Abstract
Dysregulated expression of factors that control protein synthesis is associated with poor prognosis of many cancers, but the underlying mechanisms are not well defined. Analysis of the diffuse large B-cell lymphoma (DLBCL) translatome revealed selective upregulation of mRNAs encoding anti-apoptotic and DNA repair proteins. We show that enhanced synthesis of these proteins in DLBCL is mediated by the relief of repression that is normally imposed by structure in the 5′-untranslated regions of their corresponding mRNAs. This process is driven by signaling through mammalian target of rapamycin, resulting in increased synthesis of eukaryotic initiation factor (eIF) 4B complex (eIF4B), a known activator of the RNA helicase eIF4A. Reducing eIF4B expression alone is sufficient to decrease synthesis of proteins associated with enhanced tumor cell survival, namely DAXX, BCL2 and ERCC5. Importantly, eIF4B-driven expression of these key survival proteins is directly correlated with patient outcome, and eIF4B, DAXX and ERCC5 are identified as novel prognostic markers for poor survival in DLBCL. Our work provides new insights into the mechanisms by which the cancer-promoting translational machinery drives lymphomagenesis.
Keywords: lymphoma, eIF4B, microarray analysis, apoptosis, DNA repair, mTOR
Introduction
It is well established that altered regulation at the level of mRNA translation is important in both cancer development and progression. Rates of protein synthesis are controlled both globally and specifically, and dysregulation via either route makes a significant contribution to cell transformation and tumorigenesis.1, 2, 3 In general, global protein synthesis rates are regulated by the availability of two complexes, the eukaryotic initiation factor 4F complex (eIF4F) and ternary complex.4
The eIF4F complex comprises eIF4E (the cap binding protein), eIF4G (a scaffold protein) and eIF4A (a dead box helicase). Two associated proteins, eIF4B and the functionally related eIF4H, anchor the complex to the mRNA and increase eIF4A helicase activity.5, 6 Dysregulation of the eIF4F complex members has a direct role in tumorigenesis,7, 8 in part due to increased synthesis of proteins that are normally poorly translated, particularly those encoding growth factors and oncogenes.3, 4
In diffuse large B-cell lymphoma (DLBCL), the most common lymphoma subtype, transcriptional profiling has provided important insights into lymphoma cell biology and new prognostic information.9, 10, 11, 12, 13, 14 This type of analysis identified three distinct forms: germinal center B-cell-like DLBCL, activated B-cell-like DLBCL and type III DLBCL,9, 13, 14 with a correlation between expression pattern and patient survival.9, 12, 13, 14 Recent advances in treatment have resulted in a significant improvement in outcome for patients with 5-year survival rates of ∼55%.15 However, a significant proportion of patients still succumb to the disease, and new therapies and prognostic markers to inform treatment options are required.
As transcriptional changes are not necessarily directly linked to protein expression,16, 17 we have analyzed the translatome of DLBCL cells. We show that translational dysregulation in DLBCL results in altered expression of proteins that function in apoptotic processes and in DNA repair. This results from activation of mammalian target of rapamycin (mTOR) pathway and increased eIF4B expression, which overcomes the repressive effects on the translation of specific mRNAs that is exerted by their highly structured 5′-untranslated regions (UTRs). Importantly, we demonstrate that translational dysregulation of these mRNAs is associated with poor survival of patients with DLBCL, identifying ERCC5, DAXX and eIF4B as new prognostic markers for this disease.
Materials and methods
Antibodies
Provenance and dilutions of antibodies are described in Supplementary Table 1.
Cell culture
All cells were grown in RPMI 1640 with 10–15% fetal calf serum, 5 mM L-glutamine. Cell lines used were GM01953 (control B-cell line), GM03201 (control B-cell line) and five cell lines derived from GC-DLBCL, that is, DoHH-2 (secondary DLBCL derived from follicular lymphoma (FL)), VAL (secondary DLBCL derived from FL), SuDHL-6, DB and OCI-LY19. Cell suspensions from lymph nodes were from the Faculty of Medicine Tissue Bank, University of Southampton. Approval to use patient tissue was obtained from the Leicestershire, Northamptonshire and Rutland Research Ethics Committee (LREC reference 6978).
Treatments
Inhibitors for mTOR were purchased from Selleck Chemicals (Houston, TX, USA) for AZD8055 and Sigma Aldrich (St Louis, MO, USA) for PP242. Unless stated otherwise, treatments were performed with 100 nM AZD8055 or 1 μM PP242.
SDS-polyacrylamide gel electrophoresis and western blotting
Cell line extracts were prepared as described previously.18 B cells from lymphomas or tonsils were purified using Miltenyi Biotec (Cologne, Germany) CD19 beads and MS columns following the supplier's protocol. Protein extracts were subjected to electrophoresis on SDS-polyacrylamide gels. Proteins were detected with the relevant antisera using chemiluminescent reagents.19
Pulse-labeled immunoprecipitation
For immunoprecipitations, transfected cells were grown at 5 × 105cells/ml for 24 h. They were incubated for 6 h in methionine-free medium in the presence of 0.25 mCi of 35S Met-Label (Hartmann Analytics, GmbH, Braunschweig, Germany). Cells were then lysed, and immunoprecipitation was done using protein A/G agarose beads (Santa Cruz Biotechnology, Dallas, TX, USA). Immunoprecipitated proteins were boiled in SDS containing buffer and were subjected to electrophoresis before Coomassie staining and autoradiography were done.
Sucrose density-gradient centrifugation and RNA detection
Sucrose density-gradient centrifugation was used to separate ribosomes into polysomal and subpolysomal forms. Gradients were then fractionated with continuous monitoring at 254 nm and RNA was isolated from each fraction as described previously.20
RNA analysis
Northern analysis of RNA isolated from sucrose density gradients was performed as described previously20 on at least three independent occasions.
