Abstract
The ‘Yamanaka factors' (Oct4, Sox2, Klf4 and c-Myc) are able to generate induced pluripotent stem (iPS) cells from different cell types. However, to what degree primary malignant cells can be reprogrammed into a pluripotent state has not been vigorously assessed. We established an acute myeloid leukemia (AML) model by overexpressing the human mixed-lineage leukemia-AF9 (MLL-AF9) fusion gene in mouse hematopoietic cells that carry Yamanaka factors under the control of doxycycline (Dox). On addition of Dox to the culture, the transplantable leukemia cells were efficiently converted into iPS cells that could form teratomas and produce chimeras. Interestingly, most chimeric mice spontaneously developed the same type of AML. Moreover, both iPS reprogramming and leukemia reinitiation paths could descend from the same leukemia-initiating cell. RNA-seq analysis showed reversible global gene expression patterns between these interchangeable leukemia and iPS cells on activation or reactivation of MLL-AF9, suggesting a sufficient epigenetic force in driving the leukemogenic process. This study represents an important step for further defining the potential interplay between oncogenic molecules and reprogramming factors during MLL leukemogenesis. More importantly, our reprogramming approach may be expanded to characterize a range of hematopoietic malignancies in order to develop new strategies for clinical diagnosis and treatment.
Keywords: iPS cells, MLL-AF9, primary leukemia cells, reprogramming
Introduction
Somatic cell reprogramming into a pluripotent state by the induced pluripotent stem (‘iPS') cell technology1, 2 not only holds great promise in regenerative medicine but also provides a powerful tool for studying pathological processes such as the oncogenic process. In fact, the four ‘Yamanaka' reprogramming transcription factors used for iPS induction (Oct4, Sox2 and especially Klf4 and c-Myc, also named as ‘OSKM' factors) are known for their direct or indirect oncogenic activities.3, 4, 5, 6, 7 Reciprocally, the two most well-known tumor-suppressor pathways, p53 and Rb, have also been shown to suppress iPS reprogramming.8, 9 These studies revealed some common mechanisms for tumorigenesis and somatic cell reprogramming (toward dedifferentiation).9, 10 The similarity between the two processes offers a new avenue for understanding cancer development as well as iPS generation. Therefore, defining the link between cancer cells and iPS cells is of great importance in both stem cell and cancer research fields. Previous studies demonstrated that malignant cell lines and primary cancer cells could be reprogrammed by blastocyst injection,11 nuclear transfer12, 13 or the iPS approach.14, 15, 16, 17 However, whether primary transformed cells (not established tumor cell lines) can be reprogrammed into iPS cells with a full-term in vivo developmental potential (contributing to chimeric mice or succeeding in tetraploid complementation) remains unknown. For this purpose, an animal model is required so that a full conversion between malignancy and pluripotency from the same genome by the iPS technique can be assessed. The mixed-lineage leukemia (MLL) gene-rearranged leukemia was chosen owing to the relative stability of its genome,18 thereby increasing the likelihood of successful reprogramming of leukemia cells.
In this study, we have established an acute myeloid leukemia (AML) mouse model by overexpressing the human MLL-AF9 fusion gene in hematopoietic cells harvested from ‘all-iPS' mice that carry four OSKM factors under the control of doxycycline (Dox).19, 20 On addition of Dox to the culture, the leukemia cells were efficiently converted into iPS cells that could form teratomas and produce chimeras. Interestingly, most chimeric mice spontaneously developed the same type of AML. RNA-seq analysis showed reversible global gene expression patterns between these convertible cell types, likely owing to epigenetics-driven activation or reactivation of MLL-AF9.
Materials and methods
Mice
B6-Ly5.1 and B6-Ly5.2 mice were purchased from the animal facility of State Key Laboratory of Experimental Hematology (SKLEH). The all-iPS mice were generated from tetraploid complementation as previously reported.20 The experimental protocol was approved by the Institutional Animal Care and Use Committees of SKLEH.
MLL-AF9 plasmids and virus production
MSCV-MLL/AF9-PGK-PURO was generously provided by Dr Chi Wai So. The PGK-PURO segment was replaced by IRES-green fluorescent protein (GFP) to form the MSCV-MLL/AF9-IRES-GFP construct. For retrovirus production, MSCV-MLL/AF9-IRES-GFP was transfected together with pKat and pVSVG into the 293T cell line using Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA). After 48 and 72 h of culture, supernatant was harvested and concentrated using an Amicon filter (Amicon Ultra-15 Centrifugal Filter; Merck Millipore, Billerica, MA, USA).
ES, iPS and MEF culture
Mouse embryonic stem (ES) and Ips cells were maintained in a standard mouse ES cell culture medium as previously described.20, 21 Primary mouse embryonic fibroblasts (MEFs) were obtained from 13.5-day-old embryos of Institute of Cancer Research (ICR) mouse on the basis of the protocol from Wicell (Madison, WI, USA) and cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Mouse ES and iPS cells were cultured on mitomycin C-treated MEF cells (10 μg/ml).
Leukemia mouse model
Fresh whole bone marrow (BM) cells were harvested and enriched using lineage cell depletion beads (Miltenyi, Bergisch Gladbach, Germany). Lin− stem and progenitor cells were incubated overnight in Iscove's modified Dulbecco's medium with 15% fetal bovine serum, 50 ng/ml murine stem cell factor, 10 ng/ml murine interleukin (IL)-3 and 10 ng/ml murine IL-6 to promote cell cycle entry. The prestimulated cells (5 × 105) were then spinoculated with a retroviral supernatant in the presence of 6 μg/ml polybrene (Sigma, St Louis, MO, USA) for 90 min at 1800 r.p.m. After 2 days of culture, 5 × 105 transduced cells together with 2 × 105 radioprotective cells were injected into lethally irradiated mice (9.5 Gy). Transduction efficiency was measured by Fluorescence-activated cell sorting (FACS).
Flow cytometry
BM cells were incubated with PE-CD3, PE/Cy7-Gr1, PerCP/Cy5.5-B220 and APC-Mac1 (eBioscience, San Diego, CA, USA or BD Biosciences, San Jose, CA, USA), and analyzed using LSR II (BD Biosciences). For cell sorting, leukemia cells were stained with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI), and GFP+DAPI−-live cells were sorted using a FACS Aria III sorter (BD Biosciences).
