seeded on collagen I coated six-well plates and cultured in media containing 6-bromoindirubin-3-oxime (BIO), GSK-3 specific inhibitor, as a feeder cell-free culture [31] for 4 days to maintain undifferentiated state. Before starting differentiation, cells were cultured in media without LIF and Bio for 1 day to remove the effect of BIO. The RPMI 1640 differentiation media contained B27 (2%, GIBCO), glutamine (1%, Invitrogen) and penicillin/streptomycin (1%, GIBCO) supplemented with activin A and BMP4 for 2 days. After that, cells were cultured in RPMI 1640 media containing ITS (2%, GIBCO), glutamine (1%, Invitrogen) and penicillin/streptomycin (1%, GIBCO) supplemented with activin A and bFGF for 4 days for induction of endoderm differentiation. To induce hepatocytic differentiation, mES and iPSC cells were continuously cultured in hepatocyte differentiation media [25] containing sequentially BMP4 and aFGF for 5days, HGF for 5days and OSM/Dex for 6–9 days. The supernatants were collected at day 22 or 25 for determination of albumin secretion using mouse albumin ELISA Kit (Bethyl Laboratories).
RESULTS
Generation of Lineage Stage-Specific iPS Cells
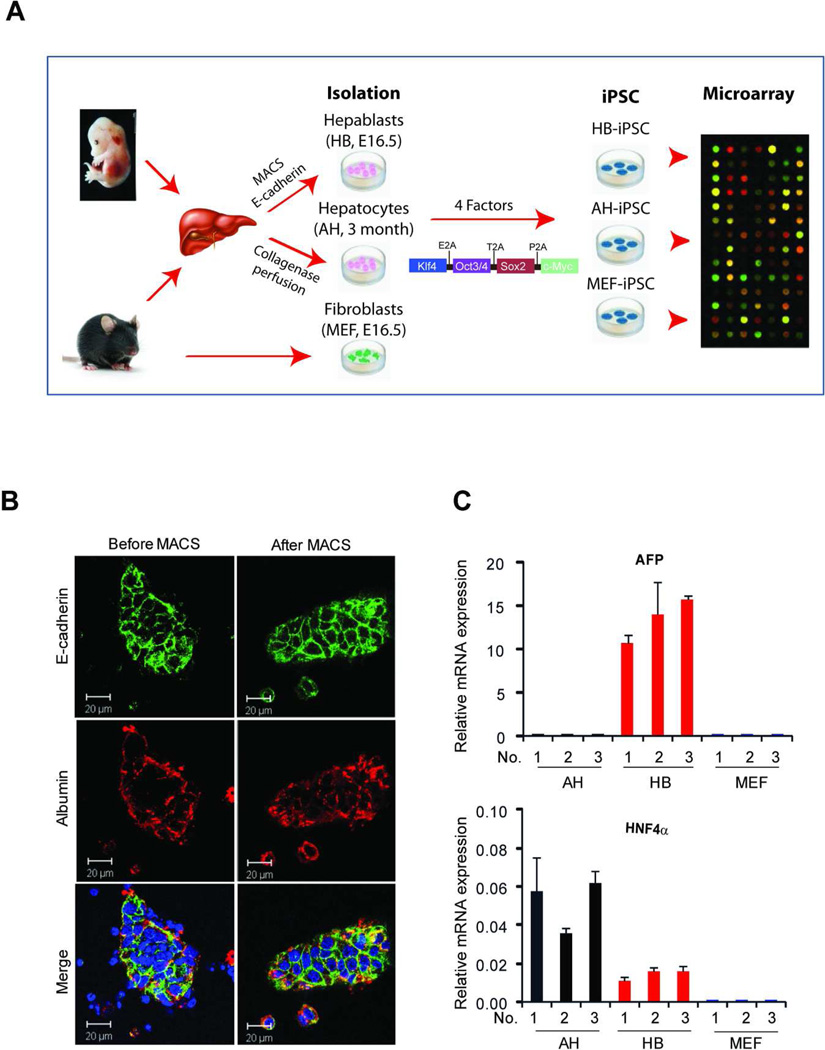
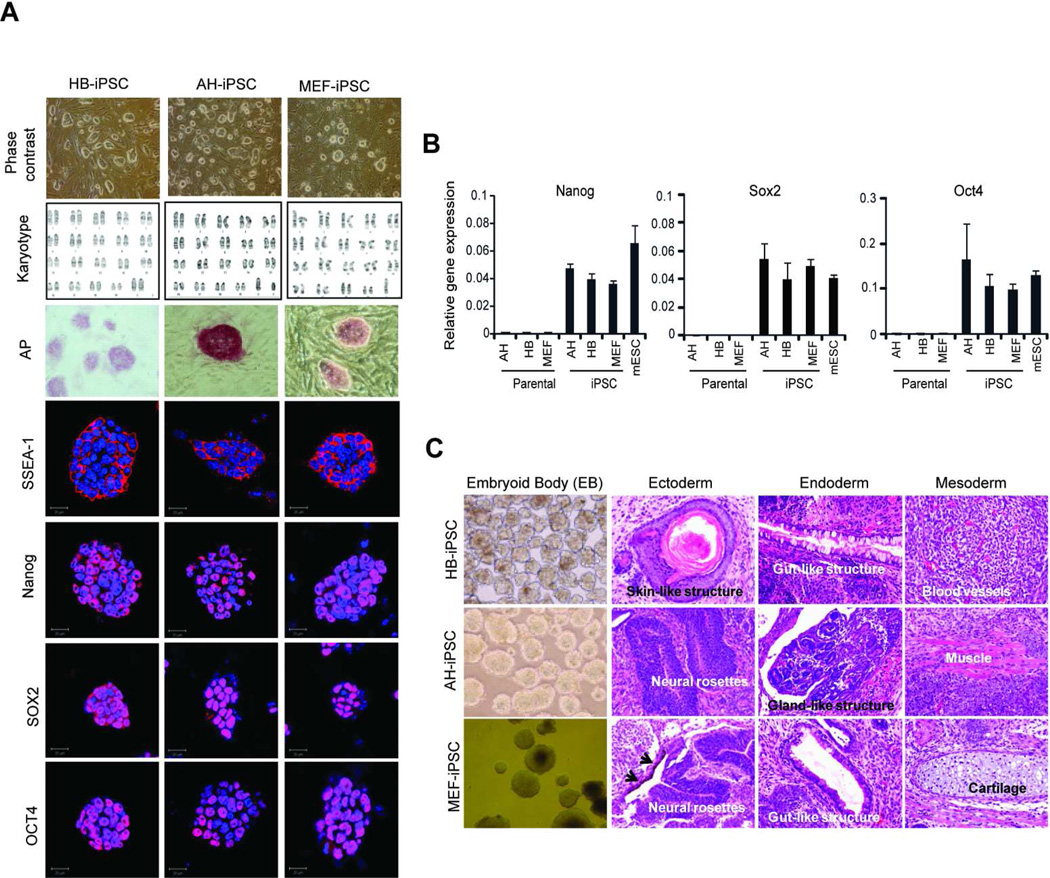
Our experimental approach was as follows: isolation of hepatic lineage cells at different stages of differentiation, followed by generation of iPSC and microarray analysis (Fig. 1A). The hepatoblasts (HB) were isolated from E16.5 fetal livers and purified by MACS sorting using anti-E-cadherin, a cell specific surface marker of immature hepatocytes [22]. Fully differentiated adult hepatocytes (AH) were isolated from 3 month-old mice by a two-step collagenase perfusion technique followed by Percoll purification [28]. The vast majority (> 99%) of the isolated HB and AH expressed E-cadherin and albumin, another specific marker of hepatic lineage cells, respectively (Fig. 1B, and not shown). Reflecting a high purity, only isolated HB expressed AFP [32], an early marker of hepatic differentiation, as measured by a highly sensitive RT-qPCR analysis, whereas adult hepatocytes exhibited the highest levels of liver specific transcription factors HNF4α [33], essential for hepatocytic differentiation (Fig. 1C). To generate iPSC, hepatoblasts, adult hepatocytes and mouse embryonic fibroblasts (MEF) were transduced with lentiviral vector pLentG-KOSM, a single polycistronic vector containing a fusion of 4 genes (Oct4, Sox2, Klf4, and c-Myc) and GFP reporter gene for easy monitoring the expression of the reprogramming factors [34]. One of the critical characteristics of complete reprogramming into iPSC is the acquisition of exogenous factor independence required for maintaining iPSC pluripotency during reprogramming [35]. In agreement with these reports, we found the ES-like colonies which lost GFP expression from passage 1 (Supporting Information Fig. S1). We further characterized the established iPSC clones by AP staining, expression of SSEA1, a surface marker of mouse ES, and karyotyping (Fig. 2A). Only clones with typical morphology of mouse ESC (a round shape, large nucleoli, and scant cytoplasm), normal karyotype and uniform expression of the major pluripotent markers, such as Oct4, Sox2 and Nanog, as judged by immunofluroescence and RT-qPCR, were selected for further analysis (Fig. 2A and B). The pluripotency of the selected iPSC was confirmed by embryonic body (EB) formation in vitro and development of teratoma upon subcutaneous transplantation into immunodeficient mice (Fig. 2C).
Figure 1. Schematic representation of the experimental design.