Reverse transcriptase-PCR and quantitative reverse transcriptase-PCR were performed as described previously.21
Preparation of fluorescently labeled cDNA for microarray hybridization and data analysis
Human cDNA microarrays were used as described previously.20 Fluorescently labeled DNA probes were generated from equal proportions of RNAs (∼7 μg) of pooled non-polysomal fractions (fractions 1–4, Cy3) and pooled polysomal fractions (fractions 6–11, Cy5). Microarray slides were scanned using a GenePix 4200A microarray scanner and GenePix Pro 5.1 software (Axon Instruments, Union City, CA, USA).
Analysis of microarray data
GenePix Pro 5.1 was used to quantify fluorescence intensities for individual spots on the microarray. The data were processed in the R platform using the Limma package.22, 23 The resulting log2-transformed ratios were as follows:
Mi (gene x)=Log2[Cy5(polysomal-associated mRNA)/Cy3(non-polysomal mRNA)] (i−replicates of experiment, i=1:3)
The log2-transformed ratios of polysomal over subpolysomal signal for each condition were then analysed by a non-parametric ranking approach using the RankProd package.24, 25 mRNAs were clustered using Gene Ontology annotations through DAVID (david.abcc.ncifcrf.gov) and IPA (Ingenuity Systems, www.ingenuity.com); pie charts were created in Microsoft Excel and heatmaps were created using MultiExperiment Viewer MeV (http://www.tm4.org/mev.html).
Death receptor-induced apoptosis
Cells were grown to 1 × 106 cells/ml and treated with different concentrations of His-tagged TRAIL (tumor necrosis factor-related apoptosis-inducing ligand) or anti-FAS antibody. Cells were collected at 4 h post treatment and stained for Annexin V and propidium iodide, and then analysed by flow cytometry.
Tissue microarrays
The two different tissue microarrays have been described previously.26 The immunostaining was performed with histostainR plus bulk kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. eIF4B staining showed a presence/absence pattern that was determined for each tumor. As expression changes were more gradual for the other proteins, tissue arrays were scored using the immunoreactive score method.27 Scoring was carried out on two independent occasions. The population was then divided into two groups using median staining intensity in the DLBCL population as a threshold. The survival of these populations was plotted using the R package ‘a package for survival analysis'28 and significance of the changes was assessed by Cox regression.
Plasmids and siRNA
5′-UTRs of ERCC5, BCL2 and DAXX were amplified by PCR (pfu DNA polymerase) and subcloned between the Spe1 and Nco1 restriction sites of a pGL3 vector. The pSV-RL vector expressing Renilla luciferase was used as an internal control. The Renilla vectors with different 5′-UTR sizes are described in Supplementary Table 2. In this case, pGL3 was used as an internal control.
eIF4B small interfering RNA (siRNA) and non-targeting control were provided as a SMART-pool of target plus siRNA (Dharmacon, Thermo Fisher Scientific, Waltham, MA, USA).
Transfection
Transfections were performed using a Nucleofector 2b TM (Lonza, Basel Switzerland), according to the manufacturer's instructions. All cell lines were transfected in the solution L using the following programs: GM01953 C-009, DoHH-2A-020 and OCI-LY19 M-013. For each transfection of 2 × 106 cells, 100 pmol of siRNA were used. For luciferase assays, 1 μg of reporter pGL3 vector and 0.3 μg of pSV-RL were added.
UTR analysis
The sequences of the 5′-UTRs were analysed for upstream open reading frames, potential internal ribosome entry segment and terminal oligopyrimidine tract motifs (in-house Perl scripts). MicroRNA target sites were predicted using TargetScan (www.targetscan.org), using conserved targets of conserved microRNA families. Structures were predicted using RNAfold.29
Results
cDNA microarray analysis shows that a subset of mRNAs are subject to differential translational regulation in DLBCL
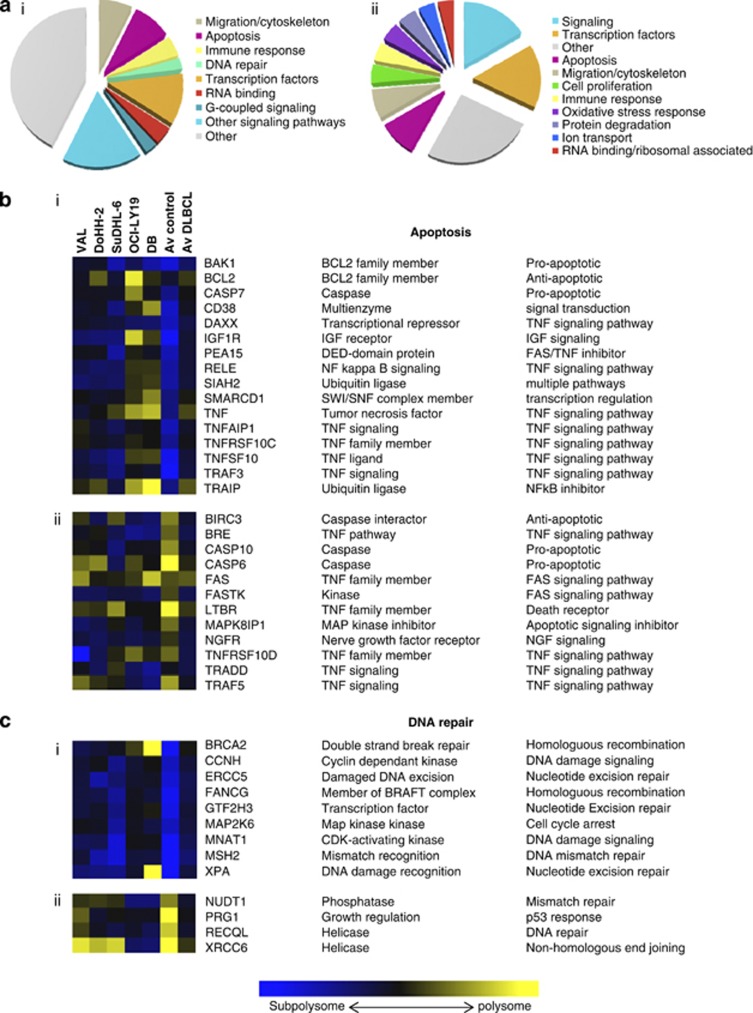
To identify selective changes in the translatome in DLBCL, polysome profiling was carried out. Cytoplasmic extracts prepared from five DLBCL cell lines (VAL, DoHH-2, SuDHL-6, OCI-LY19 and DB) and two control B-cell lines (GM03201, GM01953) derived from disease-free individuals were separated on 10–60% sucrose gradients (Supplementary Figure 1) and the RNA isolated. RNA derived from fractions 6–10 (polysomes) was compared with RNA from fractions 1–4 (subpolysomes) on microarrays to identify candidate mRNAs whose translational behavior differs significantly from the translation pattern observed in the control cells. These transcripts were then classified into different categories based on their biological function (Figure 1, Supplementary Figure 2). In DLBCL cell lines, mRNAs encoding anti-apoptotic proteins, transcription factors, proteins required for cell migration, DNA repair enzymes and subsets of RNA-binding proteins, including eIF4B (Figure 1, Supplementary Figure 2), were translationally upregulated. In contrast, mRNAs encoding pro-apoptotic proteins, cell signaling and a different subset of RNA-binding proteins were translationally downregulated (Figure 1a).