Generation of iPS cells from leukemia cells
GFP+DAPI− leukemia cells were sorted into a six-well plate (1 × 105/well) by FACS. The cells were cultured in a normal ES culture medium with 2 μg/ml Dox, 50 ng/ml murine stem cell factor, 10 ng/ml murine IL-3 and 10 ng/ml murine IL-6. Cytokines were removed from the culture system after 7 days and the cells were maintained only in the presence of Dox for another 7 days. At 1–2 days after removing Dox, ES-like colonies were individually picked up for propagation.
Karyotype analysis
The cells were cultured for 24 h and treated with colcemid (2 μg/ml) for 3.5–4 h before harvesting. The cells were washed with phosphate-buffered saline (PBS), trypsinized and transferred into 15-ml tubes for 5 min centrifugation at 1000 r.p.m. The cells were resuspended in 10 ml KCl solution (75 mM). After 10 min incubation at 37 °C, the cells were fixed by adding 2 ml fixative solution (methanol/acetic acid 3:1). The fixed cells were washed two times before mounting onto chilled slides. The slides with chromosomes were dried and treated with 0.0025% trypsin for 5 min and stained with Giemsa (1:10) for 5–10 min.
Immunofluorescence staining
Colonies were fixed in 4% paraformaldehyde for 30 min at room temperature and then incubated with 0.1% Triton X-100/PBS for 15 min at room temperature. Cells were blocked with 4% normal goat serum before incubation with a primary antibody to Oct4 (Santa Cruz, Dallas, TX, USA), SSEA-1 (Chemicon, Billerica, MA, USA), Nanog (Cosmobio, Tokyo, Japan) or Sox2 (Abcam, Cambridge, UK) at 4 °C overnight. Cells were washed three times in PBS and incubated at 37 °C for 2 h with appropriate secondary antibodies. Nuclei were identified by DAPI (Roche, Basel, Switzerland) staining at a dilution of 1:1 000 000 at room temperature for 5 min. Images were acquired using a confocal laser scanning microscope (SP2; Leica, Wetzlar, Germany).
RNA extraction, reverse transcription and PCR or real-time PCR
Total RNA was isolated using Qiagen (Hilden, Germany) RNeasy mini kit. Complementary DNA was synthesized using transcriptase (Promega, Madison, WI, USA). PCR or real-time PCR was carried out with primers listed in Supplementary Tables S4 and S5.
Teratoma formation and histological analysis
iPS cells were trypsinized and 2 × 106 iPS cells were suspended in 200 μl PBS. The cells were injected subcutaneously into the groin of severe combined immune-deficient mice. After 2–3 weeks, the mice were euthanized and the tumors were fixed and prepared for H&E staining using a standard protocol.
Chimera
To generate chimeric mice, 10–15 iPS cells were microinjected into ICR eight-cell embryos using a piezo-actuated microinjection pipette. After culture for 1 day, the embryos were transplanted into the uterus of pseudo-pregnant ICR mice.
Bisulfite genomic sequencing
Bisulfite treatment of the genomic DNA was performed with an EpiTect bisulfite kit (Qiagen). Oct4 and Nanog promoter regions were amplified with nested primers (Supplementary Table S6). The vector 3′long terminal repeat (LTR) (MLL-AF9 promoter) was amplified with two rounds of PCR using nested primers (Supplementary Table S6). The PCR products were cloned into pMD18-T vectors (Invitrogen, Life Technologies, Carlsbad, CA, USA). Ten randomly selected clones were sequenced and analyzed.
Quantitative fluorescence in situ hybridization
The iPS cells, primary leukemia cells and secondary leukemia cells were deposited on charged slides, fixed, permeabilized and stained with MLL probes (Vysis, Abbott Park, IL, USA). Slides were viewed under a Zeiss Axioplan fluorescence microscope (Oberkochen, Germany). Images were captured using Macprobe software (Applied Imaging, Santa Clara, CA, USA). At least five areas (300 cells) per sample were counted.
RNA-seq and data analysis
mRNA-seq was conducted in nine samples, including one normal hematopoietic stem and progenitor cell (HSPC), one normal-iPS (N-iPS), iPS cells from non-transduced Lin− BM cells clone, one primary leukemia cell, three leukemia-iPS (L-iPS) clones and three recurrent leukemia cells. Samples were prepared with a standard Illumina kit using the TruSeq RNA SamplePrep Guide (Illumina, San Diego, CA, USA). mRNA fragments with a length of 200–300 bp were selected for library construction. Sequencing was performed on a Hiseq2000 platform using a standard paired-end protocol. Cuffdiff V2.0.0 was used to test differentially expressed genes as described earlier.22 Data visualization and pathway enrichment were carried out using R packages and DAVID online analysis,23 respectively. Gene set enrichment analysis24 was performed in public mouse or human AML data sets.25, 26 The mRNA-seq data are available in the Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra?term=SRA062239) under accession number SRA062239.
In vitro hematopoietic differentiation
In vitro differentiation of iPS cells into the hematopoietic lineage was carried out using a standard kit (Stem Cell Technologies, Vancouver, BC, Canada). Briefly, the embryoid bodies (EBs) were formed by plating iPS cells in a 35-mm culture dish in a primary differentiation medium. At day 7, hematopoietic growth factors were added. At day 10, cells dissociated from EBs were counted, and 1 × 105/ml cells were cultured in a hematopoietic differentiation medium. Colony-forming units (CFUs) were counted after 10–14 days of culture. All the media and additional components were listed in the Stem Cell Technologies technical manual.
Statistics
GraphPad Prism software was used for statistical analyses. Unpaired Student's t test and analysis of variance were used to generate P-values for most of the data sets.