(A) Hepatoblasts (HB, E16.5); adult hepatocytes (AH) and mouse embryonal fibroblasts (MEF, E16.5) were isolated from 3-month old C57/B6 mice. iPSCs were generated by a transfection with a pLentG-KOSM vector containing a fusion of 4 genes, including Klf4, Oct3/4, Sox2 and c-Myc, and GFP, followed by global gene expression analysis. (B) Immunostaining with anti-E-cadherin before and after MACS sorting demonstrates a high purity of hepatoblasts used for HB-iPSC generation. Nuclei were counterstained with DAPI. Scale bar, 20 µm (C) Quantitative reverse transcription polymerase chain reaction analysis with primers specific to hepatic lineage cells, AFP and HNF4α, and GAPDH as a control. Only hepatoblasts expressed AFP, an early marker of hepatic lineage cells, whereas adult hepatocytes expressed higher levels of HNF4α, a marker of differentiated hepatocytes. The data are presented as mean expression levels ± SD relative to GAPDH. All experiments were performed in duplicate using 3 independent cell isolations. iPSC, induced pluripotent stem cells; HB-iPSC, hepatoblast-derived iPSC; AH-iPSC, adult hepatocyte-derived iPSC; MEF-iPSC, mouse embryonic fibroblast-derived iPSC.
Figure 2. Charaterization of iPSCs generated from hepatic lineage cells at different stages of differentiation.
(A) The morphology, karyotyping, alkaline phosphatase staining and immunostaining with antibodies against the pluripotency markers, including SSEA1, Sox2, Oct4 and Nanog in HB-iPSC, AH-iPSC and MEF-iPSC. Nuclei were counterstained with DAPI. Scale bar, 20 µm. (B) Quantitative reverse transcription polymerase chain reaction analysis with primers specific to endogenous Nanog, Sox2 and Oct4. The data are presented as mean ± SD relative to GAPDH of duplicate experiments. (C) Evidence of multi-lineage differentiation. Formation of embryoid body (BD) in vitro (left images) and teratomas in vivo (three images on the right) by HB-iPSC, AH-iPSC and MEF-iPSC. Hematoxylin and eosin staining of teratoma sections show spontaneous differentiation into all three germ layers, including ectoderm, entoderm and mesoderm. Black arrows point to pigmented epithelia. EB images, ×100 magnifications, H&E images, ×200 magnifications. iPSC, induced pluripotent stem cell; HB-iPSC, hepatoblast-derived iPSC; AH-iPSC, adult hepatocyte-derived iPSC; MEF-iPSC, mouse embryonic fibroblast-derived iPSC.
HB-iPSCs Exhibit Higher Capacity for Hepatic Differentiation than AH-iPSCs
The iPSCs derived from different cell types can differentiate more effectively into the parental rather than other cell types [16–19]. Therefore, we asked whether a lineage stage-specific differentiation state of the donor cell may be a contributing factor to the re-differentiation potency of iPSC. To address this question, we first modified the differentiation protocols developed for the ES cells [24,30] to exclude the requirement for feeder cells and embryonic body formation (Supporting Information Fig. S2A). We confirmed that the feeder-free monolayer culture of ES did not affect either the pluripotency or capacity to differentiate towards hepatocytes (Supporting Information Fig. S2B-D). In fact, hepatic differentiation in monolayer was more efficient than that driven by EB (Supporting Information Fig. S2E). Using this system and a sequential application of various combinations of growth factors and cytokines reported to be essential for targeted hepatocyte differentiation, we have shown that ES cells grown as feeder-free monolayer cultures for 4 passages gave rise to hepatocyte-like cells as judged by morphology and expression of hepatocytic specific markers, including AFP, ALB, HNF4α and G6P (Supporting Information Fig. S2).