Figure 1.
mRNAs involved in apoptosis and DNA damage response that displayed significantly different ranking in DLBCL lines compared with control cells. (a) Pie charts showing the functional distribution of proteins whose mRNA is (i) more or (ii) less polysome-associated in DLBCL cells compared with control. (b and c) A color scale was used to represent the ratio of mRNA in subpolysome to polysome fractions. Transformed from the log2[polysome:subpolysome], where blue indicates subpolysome-associated and yellow signifies that the mRNA is predominantly polysome-associated. (b(i)) mRNAs associated with apoptotic processes that are translationally upregulated. Av control represents the average value for the GM03201 and GM01953 cell lines, av DLBCL the average value for VAL, DoHH-2, Sud-HL6, OCI-LY19 and DB cell lines. (b(ii)) mRNAs associated with apoptotic processes that are translationally downregulated. (c(i) mRNAs associated with DNA repair processes that are translationally upregulated. (c(ii)) mRNAs associated with DNA repair processes that are translationally downregulated.
Changes in gene expression and the synthesis of specific proteins in DLBCL validate cDNA microarray analysis
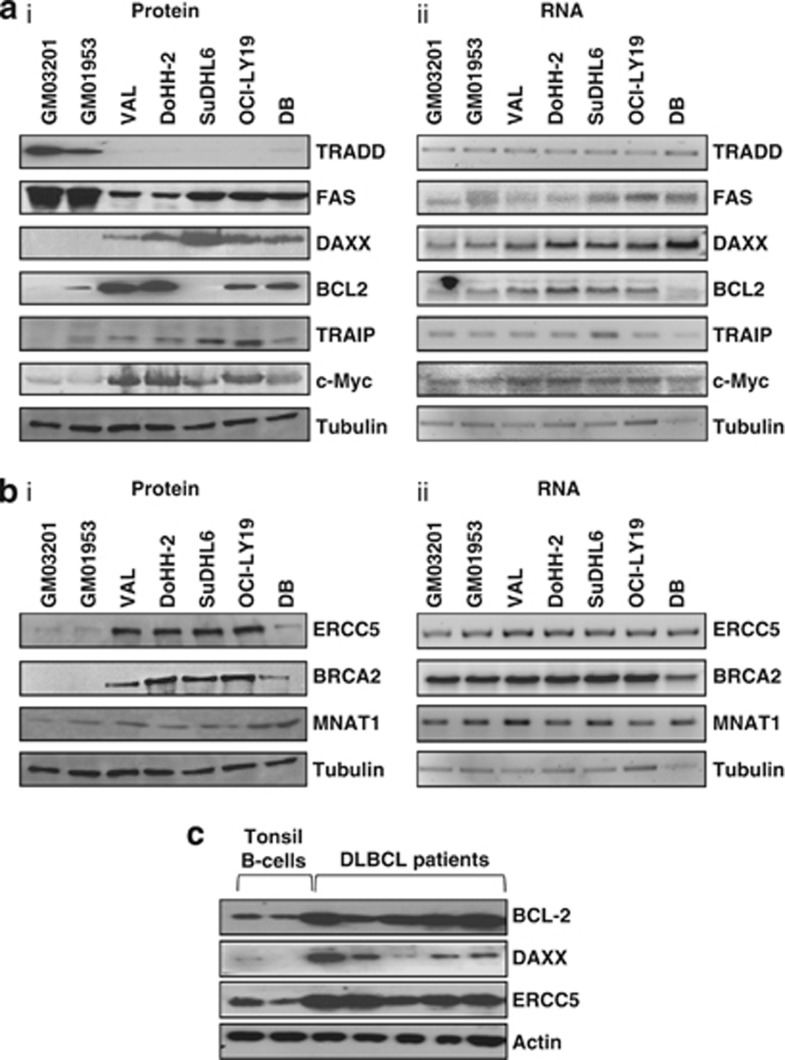
To assess the changes in expression of proteins that are associated with tumorigenesis, mRNAs whose protein products are involved in apoptosis and DNA repair were studied. Cell lysates were separated by SDS-polyacrylamide gel electrophoresis and immunoblotted for the proteins shown (Figures 2a and b, Supplementary Table 3). Differences in expression were observed in all cases, in agreement with the profiling data (Figure 1 and Supplementary Figure 2). For proteins that function in apoptosis, the data show that TRADD (tumor necrosis factor receptor type 1-associated death domain protein) and FAS were downregulated, whereas DAXX, BCL2, TRAIP and c-Myc were upregulated, albeit to different extents (Figure 2a(i)). There were no corresponding changes in the total amount of mRNA present (Figure 2a(ii), Supplementary Figure 3). For example, the level of TRADD mRNA was similar in the control cell lines and the DLBCL cell lines, and although FAS mRNA levels were more variable, neither of these correlated with the expression of the protein. The cell lines used in the study carry the t(14:18) translocation that couples the immunoglobulin heavy chain locus to the BCL2 gene resulting in transcriptional upregulation of BCL2 mRNA.30 As expected, the levels of BCL2 mRNA were higher in four out of five of the DLBCL-derived cell lines. However, this change was insufficient to account for the large increase in observed protein levels in some of the lymphoma cell lines. The differential expression of these apoptotic proteins was shown to affect cell sensitivity to FAS-mediated apoptosis (Supplementary Figure 4).