Results
Establishment of AML by MLL-AF9 overexpression
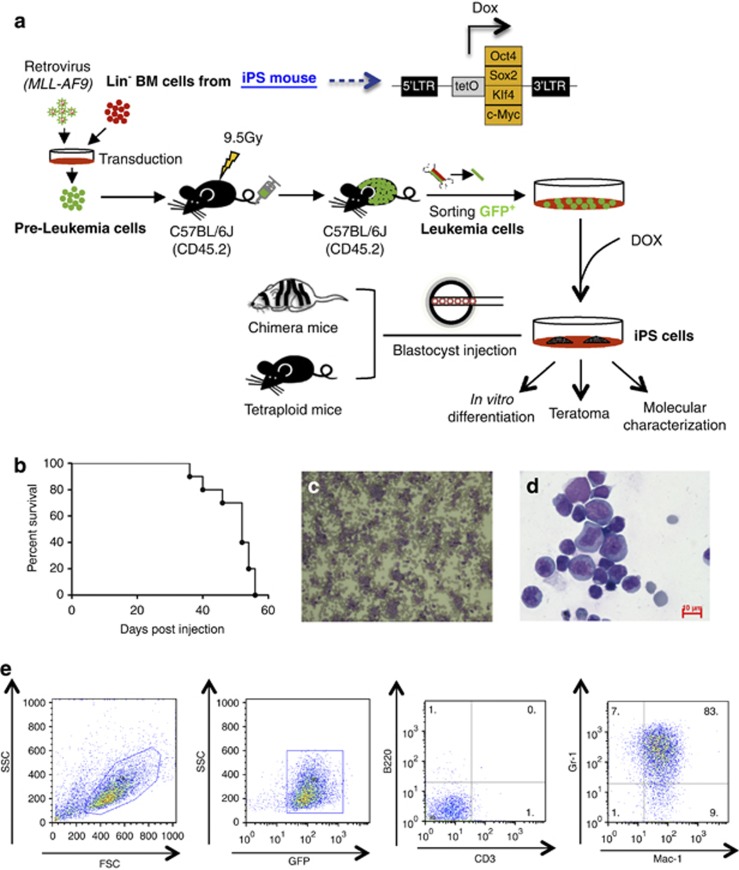
Lin− (lineage positive cell-depleted) BM HSPCs were isolated from all-iPS mice20 and transduced with a retrovirus encoding MLL-AF9 fusion gene as described before.27 OSKM factors in these HSPCs could be activated by adding Dox into the culture (Figure 1a). Transduced cells were then transplanted into lethally irradiated mice. The recipients developed leukemia within 2 months (Figure 1b). Moribund mice exhibited elevated white blood cell counts in peripheral blood (Figure 1c) and splenomegaly (Supplementary Figure S1A). Histological analysis of representative tissues and organs showed infiltration of leukemia cells (Supplementary Figure S1B). Cytospin analysis showed that the BM was completely replaced with leukemia cells (Figure 1d). FACS analysis demonstrated abundant cells positive for the GFP and myeloid lineage markers Mac-1 and Gr-1, indicating an AML phenotype (Figure 1e). Moreover, the primary leukemia cells were able to give rise to the same type of AML when injected into sublethally irradiated secondary recipients, showing the transplantablity of the leukemia cells induced by our system (Supplementary Figure S1C).
Figure 1.
Experimental design and establishment of AML. (a) Schematic representation of the strategy used in this study for reprogramming primary AML cells toward iPS. (b) A Kaplan–Meyer curve showing the survival of leukemic mice. All the mice died within 2 months (n=9). (c and d) Representative peripheral blood smear and cytospin of BM cells (Wright–Giemsa staining) showing the accumulation of immature myeloid blast cells in AML mice. Scale bars, 10 μm. (e) FACS analysis of bone marrow cells indicating the GFP+Mac-1+Gr-1+CD3−B220− phenotype of the leukemia cells.
Characterization of the iPS cell lines generated from MLL-AF9 leukemia cells
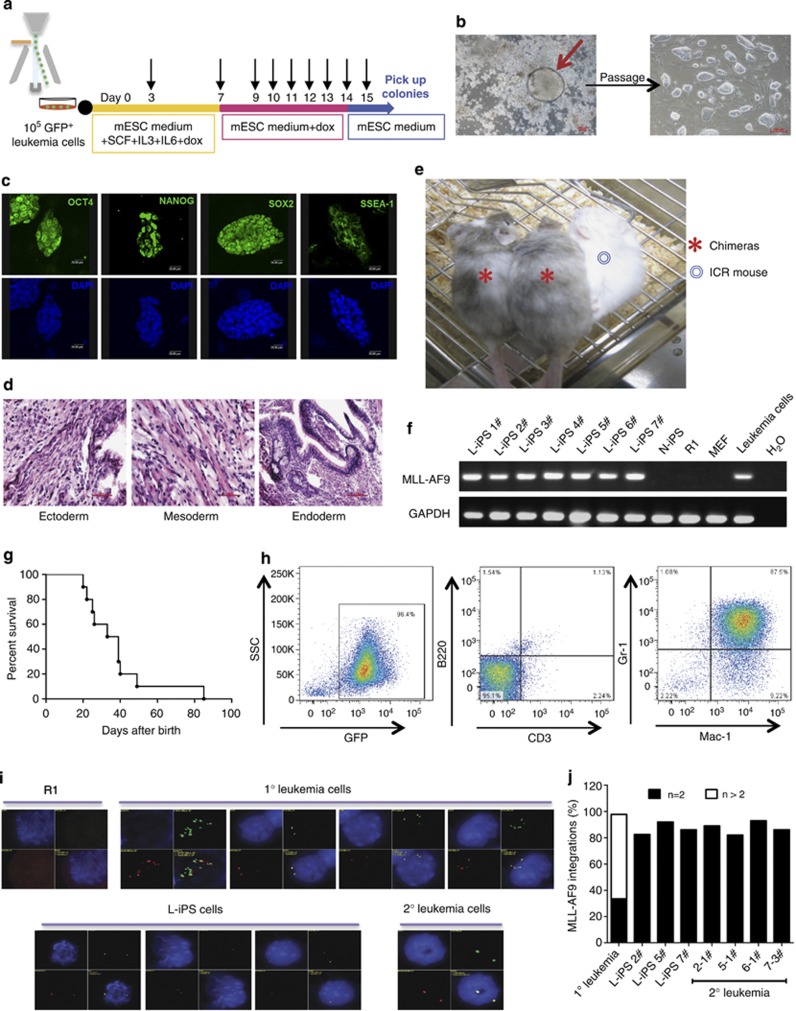
To assess whether the cells carrying the leukemic gene could be reprogrammed, GFP+ leukemia cells were sorted and plated into six-well plates coated with MEF feeder cells. The leukemia cells were cultured in mouse ES medium containing stem cell factor (50 ng/ml), IL-3 (10 ng/ml), IL-6 (10 ng/ml) and Dox (2 μg/ml) to promote leukemia cell proliferation while reactivating the four Yamanaka reprogramming factors. After 1 week in culture, the leukemia cells were maintained only in the presence of Dox until ES-like colonies appeared (typically 7 days later) before the removal of Dox. ES-like colonies were individually picked up for propagation after 1–2 days (Figure 2a). After propagation, seven L-iPS cell lines that exhibited typical morphology of ES cells (Figure 2b) were randomly selected for further characterization. iPS cells (N-iPS) from non-transduced Lin− BM cells were used as the control. Immunofluorescence experiments revealed positive staining for ES cell markers, including Oct4, Sox2, Nanog and the surface marker SSEA-1, in all seven iPS cell lines (Figure 2c). Reverse transcription-polymerase chain reaction (RT-PCR) and quantitative RT-PCR (qRT-PCR) demonstrated the expression of endogenous pluripotency genes in all seven L-iPS cells, which was comparable to that of N-iPS cells or mouse ES cells (R1) (Supplementary Figures S2A and S2B). Most L-iPS cell lines were predominantly diploid with the normal (40 XY) karyotype (Supplementary Figure S2C). Bisulfite sequencing showed higher levels of demethylation of Oct4 and Nanog promoters in L-iPS cell lines compared with the parental leukemia cells, suggesting epigenetic remodeling during reprogramming (Supplementary Figure S2D).