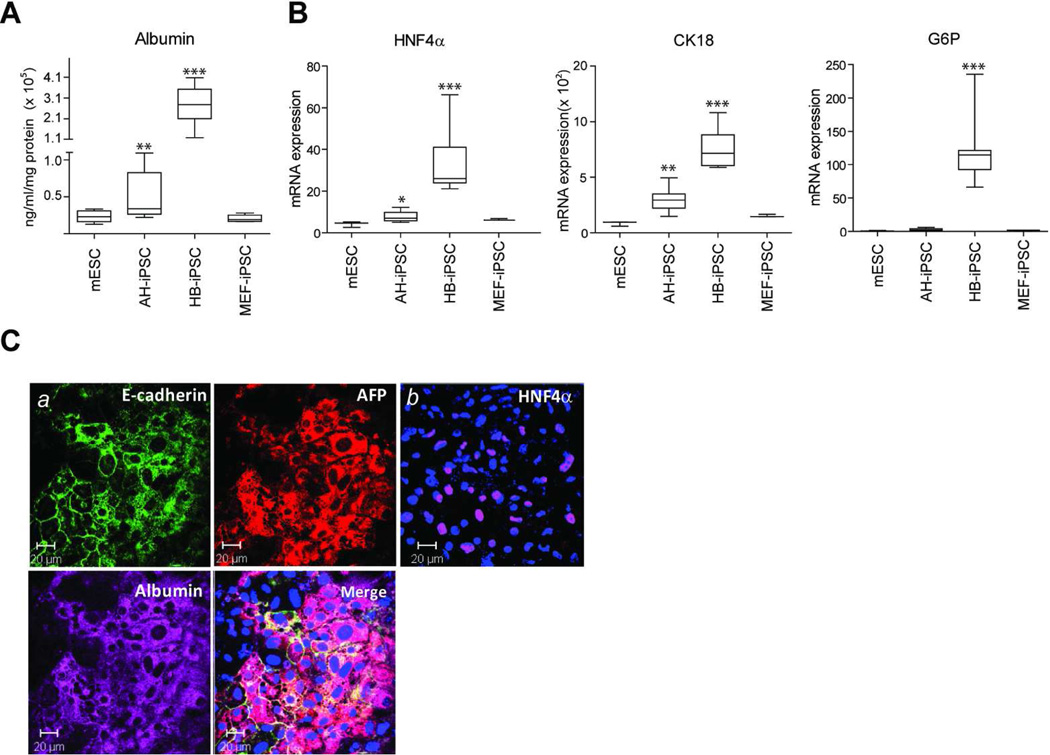
All reprogrammed cell lines were then subjected to hepatocye differentiation protocol in feeder- and serum-free conditions, and secretion of albumin into culture medium was determined as a quantitative measure of hepatocytic differentiation efficiency. The media was collected at 25 days, the peak time of hepatic differentiation as estimated by morphological criteria. As expected, iPSCs generated from early (HB-iPSC) and terminal (AH-iPSC) stages of hepatic differentiation produced more albumin than either mESCs or MEFs (Fig. 3A). The albumin secretion reached maximum values in HB-iPSC which were more than 5-fold higher as compared to the similarly treated AH-iPSC cultures. Consistent with the mouse albumin ELISA data, the HB-iPSCs also showed a considerably stronger induction of HNF4α and G6P, other known markers of hepatic differentiation as shown by real time PCR analysis (Fig. 3B) and immunoflourescence staining (Fig. 3C). Thus, the HB-iPSCs derived from the early progenitor-type cells displayed higher hepatocyte-forming potential than iPSCs derived from adult hepatocytes indicating that the differentiation potency of iPSCs may depend on the lineage stage-specific differentiation state of original cells.
Figure 3. HB-iPSCs have a greater potential to differentiate into hepatocyte-like cells in vitro.
mESC, HB-iPSC, AH-iPSC and MEF-iPSC were subjected to a hepatocyte differentiation protocol in serum- and feeder layer-free conditions as described in Material and Methods section. (A) Albumin secretion at day 25. Albumin protein levels detected in culture medium and normalized to total protein are shown in the box and whisker plots of duplicate measurements from three independent experiments for each clone (n=6 in mESC and MEF-iPSC from one clone, n=18 in AD-iPSC and HB-iPSC from 3 clones). Boxes represent upper and lower quartiles, lines within boxes represent median, and the error bars comprise the whiskers which extend to the maximum and minimum value data sets. **, P < 0.01; ***, P < 0.001; Bootstrap T-test with 10,000 random replications as compared with mESC. (B) Quantitative reverse transcription polymerase chain reaction analysis with primers specific to HNF4α, G6P, and CK18 at day 25. mRNA expression values relative to GAPDH are shown in the box and whisker plots of triplicate measurements for each clone (n=3 in mESC and MEF-iPSC from one clone, n=9 in AD-iPSC and HB-iPSC from 3 clones). Boxes represent upper and lower quartiles, lines within boxes represent median, and the error bars comprise the whiskers which extend to the maximum and minimum value data sets. *, P < 0.05; **, P < 0.01; ***, P < 0.001; Bootstrap T-test with 10,000 random replications as compared to mESC. (C) Triple immunofluorescence staining with antibodies against E-cadherin, AFP, and Albumin (a) and immunofluorescence staining with anti-HNF4α (b). Representative images of HB-iPSC cultures at day 25 are shown. Nuclei were counterstained with DAPI. Scale bar, 20 µm. iPS, induced pluripotent stem cells; HB-iPSC, hepatoblast-derived iPSC; AH-iPSC, adult hepatocyte-derived iPSC; MEF-iPSC, mouse embryonic fibroblast-derived iPSC.
Transcriptional Profiling of mES, iPS and Parental Cells
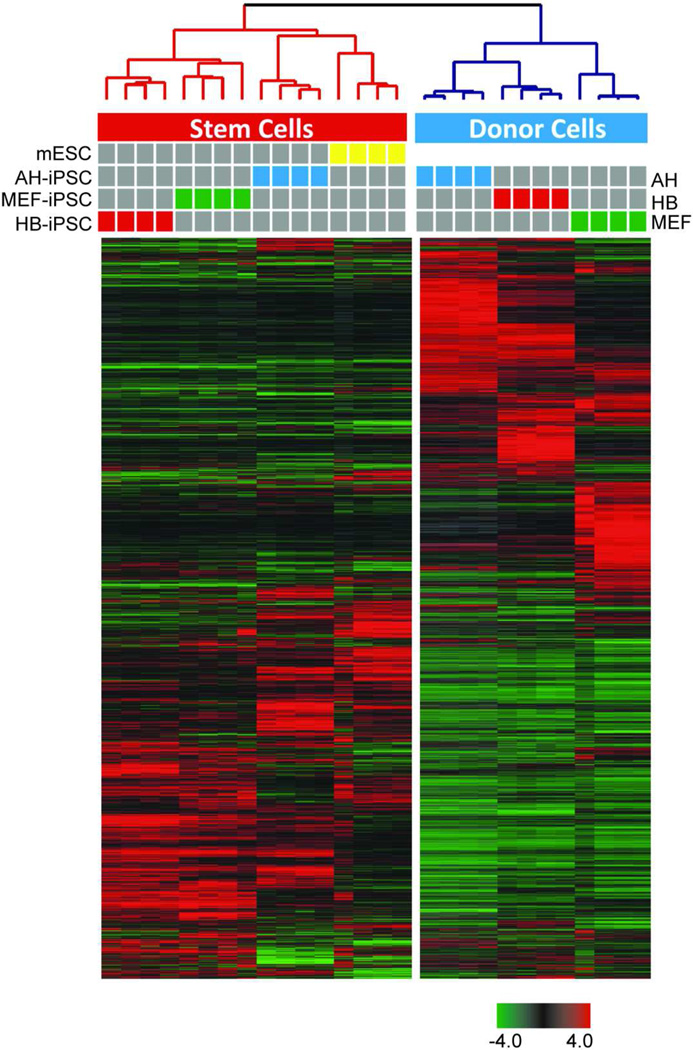
To gain a better understanding of the genomic traits of the HB-iPSC that may support the enhanced capacity for hepatocytic lineage differentiation, we performed a global gene expression analysis of parental cells (AH, HB and MEF) and the corresponding iPSCs as well as mESCs. Only low-passage iPSCs (passage 4) were used which are most likely to retain the donor memory [16–18]. To achieve a high purity, mESCs and various iPSCs were separated from the feeder cells using SSEA1-based MACS sorting (Supporting Information Fig. S3). The transcriptome profiling was performed in four replicate experiments using illumina microarrays (Fig. 4). An unsupervised hierarchical clustering analysis based on the similarity in the expression pattern of all genes in all samples showed a clear separation between the donor and stem (mES and iPS) cells. iPSCs clustered together with mESCs, indicating that reprogrammed iPSCs were closer to mES than to the parental cells (Fig. 4). However, each individual group of iPSCs (HB-iPSC, AH-iPSC and MEF-iPSC) clustered tightly together indicating that they had a unique gene expression signature (Fig. 4).