Figure 2.
mRNAs identified by the translational profiling show differential protein expression. (a(i) and b(i)) Protein extracts were generated from DLBCL cell lines or from control B cells (GM03201 and GM01953). These were subjected to SDS-polyacrylamide gel electrophoresis (PAGE), immunoblotted and probed with the antibodies indicated to assess differences in expression in the proteins from genes with a role in apoptosis (a) or DNA repair processes (b). (a(ii) and b(ii)) To measure total RNA levels, samples were taken and RNA was extracted. These samples were subjected to northern analysis using probes against DAXX, BCL2, FAS and MYC, or reverse transcriptase-PCR using primers against TRAIP, ERCC5, CCNH, BRCA2, MNAT1, actin and tubulin. Actin/tubulin were used as loading controls. The data show that there is a large increase in the expression of a number of proteins that function in apoptotic and DNA repair pathways with no net change in mRNA levels, in agreement with the array data. (c) B cells were purified from tumors obtained from individuals with DLBCL or from tonsils. Cell extracts were generated and samples separated by SDS-PAGE and immunoblotted with the antibodies shown. The data show that there is increased expression of DAXX, ERCC5 and BCL2 in patient samples in agreement with the cell line and the translational profiling data.
The translational profiling data also suggested that there would be differences in the synthesis of proteins that function in DNA repair. Therefore, western analysis was performed to examine the expression of ERCC5, BRCA2 and MNAT1 (Figure 2b(i), Supplementary Table 3). The levels of these proteins were increased with no corresponding changes in the levels of the mRNA, again strongly suggesting translational dysregulation (Figure 2b(ii), Supplementary Figure 3). Importantly, dysregulated expression of these proteins has not been reported previously in DLBCL.
Western blot analysis was carried out on patient tumor samples to examine the levels of BCL2, DAXX and ERCC5 (Figure 2c). The data show that similar changes also occur in patient samples with increased expression of these proteins detected when compared with B cells derived from control tonsils.
mRNAs with highly structured 5′-UTRs are translationally upregulated in DLBCL
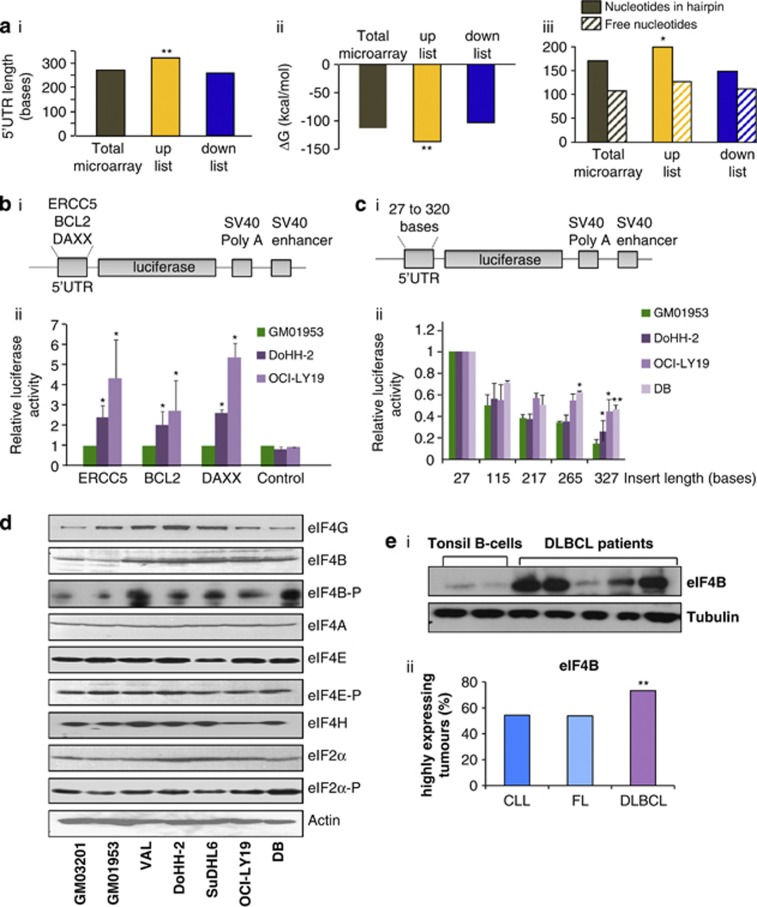
The enhanced translation of subsets of mRNAs in DLBCL could be due to unique RNA elements, as the majority of mammalian mRNAs contain regulatory RNA motifs, including upstream open reading frames, internal ribosome entry segments, terminal oligopyrimidine tracts and microRNA-binding sites.31 Therefore, the UTRs of the mRNAs in the ‘up' and ‘down' lists were examined to assess whether there were unique features that could contribute to their up- or downregulation. The proportion of mRNAs containing internal ribosome entry segments, upstream open reading frames, terminal oligopyrimidine tracts or microRNA target sites remained unchanged between the gene lists and the whole array (Supplementary Figure 5A). However, the data show a significant increase in the average 5′-UTR length and in the predicted minimum free energy (ΔG) of folding in the list of genes that were translationally upregulated in DLBCL, compared with the complete array (Figures 3a(i and ii)). Further examination of the 5′-UTR length (Supplementary Figure 5B) reveals that the cumulative distribution corresponding to the translationally upregulated group is shifted towards the longer lengths compared with the downregulated and unchanged groups; the ΔG data shows a similar pattern with a shift towards more negative free energies in the translationally upregulated group (Supplementary Figure 5C(i)). These data suggest that a population of mRNAs with long and/or structured 5′-UTRs is enriched in the translationally upregulated group.