Figure 2.
Characterization of leukemia-derived iPS cells. (a) The reprogramming procedure of AML cells. Black arrow: medium change. (b) Representative L-iPS colony derived from AML cells cultured with Dox and typical morphology of L-iPS cells after propagation. Scale bars, 100 μm. (c) Immunofluorescence staining showing the expression of pluripotency markers (OCT4, NANOG, SOX2 and SSEA-1) in L-iPS cells. The data represent one of three independent experiments. Scale bars, 20 μm. (d) Representative teratoma from L-iPS cells containing all three germ layers (ectoderm, mesoderm and endoderm). H&E staining. The data represent one of four independent experiments. Scale bars, 50 μm. (e) Blastocyst injection of L-iPS cells generated chimeric mice with high chimerism. The data represent two of ten chimeras. (f) Genomic DNA PCR showing the integration of MLL-AF9 fusion gene in L-iPS cells, demonstrating that all iPS cells tested were derived from the primary AML cells. (g) A Kaplan–Meyer curve showing the survival of chimeric mice. Most chimeras died within 2 months (n=10).(h) FACS analysis of bone marrow cells isolated from diseased chimeras showing the GFP+Mac-1+Gr-1+CD3−B220− phenotype (identical to that in primary leukemia). (i) quantitative fluorescence in situ hybridization analysis showing the MLL-AF9 integrations in R1 cells (negative control), 1° leukemia cells, L-iPS cells and 2° leukemia cells. DAPI, blue; MLL 5′, green; MLL 3′, red. (j) Percent MLL-AF9 integration in 1° leukemia cells, L-iPS cells (2#, 5# and 7#) and 2° leukemia cells (2-1#, 5-1#, 6-1# and 7-3#). 300 cells/ sample were counted.
In order to vigorously demonstrate whether reprogrammable cells are indeed derived from leukemia-initiating cells or leukemia stem cells, we conducted a series of clonal assays in vitro as well as in vivo. Leukemia cells (GFP+) from primary recipients were plated in the semisolid medium containing myeloid growth factors for 7 days, and individual AML colonies were plucked and replated for expansion (Supplementary Figure S3A). As reported previously, both type A and type B colonies were able to induce AML28 (Supplementary Figure S3B). Thus, we randomly picked up four type-A colonies and three type-B colonies for replating. After the secondary culture for 5 days, the expanded cells in each plate from one colony were collected and then separated for leukemia development in vivo and for iPS induction in vitro. Four clones of the cells (1#, 4#, 5# and 7#) were able to give rise to both AML and iPS phenotypes (Supplementary Figures S3C–S3F). To further demonstrate that fully transformed leukemia cells were able to be reprogrammed, we isolated GFP+transformed leukemia cells from secondary leukemic mice derived from 4# and 5# colony cells and separated them for leukemia development in vivo and iPS induction in vitro (Supplementary Figure S3A). The results showed that the GFP+ leukemia cells (4# and 5#) had the ability to both reinitiate leukemia and become iPS (Supplementary Figures S3G and S3H). Therefore, these cell cloning data clearly show that iPS cells can indeed be generated from a leukemia stem cell-derived clone, and the reprogramming potential was retained after secondary transplantation of leukemia.
In vivo developmental potential of L-iPS cell lines was assessed by the following experiments. On injection of several L-iPS cells lines into severe combined immune-deficient mice, teratoma consisting of all three germ layers was observed 2–3 weeks later (Figure 2d). Furthermore, L-iPS cell lines were randomly selected for injection into normal ICR blastocysts and then transferred to ICR pseudopregnant recipient females. The five L-iPS cell lines generated 10 postnatal chimeras with high chimerism as reflected by coat color (Figure 2e). We also tested whether the L-iPS cell lines could generate full-term mice by tetraploid blastocyst complementation. We selected seven iPS cell lines that were able to generate chimeras with high chimerism, and then injected totally 1106 embryos into 40 mice in total. Unfortunately, we failed to generate the tetra mice (all-iPS mice). These numbers suggest that it is unlikely we will be able to succeed in the tetraploid complementation approach based on experience. One possible reason is that the iPS cells carrying MLL-AF9 may retain some unknown negative factors that prevent the success of tetraploid complementation in the recipient mice. Importantly, PCR of genomic DNA confirmed the presence of MLL-AF9 in all the L-iPS cells, confirming that all L-iPS cells were derived from leukemia cells (Figure 2f).
Interestingly, the majority of chimeric mice developed recurrent leukemia within 2 months (Figure 2g). Its phenotype was identical to that of primary AML (Figure 2h and Supplementary Figures S4A–S4D). However, no solid tumor was found among the chimeric mice. As all male chimeras died within 40 days, and only two female chimeras survived beyond 40 days, we were not able to test the germ-line transmission using female chimeras (as all-iPS mice were generated using male mouse MEF cells, which means that the leukemia cells or L-iPS cells are male cells). Quantitative fluorescence in situ hybridization revealed only two copies of MLL-AF9 in all tested L-iPS cells, whereas multiple patterns of the integration were found in primary leukemia cells (Figures 2i–j), suggesting a highly selective process for the reprogramming in the heterogeneous leukemia cell population. The copy number of the MLL-AF9 fusion gene in recurrent leukemia cells was identical to that in the original L-iPS cells that generated the chimeric mice (Figures 2i–j), suggesting that two copies of the oncogenic MLL-AF9 gene are sufficient to initiate leukemogenesis.