Figure 4. Transcriptional profiling of mESC, iPSC and donor cells.
mESCs and early passage iPSCs (p.4) were separated from the feeder cells using SSEA1-based MACS sorting to achieve a high purity of cells used for microarray analysis. Average linkage unsupervised hierarchical clustering based on correlation coefficient of mESC, iPSCs and donor cells, including adult hepatocytes (AH), hepatoblasts (HB) and mouse embryonic fibroblasts (MEF). The dendrogram demonstrates a clear separation of Stem Cells (red) and Donor cells (blue) into two clusters. The tight cluster of Stem Cells (red) confirms that the donor cells have been successfully reprogrammed. Each cell in the matrix represents the expression level of a gene feature in an individual sample. Expression values are adjusted to the median value across all samples and genes (red and green indicate high and low expression relative to median expression). Colored bars between dendrogram and heat-map represent samples as indicated. iPS, induced pluripotent stem cells; HB-iPSC, hepatoblast-derived iPSC; AH-iPSC, adult hepatocyte-derived iPSC; MEF-iPSC, mouse embryonic fibroblast-derived iPSC.
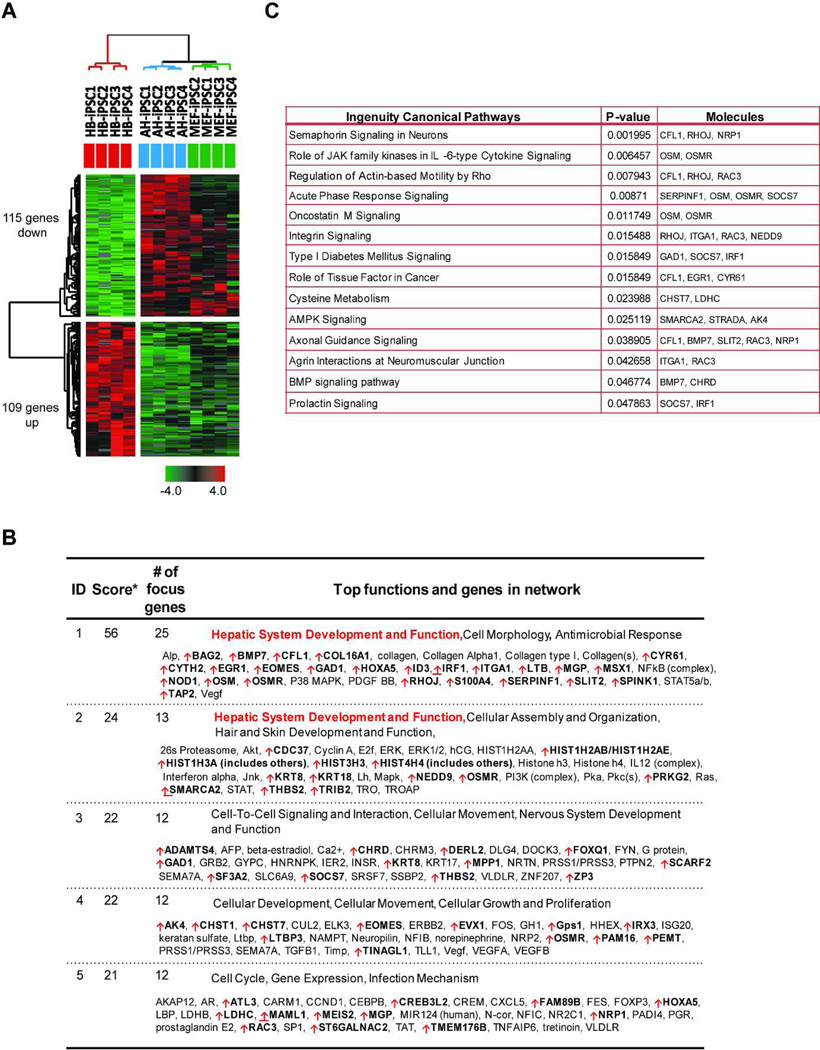
To identify genes related to the superior capacity of HB-iPSC for hepatic re-differentiation, we next analyzed genes which were differentially expressed in HB-iPSC as compared to AH-iPSC and MEF-iPSC by using Bootstrap ANOVA and contrast tests with 10,000 replications. In total, we identified 109 up- and 115 down-regulated genes using 1.5-fold expression changes and P ≤ 0.05 as selection criteria (Fig. 5A, Supporting Information Table S2). We also investigated functional networks that might be associated with hepatic differentiation. Ingenuity Pathway Analysis of HB-iPSC gene expression signature revealed that the 109 upregulated genes were significantly related to hepatic system development and function while 115 down-regulated genes were linked to embryonic, organism and tissue development (Fig. 5B, and data not shown). Among the top upregulated canonical pathways with high scores were oncostatin M and BMP signaling, known to be involved in hepatocyte maturation during liver development [36,37] (Fig. 5C). The HB-iPSC specific genes seem to be capable of facilitating rather than driving hepatic differentiation since early passage HB-iPSC maintained the pluripotency state and self-renewal under neutral conditions (Fig. 2). This notion is supported by the observation that mice deficient in oncostatin M or BMP signaling are characterized by normal or delayed liver development [38,39]. Furthermore, a comparison of the differentially expressed genes between mESCs and HB-iPSCs showed that the up-regulated HB-iPSC specific genes were expressed at low levels in mESCs.