Figure 3.
mRNAs containing highly structured 5′-UTRs are preferentially translated in DLBCL. (a) The cDNA microarray data were analysed and the average 5′-UTR lengths, and ΔG values were calculated for those mRNAs in the ‘up' and ‘down' lists, and the significance of the difference to the whole array was assessed by Student's t-tests. The data show that the mRNAs in the upregulated list have longer 5′-UTRs (P=0.002; i), have greater −ΔG values (P=0.001; ii) and have more nucleotides incorporated into hairpins (P=0.006; iii); all three criteria would be expected to decrease the translatability of mRNAs in these groups. (b(i)) The cDNAs corresponding to the 5′-UTRs of DAXX, ERCC5 and BCL2 were subcloned into a luciferase reporter vector based on pGL3. (b(ii)) The reporter constructs were transfected into cell lines derived from DLBCL and control B cell, and luciferase activity determined. The data show that there is greater expression of luciferase in the DLBCL-derived cell lines than in the control cell lines for all plasmids, suggesting that the 5′-UTRs are less inhibitory in these cell lines. Significance was assessed by paired Student's t-tests on three independent experiments (*P<0.05, **P<0.01 and ***P<0.001). (c(i)) Unrelated DNA sequences of increasing length (Supplementary Table 2) were subcloned into the 5′-UTR of pGL3 upstream of the luciferase cistron. (c(ii)) These plasmids were transfected into the control cell line (GM01953) and cell line derived from DLBCL (DoHH-2, OCI-LY19 and DB), and luciferase activity determined. (d) Protein extracts derived from cell lines representative of either DLBCL (Val, DoHH-2, SuDHL-6, OCI-LY19 and DB) and two control B-cell lines (GM03201 and GM01953) were separated by SDS-polyacrylamide gel electrophoresis (PAGE). Samples were immunoblotted with eukaryotic initiation factor (eIFs)-specific antibodies. The data show that only eIF4B levels were increased in these cell lines. (e(i)) Extracts were generated from B cells purified from tonsils of tumor-free individuals or tumors from patients with DLBCL, and separated by SDS-PAGE and immunoblotted to examine eIF4B levels. Tubulin was used as a loading control. The data show that eIF4B levels were elevated in the five DLBCL samples tested. (e(ii)) Tissue microarrays that contained tissues from 362 lymphomas, including 196 DLBCL, 76 FL and 52 chronic lymphocytic leukemias (CLLs) were probed by an antibody against eIF4B and scored as described in the experimental procedures section. These data show that there is significantly higher expression of eIF4B in DLBCL compared with that in CLL and FL. Significance between the different categories was assessed by χ2-test (*P<0.05).
To determine whether the higher overall ΔG observed in the translationally upregulated mRNAs was simply a consequence of the longer 5′-UTR lengths, a number of criteria were examined, including the distributions of the number of nucleotides that were either paired (Supplementary Figure 5C(ii)) or free (Supplementary Figure 5C(iii)), and the longest string of nucleotides involved in a structure (Supplementary Figure 5C(iv)) or unstructured regions (Supplementary Figure 5C(v)). These analyses show that there is a significant enrichment of mRNAs containing structure in translationally upregulated mRNAs in DLBCL, whereas the number of free nucleotides and the size of unstructured regions is similar in all three groups. Taken together, these data suggest that the degree of structure is more important than length in determining enrichment in the ‘up' list.
To investigate this finding further, the 5′-UTRs of three of the mRNAs that were shown to be translationally upregulated (ERCC5, BCL2 and DAXX) were subcloned into luciferase reporter vectors (Figure 3b(i)) and transfected into DLBCL-derived cell lines (Figure 3b(ii)). There was an increase in the synthesis of the luciferase reporter enzyme from all constructs in the DLBCL-derived cell lines when compared with the control, suggesting that enhanced translation of those mRNA in DLBCL was dependent upon features in their 5′-UTRs. Therefore, vector constructs were generated to examine the effect of length/structure on reporter protein synthesis in DLBCL (Figure 3c(i), Supplementary Table 2). These constructs were transfected into the control and DLBCL-derived cell lines and luciferase expression was determined. The data show that constructs with longer 5′-UTRs were repressed in the control cells, and that this repression is significantly less in DLBCL cell lines (Figure 3c(ii)). It has been shown in other systems that modifying the protein components of the translation apparatus has the net effect of altering the dynamic range of translation efficiencies across the entire transcriptome, so that those mRNAs that are normally poorly translated, for example, those that contain highly structured 5′-UTRs as shown here, are selectively upregulated.4, 32 Therefore, the levels and phosphorylation status of the translational apparatus in DLBCL were assessed (Figure 3d). Interestingly, the data show that whereas there was no difference in the expression of the eIFs that have been shown previously to be associated with a transformed phenotype, namely eIF4E, eIF4G and eIF4A,3, 33 there was an increase in the expression of eIF4B and a relative increase in eIF4B-P (Figure 3d). eIF4B stimulates translation by increasing the activity of the helicase eIF4A,34 it is required for 48S complex formation via its interaction with poly(A)-binding protein and contributes to selective translation of mRNAs with highly structured 5′-UTRs.3, 5, 35 Importantly, eIF4B expression was also increased in DLBCL patient samples, albeit to differing extents (Figure 3e(i)). To analyze eIF4B expression from a larger number of patient samples simultaneously, tissue microarrays were used (Supplementary Figure 6A, Supplementary Table 4). These contained tissues from 362 lymphomas, including 196 DLBCL, 76 FLs and 52 chronic lymphocytic leukemias. In agreement with the cell line data, there was a significant increase in eIF4B expression in the DLBCL tumors compared with chronic lymphocytic leukemia and FL (Figure 3e(ii)). Interestingly, there was a correlation between eIF4B overexpression and the levels of DAXX and ERCC5 in the tumor samples (Figure 2c) and in tissue microarrays (Supplementary Figure 6B).