In short, we have demonstrated the following: (1) despite the presence of a leukemogenic gene, MLL-AF9 leukemia cells can be directly converted into iPS cells in vitro; (2) MLL-AF9 leukemia-derived iPS cells have the potential to develop into most tissue types in vivo; and (3) recurrent leukemia may occur in L-iPS derived chimeric mice.
Silence of the leukemogenic gene in the iPS cells
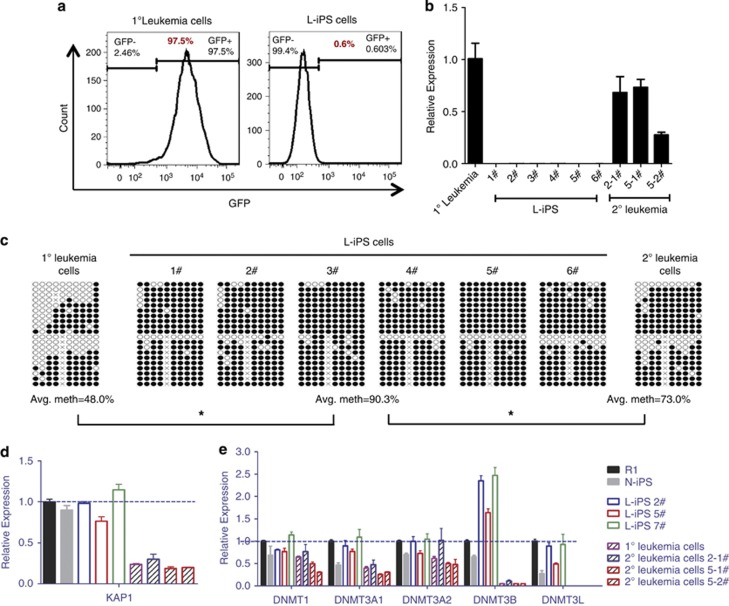
The convertible phenotypes between leukemia and iPS normalcy prompted us to further obtain a molecular basis underlying leukemogenesis versus reprogramming. MLL-AF9 is driven by the LTR promoter in the retroviral vector.29, 30, 31 LTR promoter activity can be silenced in ES cells mainly by KAP1 (KRAB-associated protein 1)- or Dnmts (DNA methyltransferases)-mediated methylation.32, 33, 34 As expected, GFP was not expressed in L-iPS cells (Figure 3a), indicating that the retroviral vector was silenced in L-iPS cells. qRT-PCR and RNA-Seq analyses demonstrated that MLL-AF9 was not expressed in all tested L-iPS cells (Figure 3b and Supplementary Figure S5A). In contrast, it was expressed in most (five/eight) chimeras on day 30 (Supplementary Figure S5B) and in all mice when leukemia was fully developed (Figure 3b and Supplementary Figure S5A). Bisulfite genomic sequencing of the 3′LTR of the MLL-AF9 vector demonstrated a higher level of methylation of the vector in L-iPS cells than in primary leukemia cells or recurrent leukemia cells (Figure 3c), which confirmed that MLL-AF9 silencing was due to methylation of the retroviral vector. Furthermore, an analysis of KAP1 expression by qRT-PCR revealed that ES, N-iPS and L-iPS all highly expressed the KAP1 gene compared with leukemia cells (Figure 3d), consistent with its role in silencing the retroviral vector in ES cells. Examination of Dnmts (including Dnmt1, Dnmt3a, Dnmt3b and Dnmt3l) using qRT-PCR revealed a higher expression of Dnmt3b in L-iPS cells than in ES, N-iPS and leukemia cells (Figure 3e), suggesting a more dominant role of Dnmt3b compared with other Dnmts in silencing the MLL-AF9 transgene.
Figure 3.
Mechanism of MLL-AF9 gene silencing. (a) FACS analysis of GFP expression in 1° leukemia cells and L-iPS cells, revealing that GFP was not expressed in L-iPS cells. (b) qRT-PCR analysis of the MLL-AF9 fusion gene expression, showing that the fusion gene was silenced in L-iPS cells but not in 1° and 2° leukemia cells (n=3). Bars represent mean±s.d. (c) Bisulfite genomic sequencing of the 3′LTR of MLL-AF9 vector in 1° leukemia cells, L-iPS cells and 2° leukemia cells. Black and white circles indicate methylated and unmethylated cytosine-phosphate-guanine (CpGs), respectively. Differences in methylation between each type of cell that were statistically significant are indicated (n⩾2), *P<0.05. (d) qRT-PCR analysis of KAP1 gene expression in different types of cells (n=3). Bars represent mean±s.d. (e) qRT-PCR analysis of Dnmts expression in different types of cells (n=3). Bars represent mean±s.d.
Hematopoietic differentiation of iPS cells in vitro
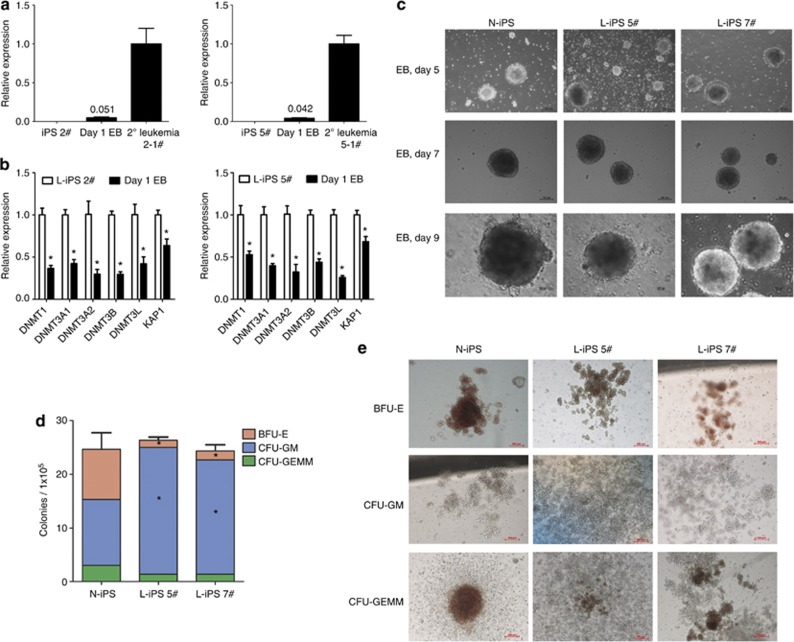
In the next set of experiments, we cultivated iPS cells into EBs, and the colony-forming cells were subsequently assayed to quantify hematopoietic cell differentiation upon reactivation of MLL-AF9. The expression of MLL-AF9 in EBs occurred as early as day 1 (Figure 4a). Moreover, Dnmts and KAP1 genes were downregulated on day 1 in EBs compared with L-iPS cells (Figure 4b). Therefore, the lower expression of Dnmts and KAP1 may be involved in the activation of the fusion gene. The N-iPS cells and the L-iPS cells generated identical EB morphology (Figure 4c) and comparable yields of total colony-forming cells (Figure 4d). All three iPS cell lines could differentiate into blast-forming unit-erythrocytes (BFU-Es), CFU-granulocyte-macrophages (GMs) and CFU-granulocyte-erythrocyte-macrophage-megakaryocytes (GEMMs) (Figure 4e). However, L-iPS cells formed more CFU-GM colonies and fewer BFU-E and CFU-GEMM colonies compared with N-iPS cells (Figure 4d). Thus, the favored differentiation toward myeloid lineage was consistent with the phenotype of resulting leukemia in the mice.