Figure 5. Gene network analysis of HB-iPSC gene signature.
(A) Unsupervised hierarchical clustering of HB-iPSC, AH-iPSC, and MEF-iPSC: The gene expression signature of 224 differentially expressed genes (109 up and 115 down) from HB-iPSC identified by Bootstrap ANOVA and contrast test with 10,000 random replications (n=4, P ≤ 0.05 and fold changes ≥ 1.5) (B) Ingenuity Pathway Analysis of 109 up-regulated genes in HB-iPSC. The top 5 gene networks include hepatic system development and function. *, The score represents the likelihood that the focus genes within the network are found therein by random chance. Upregulated genes are marked with red arrow. (C) Canonical pathways associated with 109 upregulated genes in HB-iPSC as defined by Ingenuity Pathway Analysis tools (P ≤ 0.05, Fisher’s exact test). iPS, induced pluripotent stem cells; HB-iPSC, hepatoblast-derived iPSC; AH-iPSC, adult hepatocyte-derived iPSC; MEF-iPSC, mouse embryonic fibroblast-derived iPSC.
HB-iPSC Retain the Hepatoblast-Specific Donor Memory Genes
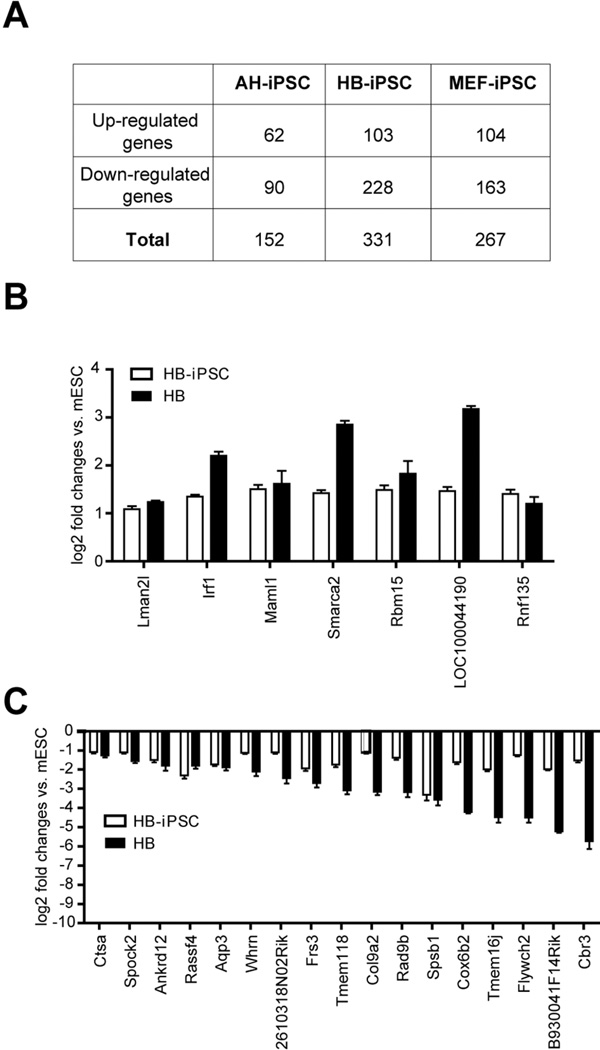
Since the gene expression signature of iPSC may represent a transcriptional donor memory [18,40,41], we searched for the donor memory genes in each iPSC group. For this we have adopted a strategy described by Zhumur Ghosh et al. [41] which is based on the sequential comparisons of the commonly differentially expressed genes (either upregulated or downregulated, P≤0.05, at least 2 fold expression changes) between parental (donor) cells and mESCs, defined as gene set A, and between iPSCs and mESCs, defined as gene set B. This strategy allowed for identifying transcriptional donor memory genes in each category of iPSCs (Fig. 6A). Noteworthy, among these genes, we did not find those encoding liver-enriched transcription factors (e.g. HNFα) or hepatocyte specific proteins (e.g. AFP and albumin) supporting a proposed suppression of master transcriptional regulators during cellular reprogramming [18]. By employing Venn diagram comparisons of donor memory genes in HB-iPSC, AH-iPSC, and MEF-iPSC, we then identified 204 genes (62 up- and 142 down-regulated genes) as donor memory genes unique for HB-iPSC (Supporting Information Fig.S4 and Table S3). Since donor memory genes are thought to facilitate the efficiency of re-differentiation to the original cells [16,17,19], we sought to compare the commonly dysregulated genes in the HB-iPSC gene signature (Supporting Information Table S2) and the unique donor memory genes in HB-iPSC (Supporting Information Table S3). This strategy permitted identification of the donor memory genes in HB-iPSC retained in HB-iPSC gene signature (herein referred to as HB-iPSC specific donor memory genes), which may support a greater hepatocytic-differentiation potential of HB-iPSCs as compared to AD-iPSCs or MEF-iPSCs. In total, 24 donor memory genes (including 7 up-regulated genes and 17 down-regulated genes) were identified in HB-iPSC (Fig. 6B and C). Although some of these genes were also expressed at similar levels in other parental cells, such as AD and MEF (Supporting Information Fig. S6A and C), only HB-iPSC retained their stable expression pattern consistent with a cell-specific donor memory preserved in the HBbut not AD- or MEF-derived iPSCs (Supporting Information Fig. 6B and D). This result is an agreement with a previous report in human iPSC [18]. It has been suggested that incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells and may be explained by the differences in DNA methylation and/or chromatin modifications during cellular reprogramming.
Figure 6. HB-iPSC specific donor memory.
(A) The number of donor memory genes in AH-iPSC, HB-iPSC, and MEF-iPSC. The donor memory genes, as described by Zhumur Ghosh et al. [41], are identified based on the sequential comparisons of the commonly differentially expressed genes (P ≤ 0.05, at least 2 fold expression changes) between parental (donor) and mES cells and between iPS and mES cells (B, C) 7 up- and 17 down-regulated HB-iPSC specific donor memory genes. The data are shown as means of log2 fold changes relative to mES cells ± SEM (n=4). iPS, induced pluripotent stem cells; HB, hepatoblast; HB-iPSC, hepatoblast-derived iPSC; AH-iPSC, adult hepatocyte-derived iPSC; MEF-iPSC, mouse embryonic fibroblast-derived iPSC.