Translational dysregulation of ERCC5, DAXX and BCL2 correlates with enhanced eIF4B expression
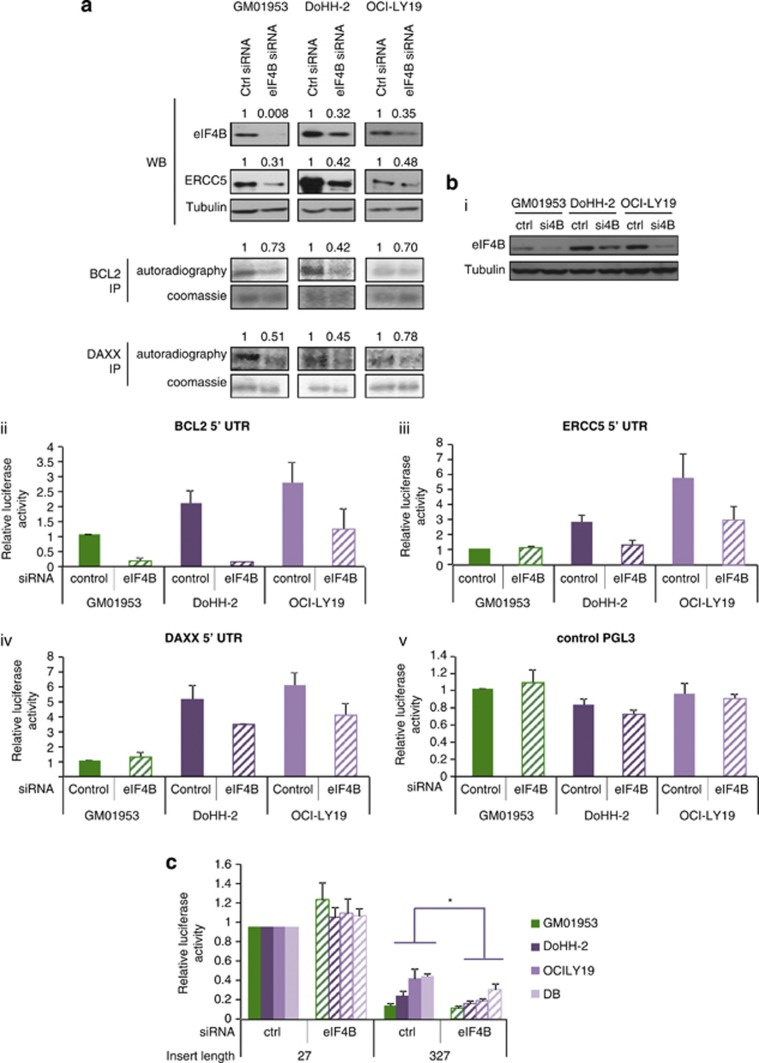
To test the hypothesis that increased eIF4B levels directly elevated the synthesis of subsets of proteins, including DAXX and BCL2 in DLBCL, cells were transfected with siRNA directed against eIF4B mRNA and potential target proteins were analyzed, either by western blotting (ERCC5) or pulse-labeling and immunoprecipitation (BCL2 and DAXX, due to the long half-lives of these proteins, which would mask changes in net synthesis rates). When eIF4B levels were reduced, there was a decrease in the expression of ERCC5, BCL2 and DAXX (Figure 4a).
Figure 4.
eIF4B modulation restores repression of translation on mRNAs that is exerted by structured 5′-UTRs. (a) Cell lines derived from patients with DLBCL (DoHH-2 and OCI-LY19) and control B cells (GM01953) were transfected with siRNA to eIF4B. After 48 h, cell extracts were generated, which were separated by SDS-polyacrylamide gel electrophoresis and immunoblotted for eIF4B and ERCC5 levels. Pulse-labeled immunoprecipitation was used to measure the levels of BCL2 and DAXX after 24 h of eIF4B siRNA treatment, as these proteins have long half-lives. The gels were quantified using Image J software. The numbers shown represent expression normalized to tubulin (eIF4B and ERCC5) or autoradiography normalized to Coomassie staining (BCL2 and DAXX). The data show that in DLBCL-derived cells reducing eIF4B expression decreased the expression of DAXX, ERCC5 and BCL2. (b) The levels of eIF4B were reduced by siRNA (i). The reporter plasmids containing the 5′-UTRs of BCL2, ERCC5, DAXX or a control pGL3 (ii–v) were transfected into the cell lines ±siRNA to eIF4B. After 48 h, luciferase activity was determined. The data show that there was a reduction in luciferase activity when the level of eIF4B was reduced for all the 5′-UTRs examined. Significance was assessed by paired Student's t-tests on three independent experiments (*P<0.05, **P<0.01, ***P<0.001). (c) Plasmids containing unrelated DNA sequences of 27 or 327 nucleotides upstream of the luciferease cistron were transfected into the control cell line (GM01953) or cell lines derived from DLBCL (DoHH-2, OCI-LY19 and DB) ±siRNA to eIF4B, and luciferase activity determined.
To examine whether the regulation eIF4B target mRNAs was mediated by elements in their 5′-UTRs, the level of eIF4B was reduced by siRNA (Figure 4b(i)) and cells were transfected with reporter plasmids (Figure 4b(ii–v)). There were decreases in the expression of luciferase from the reporter plasmids that contained the ERCC5, BCL2 and DAXX 5′-UTRs in the lymphoma-derived cell lines when eIF4B was reduced (Figure 4b(iii–v)). These data suggest that high eIF4B levels could contribute to the enhanced synthesis of subsets of proteins, including DAXX, BCL2 and ERCC5, by unwinding structured 5′-UTRs. To confirm that the relief of repression on structured mRNAs was directly due to the higher levels of eIF4B in these cells and independent of sequence, cells were co-transfected with siRNAs against eIF4B and constructs containing structured 5′-UTRs of unrelated sequence (Supplementary Table 2). In this case, the enhanced synthesis that was observed in DLBCL-derived cells when compared with the control lines was ablated (Figure 4c).