Figure 4.
Hematopoietic colony formation of L-iPS cells and N-iPS cells. (a) qRT-PCR analysis of MLL-AF9 gene expression in L-iPS (2#, 5#) and EB cells on day 1 and in 2° leukemia cells (n=3). Bars represent mean±s.d. of EBs. (b) qRT-PCR analysis of Dnmts and KAP1 genes expression in L-iPS (2#, 5#) and EB cells at day 1 (n=3). Bars represent mean±s.d. *P<0.05. (c) Representative photomicrographs of EBs derived from N-iPS, L-iPS 5# and L-iPS 7# on days 5, 7 and 9. Scale bars, 100 μm. (d) The number of BFU-E, CFU-GM and CFU-GEMM colonies derived from 1 × 105 cells of day 10 EBs of N-iPS, L-iPS 5# and L-iPS 7#. Bars represent mean±s.d. (e) Representative images of BFU-E, CFU-GM and CFU-GEMM colonies derived from day 10 EBs of N-iPS, L-iPS 5# and L-iPS 7#. Scale bars, 100 μm. The data represent one of three independent experiments.
Reversible transcriptome underlying the conversion between leukemia cells and iPS cells
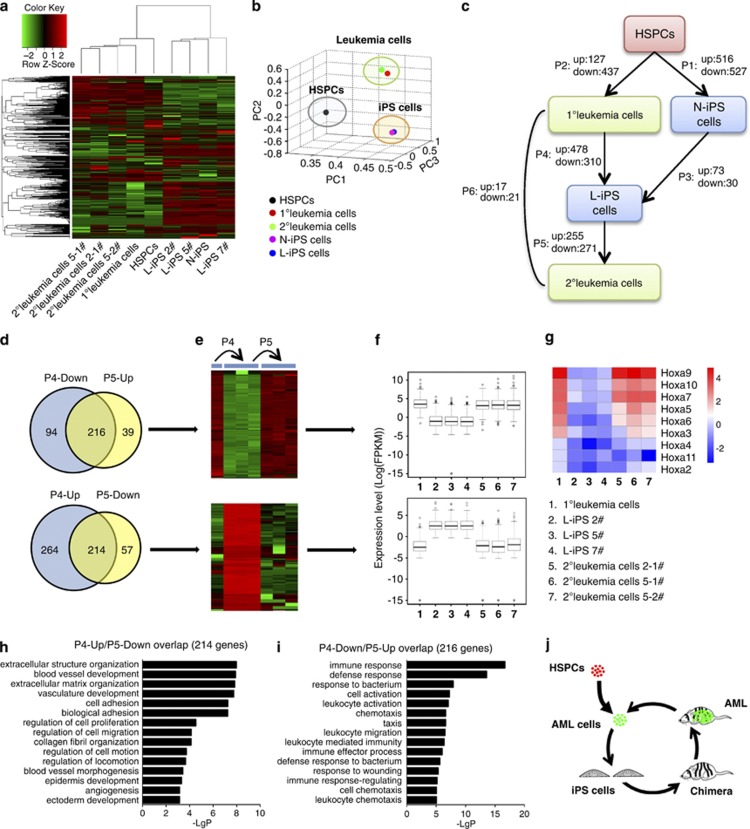
RNA-Seq was applied to show the gene expression changes along with the conversion between leukemia and iPS cells. Clustering and principal component analysis of the whole-gene expression revealed that samples segregated into three transcriptionally distinct groups (Figures 5a–b; Supplementary Table S1). N-iPS and L-iPS cells clustered together, and only 103 differentially expressed genes were found between these two groups (73 upregulated and 30 downregulated genes in L-iPS versus N-iPS, Figure 5c, Supplementary Table S2), indicating that N-iPS and L-iPS had similar gene expression patterns despite different origins (normal versus malignant states). In addition, gene expression patterns of the 1° and 2° leukemia cells seemed to be more overlapped (only 38 differentially expressed genes, Figure 5c, Supplementary Table S2).
Figure 5.
Molecular profiles indicating the conversion of leukemia and iPS cells. (a) Unsupervised hierarchical clustering showing gene expression profiles in different samples. Color indicates normalized expression level (log); red corresponds to higher expression. Row and column represent gene and sample, respectively. (b) Principal component analysis of gene expression profiles of different samples indicating similarity between cells from different origins. (c) A diagram showing the relationship between each sample and the numbers of differentially expressed genes in each process. Up: genes upregulated in arrow-pointing cells; down: genes downregulated in arrow-pointing cells. P1∼P6: differentially expressed genes in these processes. (d–f) Venn diagram detailing shared and distinct genes between P4 and P5. Heatmap and box plots showing expression profiles of these genes in 1° leukemia cells, L-iPS cells and 2° leukemia cells. X axis: sample; y axis: expression level of each gene, log transformed. (g) Heatmap showing the Hoxa gene-expression level in 1° leukemia cells, L-iPS cells and 2° leukemia cells. Color indicates normalized expression level (log); Red corresponds to higher expression. Row and column represent gene and sample, respectively. (h and i) Gene ontology analysis of biological functions of the two gene sets (P4-up/P5-down and P4-down/P5-up, Supplementary Table S3), showing the P-values of the first 15 categories. (j) Schematic presentation of the reversible process of ‘leukemia-iPS-leukemia'.