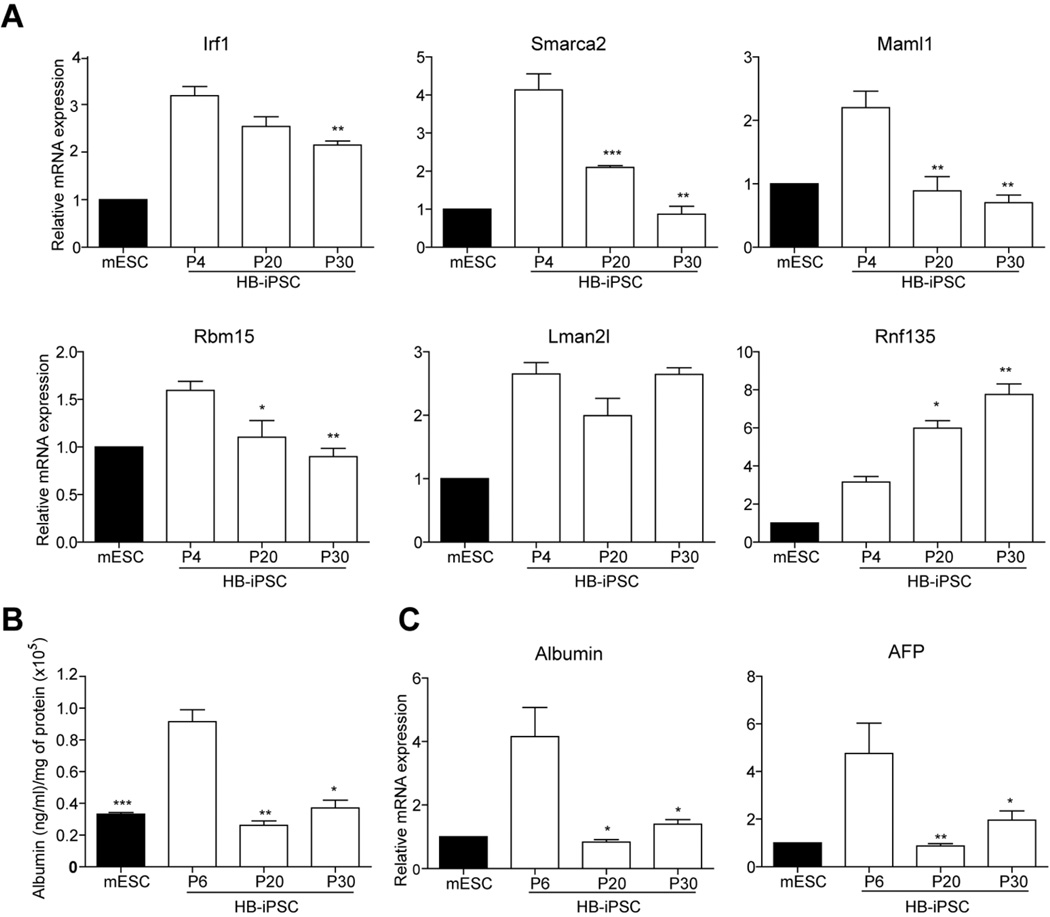
We next assessed if this small number of donor memory genes is functionally relevant. To do so, we correlated the persistence of the donor cell gene expression with the capacity of HB-iPSCs to undergo directed differentiation into hepatocytes upon extended passaging. RT-qPCR confirmed up-regulation of the HB-iPSC specific donor memory in early passage HB-iPSC (p4) as compared to the mESCs (Fig. 7A). However, in late passage HB-iPSCs (p20 and p30), the expression levels of four out of six annotated up-regulated genes, including Smarca2 and Maml1 which belong to the top functional networks involved in hepatic system development and function as well as regulation of cell cycle and gene expression, were gradually decreasing albeit with a different kinetics (Fig. 7A). The transcript levels of Lmna2l did not change whereas Rnf135 levels continued to increase. Furthermore, HB-iPSC at passages 20 and 30 lost the advantage of the early passage HB-iPSC for hepatocytic differentiation (Fig.7B and C). Together these findings suggest that the HB-iPSC specific donor memory could contribute to facilitating hepatic differentiation.
Figure 7. The reduced capacity of HB-iPSCs to undergo directed differentiation into hepatocytes upon extended passaging.
(A) Quantitative reverse transcription polymerase chain reaction analysis with primers specific to the HB-iPSC specific donor memory , including Irf1, Smarca2, Maml1, Rbm15, Lman2l, Rnf135. The mRNA expression values were normalized to GAPDH and shown as means ± SEM of triplicate measurements expressed relative to mESC. (n=3, Bootstrap T-test with 10,000 random replications. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 as compared to HB-iPSC at passage 4. (B) Albumin secretion in culture medium at day 25 during in vitro hepatic differentiation of mESC and HB-iPSC at indicated passages. Albumin protein was detected in culture medium using enzyme-linked immunosorbent assay (ELISA) and normalized to total protein. The data are shown as means ± SEM of duplicate measurements from two independent experiments (n=4, Bootstrap T-test with 10,000 random replications, *, P < 0.05; **, P < 0.01; and ***, P < 0.001, as compared to HB-iPSC at passage 6). (C) Quantitative reverse transcription polymerase chain reaction analysis with primers specific to hepatic specific markers Albumin and AFP at day 25. The data are shown as means ± SEM of triplicate measurements normalized to GAPDH and expressed relative to the mESC levels. (n=3, Bootstrap T-test with 10,000 random replications, *P < 0.05; and **P < 0.01 as compared to HB-iPSC at passage 6. HB-iPSC, hepatoblast-derived iPSC. iPS, induced pluripotent stem cells; P, passage.
DISCUSSION
Current research suggests that cellular origin may affect in vitro differentiation capacity of iPSCs [16,17,19]. Here, we have extended these studies to demonstrate that differentiation potential of iPSCs may depend on the lineage stage-specific differentiation state of donor cells. Using hepatic lineage as a well defined experimental model, we have generated iPSC clones from cells at early (HB-iPSC) and end (AH-iPSC) stages of hepatocyte differentiation through lentiviral expression of Oct4, Sox2, Nanog and c-Myc. All reprogrammed cells expressed known pluripotency markers, formed EB in vitro, gave rise to the cells from three embryonic germ layers in vivo and maintained a typical ESC-like morphology for 30 passages demonstrating their complete reprogramming (Fig.2, Fig. S1). Nevertheless, despite the global similarities with ES cell transcriptome (Fig. 4), the iPSCs generated from early hepatic progeny cells (HB-iPSCs) retained a transient transcriptional memory characteristic of donor cells as previously reported [9–11,16–18,40–42] and displayed a greater hepatocyte-forming potential in vitro as compared to either mESCs or iPSCs obtained from terminally differentiated adult hepatocytes and MEF (Fig. 3).