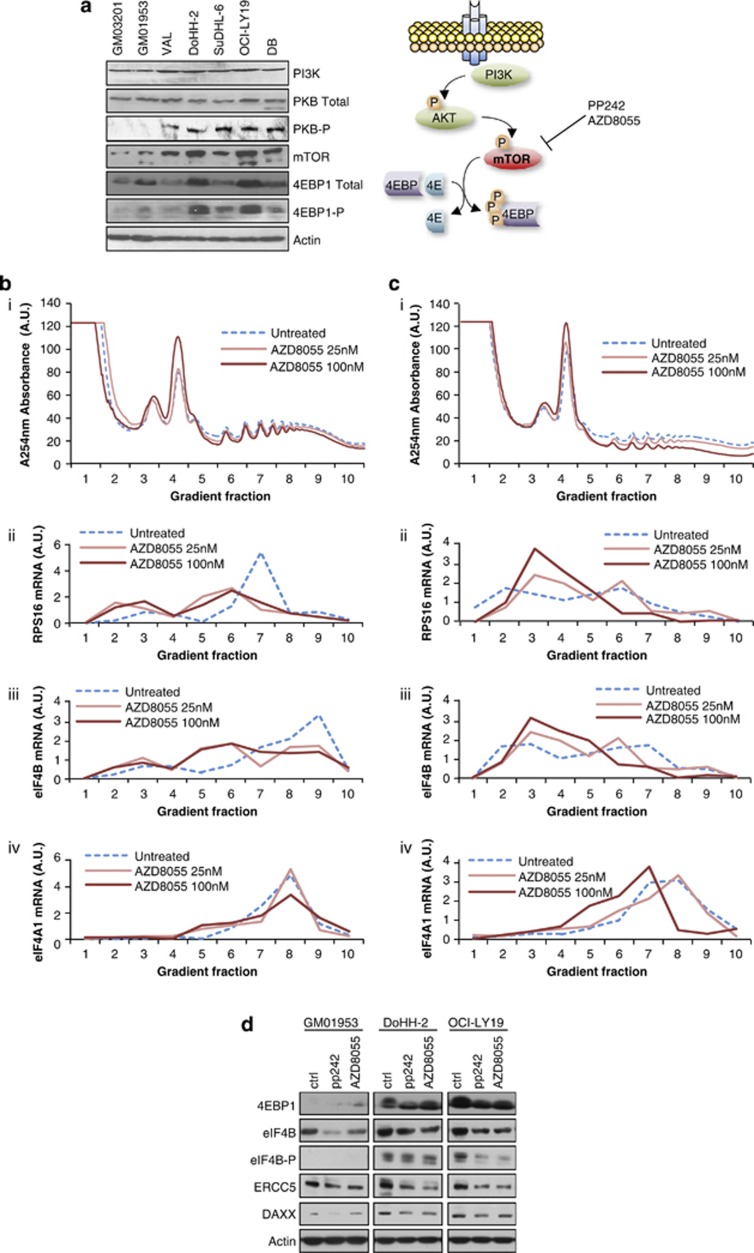
Signaling via the mTOR signaling pathway upregulates eIF4B expression
The profiling data (Figure 1, Supplementary Figure 2) indicated that eIF4B was also dysregulated in DLBCL at the level of translation. Several lines of evidence suggest that the translational dysregulation of eIF4B could result from aberrant signaling through the mTOR pathway. Activation of the mTOR pathway has been shown previously in DLBCL, although not in the cell lines used herein, and a recent study has also reported activation of mTOR in a subset of DLCBL patients.36 Moreover, eIF4B was present on two lists of genes whose expression changed following mTOR inhibition.37, 38 Therefore, DLBCL and control cells were immunoblotted to assess the expression and phosphorylation status of protein kinase B/AKT, mTOR and 4EBP1. There was an increase in expression and phosphorylation of these proteins in the DLBCL-derived cell lines (Figure 5a). Although mTOR activation induces phosphorylation of eIF4B,35 our data show that there are changes in the total level of eIF4B protein in DLBCL. To assess whether mTOR inhibition decreased the translation of eIF4B in DLBCL, cells were incubated with the inhibitor AZD8055. Polysomes were separated by sucrose density-gradient centrifugation, and quantitative reverse transcriptase-PCR was carried out to assess changes in polysomal distribution of mRNAs encoding a known mTOR target, RPS16 and eIF4B (Figures 5b(i–iv) and c(i–iv)). eIF4A was used as a control. The data showed a shift of RPS16 and eIF4B mRNAs from the polysomes following incubation with AZD8055 (Figures 5b(ii, iii) and c(ii, ciii)), but little or no effect on eIF4A mRNA (Figures 5b(iv) and c(iv)), which again suggests that eIF4B expression is regulated via signaling through mTOR.37, 38
Figure 5.
Signaling through mTOR links eIF4B expression to DAXX and ERCC5. (a(i)) Cell lines derived from DLBCL or control cells, as shown, were lysed, separated by SDS-polyacrylamide gel electrophoresis (PAGE), and immunoblottings were probed with antibodies against phosphoinositide 3-kinase (PI3K), protein kinase B (PKB), mTOR and 4EBP1. The data suggest that the mTOR signaling pathway is upregulated in these cells when compared with that in the controls. (a(ii)) Schematic to illustrate signaling through the mTOR pathway. (b and c) Two DLBCL cell lines DoHH-2 (b) and OCI-LY19 (c) were treated with the mTOR inhibitor AZD8055 at 25 or 100 nM. After 2 h, cells were collected and post-nuclear fractions were applied to sucrose density gradients to separate the actively translating polysomes from the subpolysomal material (b(i) and c(i)). RNA was isolated from each fraction and quantitative reverse transcriptase-PCR was used to assess the proportion of RPS16 (b(ii) and c(ii)), eIF4B (b(iii) and c(iii)) and eIF4A (b(iv) and c(iv)) associated with each fraction. (d) Control cell line (GM01953) or DLBCL lines (DoHH-2 and OCI-LY19) were treated with PP242 or AZD8055, cell extracts were generated, separated by SDS-PAGE and immunoblotted for 4EBP1-P, eIF4B, eIF4B-P, ERCC5 and DAXX. The data show that the reduction in eIF4B as a result of exposure to mTOR inhibitors also reduces the expression of its downstream targets ERCC5 and DAXX.