In order to document the reversible process (P4 and P5) between leukemia cells and iPS cells by molecular profiling (Figure 5c), differentially expressed genes among 1° leukemia cells, L-iPS cells and 2° leukemia cells were compared. Downregulated genes in L-iPS cells versus 1° leukemia cells were referred to as ‘P4-down'. By analogy, other differentially expressed gene sets were ‘P4-up', ‘P5-down' and ‘P5-up'. If the process of L-iPS–leukemia was reversible, most genes in P4-down and P5-up or in P4-up and P5-down. Our experiments demonstrated that there was more than 78% overlap of the P5 genes with the P4 genes. A comparison of overall differentially expressed genes in these three types of cells demonstrated opposite gene variation between P4 and P5 and similar gene profiles between 1° leukemia cells and 2° leukemia cells (Figures 5d–f, Supplementary Table S3). Gene set enrichment analysis revealed that MLL-AF9 signature genes were enriched in P4-down and P5-up, whereas the reverse enrichment was observed in P4-up and P5-down (Supplementary Figures S6A and S6B). This result demonstrated P4 as a reverse process and P5 as a forward process for leukemogenesis. Previous studies found that multiple HoxA cluster genes were highly expressed in MLL-rearranged leukemias (especially Hoxa7, Hoxa9 and Hoxa10),35 and this was also observed in our study (Figure 5g). Gene ontology analyses identified processes such as ‘extracellular structure organization', regulation of ‘cell adhesion' and ‘development' as being significantly enriched in the P4-up/P5-down overlapped gene set (Figure 5h). These genes are probably important for cell remodeling. Interestingly, ‘immune response'-related catalogs were highly enriched in the P4-down/P5-up overlapped gene set (Figure 5i), indicating that immunogenicity was diminished to a certain extent in iPS status. Therefore, the reversible phenotypes between leukemia cells and iPS cells are largely based on traceable changes in global gene expression (Figure 5j).
Discussion
This study demonstrates for the first time that primary leukemia cells can be fully reprogrammed into iPS cells with the potential of developing into chimeric mice. The chimeric mice could develop spontaneous leukemia due to reactivation of MLL-AF9. iPS cells derived from normal and leukemic origins share a nearly identical gene expression profile. Convertible phenotypes between MLL-AF9 leukemia cells and their derived iPS cells are largely associated with MLL-AF9-dependent gene expression patterns that are likely governed by epigenetic regulators.
The developmental potential of cancer cells is a classical topic in cancer biology as well as in developmental biology. Earlier studies in the 70s showed that certain chromosomal abnormalities in cancer cells do not act as barriers for revealing their developmental potential, as mouse embryonic carcinoma cell lines bearing chromosomal changes were able to contribute to chimeras when the cells were injected into a blastocyst.11, 36 A definitive evidence for the reprogramming of mouse embryonic carcinoma nuclei into a pluripotent state was later obtained by the nuclear transfer technique in Jaenisch's group.12, 13 Meanwhile, his group also succeeded in nuclear cloning of mouse melanoma genome.12, 13 Interestingly, the embryonic carcinoma-derived chimeric mice developed melanoma as well as other types of cancer upon induction of Dox. Overall, however, the efficiency of using nuclear transfer to reprogram cancer cell nuclei was extremely low, which was presumably because of technical difficulties and genomic instability of the cancer cells. The iPS technology may potentially offer a much more efficient method for reprogramming a broad range of cell types including cancer cells. There have been three reports on the iPS induction with a human colon cancer cell line, a human chronic myeloid leukemia cell line and chronic myeloid leukemia primary cells.14, 15, 16 However, owing to the limitation of assessing the pluripotency of these human iPS-like cells in vivo, it is not clear whether the iPS approach is able to reprogram cancer cells into the truly pluripotent state as assessed by chimeric assay or tetraploid complementation. To this end, we chose a murine system to vigorously assess the convertibility of malignancy and pluripotency, which cannot be readily achieved with human cells. Admittedly, although we were able to obtain chimeric mice from the leukemia-derived iPS cells, we did not succeed in the tetraploid complementation. Thus, future efforts are required to reach this goal by testing different types of mouse malignant cells.
The reprogramming approach as applied in our current study may provide a unique angle for interrogating a specific type of cancer of interest. MLL leukemia is a unique type of aggressive acute leukemia with poor prognosis. Rearrangements of MLL with about 70 known fusion partners are found in more than 70% of infant leukemia cases, 10% of adult AML cases and in many cases of secondary acute leukemia.35, 37 In contrast to most other translocations that usually cause hematopoietic malignancies only within a specific lineage, MLL translocations, can result in AML, acute lymphoblastic leukemia, mixed-phenotype acute leukemia and myelodysplastic syndrome, depending on the partner fusion gene, patient age and drug treatment regime. Despite this fact, there is a shared core gene expression profile as exemplified by higher expression of the HoxA genes among these different types of MLL leukemia.25, 37 This signature is also recaptured in our current study (Figure 5g). Moreover, it appears that very few chromosomal alterations and additional genomic mutations have been found in MLL leukemia cells,18 and our own analysis on the whole-exome sequencing of iPS versus leukemia cells in our model did not reveal more mutations in the recurrent leukemia cells (data not shown), suggesting that MLL leukemias are largely driven by epigenetic dysregulation.37 Given the greater similarity of the transcriptomes between primary and recurrent leukemia cells (Figures 5a–c), our model system reinforces the notion that epigenetics is sufficient to drive the development of MLL-AF9 leukemia in chimeric mice. Further bioinformatic analyses on RNA-seq, exome sequencing and epigenetic profiling are still needed to delineate the circuitry between genetic and epigenetic elements during the transition between malignancy and pluripotency. Conversely, our current work also implies that the epigenetic regulators during reprogramming process toward a pluripotent state are able to silence certain oncogenic genes (for example, MLL-AF9) and that such a tumor suppressor effect can be reversed once an oncogenic gene is turned on by distinct epigenetic regulators during tissue/organ development, although demonstrated in an artificial system.
Arguably, it should be noted that the reversibility of malignancy and pluripotency as demonstrated here is not a generic phenomenon among different types of cancer. In fact, in our preliminary study, we found that the Notch1-initiated T-acute lymphoblastic leukemia state via retroviral mediation could not be fully reprogrammed into iPS cells (Supplementary Figures S7A–S7C), suggesting that upregulation of the Yamanaka factors is not sufficient to silence the retroviral promoter in at least some types of hematopoietic malignancies. Because our retroviral model is not an ideal mimic for pathogenesis of human leukemia, experiments with a non-retroviral strategy (for example, MLL-AF9 Knock-in model) is certainly needed in our further investigation. However, despite the disadvantage of using the retroviral model as compared with the ‘knock-in' genetic approach, the reprogramming potential of malignant cells by the current approach offers a new opportunity for characterizing a range of malignancies and for developing novel therapeutic strategies for at least certain types of cancer by manipulating different pluripotent genes. The retroviral approach in conjunction with the ‘Tet-on' iPS mice with different pluripotent genes may provide a more robust way for dissecting, and a better understanding of, the reprogramming potential of a variety of hematological malignancies toward our ultimate goals at this stage of the research.