Consistent with being more proficient in generating albumin-producing hepatocyte-like cells, HB-iPSCs also exhibited a unique gene expression signature which clearly differentiated them from both AH-iPSC and MEF-iPSC (Fig.5). The stem cells used for microarray analysis were separated from the contaminating feeder cells by MACS sorting using a surface marker of pluripotent cells SSEA1 mES and iPS cells (Fig. S3). This approach yielded on average 93–97% enrichment of stem cells and allowed a high reproducibility of gene expression data (Fig.4). Although no differences in the expression of lineage-specific transcription factors were found, analysis of the gene expression networks in iPSCs reprogrammed from hepatoblasts revealed a significant association of HB-iPSC gene expression signature with hepatic system development and function (Fig.5). Among the top upregulated canonical pathways were oncostatin M and BMP signaling controlling hepatocyte maturation [36] and induction of liver budding [37] during liver development. These results suggest that the gene expression differences between HB-iPSC and AD-iPSC may be responsible for the variability in their differentiation potency as compared to both ESCs and other iPSCs.
This notion was further supported by identification of donor memory genes (Fig.S4) postulated to predispose iPSC to differentiate more effectively into original cells [16,17,19,42]. Using a strategy first described by Ghosh and colleagues [41], we compared the gene expression profiles of the generated iPSCs and donor cells with respect to mESC. These analyses allowed categorizing 24 genes as the HB-iPSC specific donor memory including seven upregulated genes (Fig. 6B & C and Supporting Information Fig. S6A-D). Among the genes with annotated functions, was Smarca2, SWI/SNF related matrix associated regulator of chromatin also known as BRM [43]. The Smarca2 plays an important role in enhancing expression of albumin [44] and hepatic differentiation [45] during liver development as well as in germ-layer formation in mESC in vivo [46]. Noteworthy, two of the upregulated genes (i.e.Maml1 and Rbml5) are implicated in cell fate decisions via modulation of Notch signaling. Thus, Maml1, a mastermind-like 1 transcriptional co-activator of Notch signaling [47], has been shown to prevent the development of erythroid/megakaryocytic cells [48,49] as well as pancreatic acinar cells [50], whereas enforced expression of Rbm15 has been found to inhibit myeloid differentiation [51]. This gene set also contained Irf1, a transcriptional regulator of type 1 interferons and interferoninducible genes required for DNA damage-induced growth arrest and apoptosis [52]; Rnf135, a widely expressed gene containing a RING finger domain at the N terminus and a B30.2/SPRY domain at the C terminus implicated in diverse functions [53] as well as Lman2l and LOC100044190 with little known or unidentified functions.
The expression of 4/6 upregulated genes, including Smarca2, Maml2l, Irf1, and Rbm15, was progressively decreasing to the levels similar to the ES cell state upon extended passaging of HB-iPS cells (p30) (Fig.7) in agreement with evidence that genes reflecting the cell of origin do not represent a permanent feature of iPSC signature [9–11,16–18,35].This was paralleled by a remarkable reduction in hepatocyte-forming potential of the late passage HB-iPSCs which did not exceed that of ESCs or iPSCs obtained from terminally differentiated adult hepatocytes and MEF (Fig. 3, 7). Although the functional relevance of the HB-iPSC specific donor memory awaits further investigation, these data suggest that they may enhance the efficiency of hepatic differentiation via direct (e.g.Smarca2) or indirect (e.g.Maml1, Rbm15) effects.
The transcriptional differences between iPS and ES cells have be explained by epigenetic memory inherited form parental cells which facilitates differentiation into the donor cells [16]. In support of importance of DNA methylation during reprogramming, DNA methyltransferase (DNMT) 1 was strongly and uniformly upregulated [1,54] across all generated iPS cell lines to the levels of ESCs as compared to the parental cells. Furthermore, all reprogrammed cells showed a significantly higher albeit variable expression of DNMT3b, the de novo methyltransferase induced upon cellular reprogramming [9,55], than found in ESCs and original cells (Fig.S5).
CONCLUSION
Our findings demonstrate that iPS cells generated from progenitor-like hepatoblasts retained a transient transcriptional memory characteristic of donor cells and were more efficient to differentiate into hepatocytes in vitro as compared to either mESCs or iPSCs obtained from terminally differentiated adult hepatocytes and MEF. This is the first example of lineage stage-specific donor memory genes to be associated with facilitating targeted differentiation of iPS cells. The identification of donor memory genes which are functionally relevant for induced differentiation of iPSCs to hepatocytic lineage may help to advance therapeutic application and contribute to a better understanding of liver development. It remains to be explored whether these findings are relevant for iPSC generated from other cell lineages.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Susan Garfiled and Langston Lim for the assistance with confocal imaging and Barbara Taylor for the help with FACS analysis.
Financial Support: This research was supported by the Intramural Research Program of the US National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
Author Contribution
Seung Bum Lee: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing
Daekwan Seo: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing
Dongho Choi: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing
Kye-yoon Park: collection and/or assembly of data
Agnes Holczbauer: collection and/or assembly of data
Jens U. Marquardt: collection and/or assembly of data
Elizabeth A. Conner: administrative support, collection and/or assembly of data
Valentina M. Factor: data analysis and interpretation, manuscript writing
Snorri S. Thorgeirsson: conception and design, financial support, provision of study material or patients, data analysis and interpretation, manuscript writing, final approval of manuscript
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors indicate no potential conflicts of interest.
REFERENCES
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Aoi T, Yae K, Nakagawa M, et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 4.Liu H, Ye Z, Kim Y, et al. Generation of endoderm-derived human induced pluripotent stem cells from primary hepatocytes. Hepatology. 2010;51:1810–1819. doi: 10.1002/hep.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park IH, Arora N, Huo H, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghodsizadeh A, Taei A, Totonchi M, et al. Generation of liver disease-specific induced pluripotent stem cells along with efficient differentiation to functional hepatocyte-like cells. Stem Cell Rev. 2010;6:622–632. doi: 10.1007/s12015-010-9189-3. [DOI] [PubMed] [Google Scholar]
- 8.Marchetto MC, Carromeu C, Acab A, et al. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doi A, Park IH, Wen B, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin MH, Mason MJ, Xie W, et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pick M, Stelzer Y, Bar-Nur O, et al. Clone- and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells. 2009;27:2686–2690. doi: 10.1002/stem.205. [DOI] [PubMed] [Google Scholar]
- 12.Mayshar Y, Ben-David U, Lavon N, et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 13.Lister R, Pelizzola M, Kida YS, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu BY, Weick JP, Yu J, et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci USA. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Q, Lu SJ, Klimanskaya I, et al. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells. 2010;28:704–712. doi: 10.1002/stem.321. [DOI] [PubMed] [Google Scholar]
- 16.Polo JM, Liu S, Figueroa ME, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim K, Doi A, Wen B, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohi Y, Qin H, Hong C, et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13:541–549. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bar-Nur O, Russ HA, Efrat S, et al. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet Beta cells. Cell Stem Cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 20.Zaret KS. Regulatory phases of early liver development: paradigms of organogenesis. Nat Rev Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 21.Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev Cell. 2010;18:175–189. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 22.Nitou M, Sugiyama Y, Ishikawa K, et al. Purification of fetal mouse hepatoblasts by magnetic beads coated with monoclonal anti-E-cadherin antibodies and their in vitro culture. Exp Cell Res. 2002;279:330–343. doi: 10.1006/excr.2002.5615. [DOI] [PubMed] [Google Scholar]
- 23.Gai H, Nguyen DM, Moon YJ, et al. Generation of murine hepatic lineage cells from induced pluripotent stem cells. Differentiation. 2010;79:171–181. doi: 10.1016/j.diff.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Si-Tayeb K, Noto FK, Nagaoka M, et al. Highly efficient generation of human hepatocytelike cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heo J, Factor VM, Uren T, et al. Hepatic precursors derived from murine embryonic stem cells contribute to regeneration of injured liver. Hepatology. 2006;44:1478–1486. doi: 10.1002/hep.21441. [DOI] [PubMed] [Google Scholar]
- 26.Dhawan A, Puppi J, Hughes RD, et al. Human hepatocyte transplantation: current experience and future challenges. Nat Rev Gastroenterol Hepatol. 2010;7:288–298. doi: 10.1038/nrgastro.2010.44. [DOI] [PubMed] [Google Scholar]
- 27.Ochiya T, Yamamoto Y, Banas A. Commitment of stem cells into functional hepatocytes. Differentiation. 2010;79:65–73. doi: 10.1016/j.diff.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 28.Kao CY, Factor VM, Thorgeirsson SS. Reduced growth capacity of hepatocytes from c-myc and c-myc/TGF-alpha transgenic mice in primary culture. Biochem Biophys Res Commun. 1996;222:64–70. doi: 10.1006/bbrc.1996.0698. [DOI] [PubMed] [Google Scholar]
- 29.Conner DA. Mouse embryo fibroblast (MEF) feeder cell preparation. Curr Protoc Mol Biol. 2001 doi: 10.1002/0471142727.mb2302s51. Chapter 23:Unit 23.2. [DOI] [PubMed] [Google Scholar]
- 30.Morrison GM, Oikonomopoulou I, Migueles RP, et al. Anterior definitive endoderm from ESCs reveals a role for FGF signaling. Cell Stem Cell. 2008;3:402–415. doi: 10.1016/j.stem.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 31.Sato N, Meijer L, Skaltsounis L, et al. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 32.Jones EA, Clement-Jones M, James OF, et al. Differences between human and mouse alphafetoprotein expression during early development. J Anat. 2001;198:555–559. doi: 10.1046/j.1469-7580.2001.19850555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 2000;14:464–474. [PMC free article] [PubMed] [Google Scholar]
- 34.Shao L, Feng W, Sun Y, et al. Generation of iPS cells using defined factors linked via the self-cleaving 2A sequences in a single open reading frame. Cell Res. 2009;19:296–306. doi: 10.1038/cr.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamiya A, Kinoshita T, Ito Y, et al. Fetal liver development requires a paracrine action of oncostatin M through the gp130 signal transducer. EMBO J. 1999;18:2127–2136. doi: 10.1093/emboj/18.8.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duncan SA, Watt AJ. BMPs on the road to hepatogenesis. Genes Dev. 2001;15:1879–1884. doi: 10.1101/gad.920601. [DOI] [PubMed] [Google Scholar]
- 38.Hirabayashi Y, Sekiguchi T, et al. Targeted disruption of oncostatin M receptor results in altered hematopoiesis. Blood. 2003;102:3154–3162. doi: 10.1182/blood-2003-02-0367. [DOI] [PubMed] [Google Scholar]
- 39.Rossi JM, Dunn NR, Hogan BL, et al. Distinct mesodermal signals, including BMPs from the septum transversum mesenchyme, are required in combination for hepatogenesis from the endoderm. Genes Dev. 2001;15:1998–2009. doi: 10.1101/gad.904601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchetto MC, Yeo GW, Kainohana O, et al. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS One. 2009;4:e7076. doi: 10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghosh Z, Wilson KD, Wu Y, et al. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS One. 2010;5:e8975. doi: 10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barrero MJ, Izpisua Belmonte JC. iPS cells forgive but do not forget. Nat Cell Biol. 2011;13:523–525. doi: 10.1038/ncb0511-523. [DOI] [PubMed] [Google Scholar]
- 43.Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell. 2003;11:377–389. doi: 10.1016/s1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 44.Inayoshi Y, Miyake K, Machida Y, et al. Mammalian chromatin remodeling complex SWI/SNF is essential for enhanced expression of the albumin gene during liver development. J Biochem. 2006;139:177–188. doi: 10.1093/jb/mvj015. [DOI] [PubMed] [Google Scholar]
- 45.Gresh L, Bourachot B, Reimann A, et al. The SWI/SNF chromatin-remodeling complex subunit SNF5 is essential for hepatocyte differentiation. EMBO J. 2005;24:3313–3324. doi: 10.1038/sj.emboj.7600802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao X, Tate P, Hu P, et al. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci U S A. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu L, Aster JC, Blacklow SC, et al. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- 48.Poirault-Chassac S, Six E, Catelain C, et al. Notch/Delta4 signaling inhibits human megakaryocytic terminal differentiation. Blood. 2010;116:5670–5678. doi: 10.1182/blood-2010-05-285957. [DOI] [PubMed] [Google Scholar]
- 49.Ishiko E, Matsumura I, Ezoe S, et al. Notch signals inhibit the development of erythroid/ megakaryocytic cells by suppressing GATA-1 activity through the induction of HES1. J Biol Chem. 2005;280:4929–4939. doi: 10.1074/jbc.M406788200. [DOI] [PubMed] [Google Scholar]
- 50.Hald J, Hjorth JP, German MS, et al. Activated Notch1 prevents differentiation of pancreatic acinar cells and attenuate endocrine development. Dev Biol. 2003;260:426–437. doi: 10.1016/s0012-1606(03)00326-9. [DOI] [PubMed] [Google Scholar]
- 51.Ma X, Renda MJ, Wang L, et al. Rbm15 modulates Notch-induced transcriptional activation and affects myeloid differentiation. Mol Cell Biol. 2007;27:3056–3064. doi: 10.1128/MCB.01339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi L, Perin JC, Leipzig J, et al. Genome-wide analysis of interferon regulatory factor I binding in primary human monocytes. Gene. 2011;487:21–28. doi: 10.1016/j.gene.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Douglas J, Cilliers D, Coleman K, et al. Mutations in RNF135, a gene within the NF1 microdeletion region, cause phenotypic abnormalities including overgrowth. Nat Genet. 2007;39:963–965. doi: 10.1038/ng2083. [DOI] [PubMed] [Google Scholar]
- 54.Mikkelsen TS, Hanna J, Zhang X, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deng J, Shoemaker R, Xie B, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.