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology of China (2010CB945204, 2011CB964800, 2012CB966600 and 2011ZX09102-010-04) and from the National Nature Science Foundation of China (81090410, 30825017, 90913018 and 81130074).
Author contributions
YL and HC contributed equally to this study. YL and HC designed and performed the experiments, analyzed the data and wrote the paper. SG performed the chimera and tetraploid complementation experiments. ZZ, FH and XL contributed to RNA-seq and data analyses. LH performed quantitative fluorescence in situ hybridization experiments. DH performed the FACS analysis. YL participated in reprogramming of ICN1-overexpressed cells. HZ and JX helped culture MEF cells. LK provided all-iPS mice. QW and WY conducted research, analyzed the data and assisted with the manuscript. SG provided the all-iPS mice and helped establish the iPS and chimeric protocols. TC conceived the study, designed the experiments, interpreted the results, wrote the paper and oversaw the research project.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Daley GQ. Common themes of dedifferentiation in somatic cell reprogramming and cancer. Cold Spring Harb Symp Quant Biol. 2008;73:171–174. doi: 10.1101/sqb.2008.73.041. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Chen Y, Shi L, Zhang L, Li R, Liang J, Yu W, et al. The molecular mechanism governing the oncogenic potential of SOX2 in breast cancer. J Biol Chem. 2008;283:17969–17978. doi: 10.1074/jbc.M802917200. [DOI] [PubMed] [Google Scholar]
- Wang J, Xie LY, Allan S, Beach D, Hannon GJ. Myc activates telomerase. Genes Dev. 1998;12:1769–1774. doi: 10.1101/gad.12.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Kanai M, Huang S, Xie K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis. 2006;27:23–31. doi: 10.1093/carcin/bgi243. [DOI] [PubMed] [Google Scholar]
- Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapia N, Scholer HR. p53 connects tumorigenesis and reprogramming to pluripotency. J Exp Med. 2010;207:2045–2048. doi: 10.1084/jem.20101866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz B, Illmensee K. Normal genetically mosaic mice produced from malignant teratocarcinoma cells. Proc Natl Acad Sci USA. 1975;72:3585–3589. doi: 10.1073/pnas.72.9.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blelloch RH, Hochedlinger K, Yamada Y, Brennan C, Kim M, Mintz B, et al. Nuclear cloning of embryonal carcinoma cells. Proc Natl Acad Sci USA. 2004;101:13985–13990. doi: 10.1073/pnas.0405015101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Blelloch R, Brennan C, Yamada Y, Kim M, Chin L, et al. Reprogramming of a melanoma genome by nuclear transplantation. Genes Dev. 2004;18:1875–1885. doi: 10.1101/gad.1213504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Pruszak J, Varadarajan M, Blomen VA, Gokhale S, Camargo FD, et al. Generation of iPSCs from cultured human malignant cells. Blood. 2010;115:4039–4042. doi: 10.1182/blood-2009-07-231845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi N, Ishii H, Nagai K, Hoshino H, Mimori K, Tanaka F, et al. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc Natl Acad Sci USA. 2010;107:40–45. doi: 10.1073/pnas.0912407107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano K, Arai S, Hosoi M, Taoka K, Takayama N, Otsu M, et al. Generation of induced pluripotent stem cells from primary chronic myelogenous leukemia patient samples. Blood. 2012;119:6234–6242. doi: 10.1182/blood-2011-07-367441. [DOI] [PubMed] [Google Scholar]
- Hu K, Yu J, Suknuntha K, Tian S, Montgomery K, Choi KD, et al. Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood. 2011;117:e109–e119. doi: 10.1182/blood-2010-07-298331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Takeda S, Kumar R, Westergard TD, Brown EJ, Pandita TK, et al. Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint. Nature. 2010;467:343–346. doi: 10.1038/nature09350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambrink T, Foreman R, Welstead GG, Lengner CJ, Wernig M, Suh H, et al. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang L, Wang J, Zhang Y, Kou Z, Gao S. iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell Stem Cell. 2009;5:135–138. doi: 10.1016/j.stem.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Li Y, Liu Y, He J, Wang F, Liu S, Zhang Y, et al. Long-term survival of exogenous embryonic stem cells in adult bone marrow. Cell Res. 2011;21:1148–1151. doi: 10.1038/cr.2011.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross ME, Mahfouz R, Onciu M, Liu HC, Zhou X, Song G, et al. Gene expression profiling of pediatric acute myelogenous leukemia. Blood. 2004;104:3679–3687. doi: 10.1182/blood-2004-03-1154. [DOI] [PubMed] [Google Scholar]
- Li Z, Huang H, Chen P, He M, Li Y, Arnovitz S, et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat Commun. 2012;3:688. doi: 10.1038/ncomms1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs MC, Kim YM, Krivtsov AV, Wright RD, Feng Z, Agarwal J, et al. MLL-AF9 and FLT3 cooperation in acute myelogenous leukemia: development of a model for rapid therapeutic assessment. Leukemia. 2008;22:66–77. doi: 10.1038/sj.leu.2404951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell. 2006;10:257–268. doi: 10.1016/j.ccr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Jahner D, Stuhlmann H, Stewart CL, Harbers K, Lohler J, Simon I, et al. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298:623–628. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- Challita PM, Kohn DB. Lack of expression from a retroviral vector after transduction of murine hematopoietic stem cells is associated with methylation in vivo. Proc Natl Acad Sci USA. 1994;91:2567–2571. doi: 10.1073/pnas.91.7.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Illmensee K, Mintz B. Totipotency and normal differentiation of single teratocarcinoma cells cloned by injection into blastocysts. Proc Natl Acad Sci USA. 1976;73:549–553. doi: 10.1073/pnas.73.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernt KM, Armstrong SA. Targeting epigenetic programs in MLL-rearranged leukemias. Hematology Am Soc Hematol Educ Program. 2011;2011:354–360. doi: 10.1182/asheducation-2011.1.354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.