To assess the effect of mTOR inhibitors PP242 and AZD8055 on eIF4B protein levels, cells were incubated with these compounds and immunoblottings carried out. As expected, there were changes in the degree of phosphorylation of 4EBP1 and eIF4B35 but, importantly, also a decrease in expression of eIF4B, and two of its downstream targets, ERCC5 and DAXX, albeit to different extents (Figure 5d).
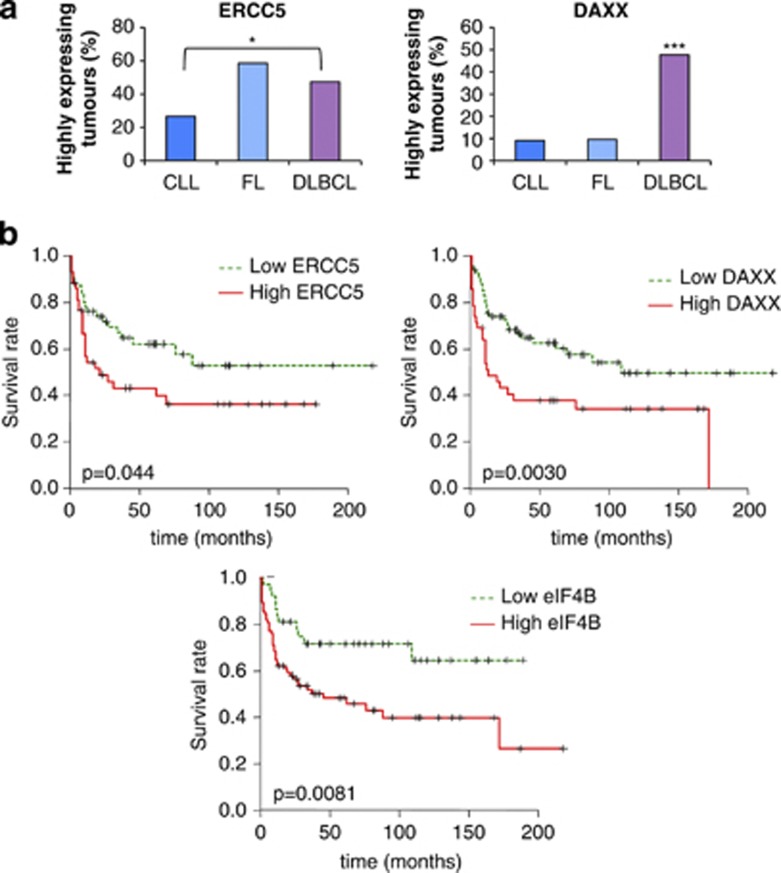
Differences in eIF4B, ERCC5 and DAXX expression in cell lines correlate with alterations in B-cell lymphoma tumor samples and patient survival
It was then important to assess whether the differences in protein expression were also present in patients with DLBCL and whether this was associated with patient survival. It is well established that BCL2 is upregulated in different non-Hodgkin lymphoma types,39, 40, 41 and its elevated expression is associated with poor prognosis in DLBCL;41 hence, the role of this protein in patient survival was not examined further. However, DAXX and ERCC5 expression have not been studied previously in DLBCL in this regard. Therefore, to determine the expression of DAXX and ERCC5 in a large number of patient samples simultaneously, tissue microarrays were used (Supplementary Figure 6, Supplementary Table 4). ERCC5 expression was significantly higher in DLBCL when compared with that in chronic lymphocytic leukemia (Figure 6a(i)). DAXX protein expression in DLBCL was increased significantly when compared with that in FL and chronic lymphocytic leukemia (Figure 6a(ii)). Kaplan–Meier analysis was performed to determine whether there were differences in survival associated with the increased expression of DAXX, ERCC5 and eIF4B (Figures 6b(i–iii)). Importantly, the data show that high DAXX, ERCC5 and eIF4B expression correlated with poor overall survival for all subtypes of DLBCL, providing three new prognostic markers that would identify those with worse outcome for this disease (Figures 6b(i–iii)).
Figure 6.
Tissue microarray shows that altered expression of the proteins identified by translational profiling in tumors from patients with B-cell lymphoma corresponds with survival. (a) Tissue microarrays that contained tissues from 362 lymphomas, including 196 DLBCL, 76 FL and 52 chronic lymphocytic leukemias (CLLs) were probed with antibodies against ERCC5 (i) and DAXX (ii), and scored as described in the experimental procedures section. The graphs represent percentage of tumors from each type expressing high levels of the scored protein. The data show that there are differences in expression of DAXX in the DLBCL patient samples compared with the other tumor types, and for ERCC5 a difference in the expression in FL and DLBCL when compared with CLL. Significance between the different categories was assessed by χ2-test (*P<0.05, **P<0.01, ***P<0.001). (b) Patients were separated into groups that have low or high expression of ERCC5 (i) or DAXX (ii), or eIF4B (iii). Survival curves were generated using Kaplan–Meier approximation to compare overall survival in different groups of patients. The significance of the changes in survival was assessed using log-rank P-value. The data show that there are significant decreases in survival in patients that have high expression for ERCC5 (P=0.044), DAXX (P=0.003) and eIF4B (P=0.0081).
Discussion
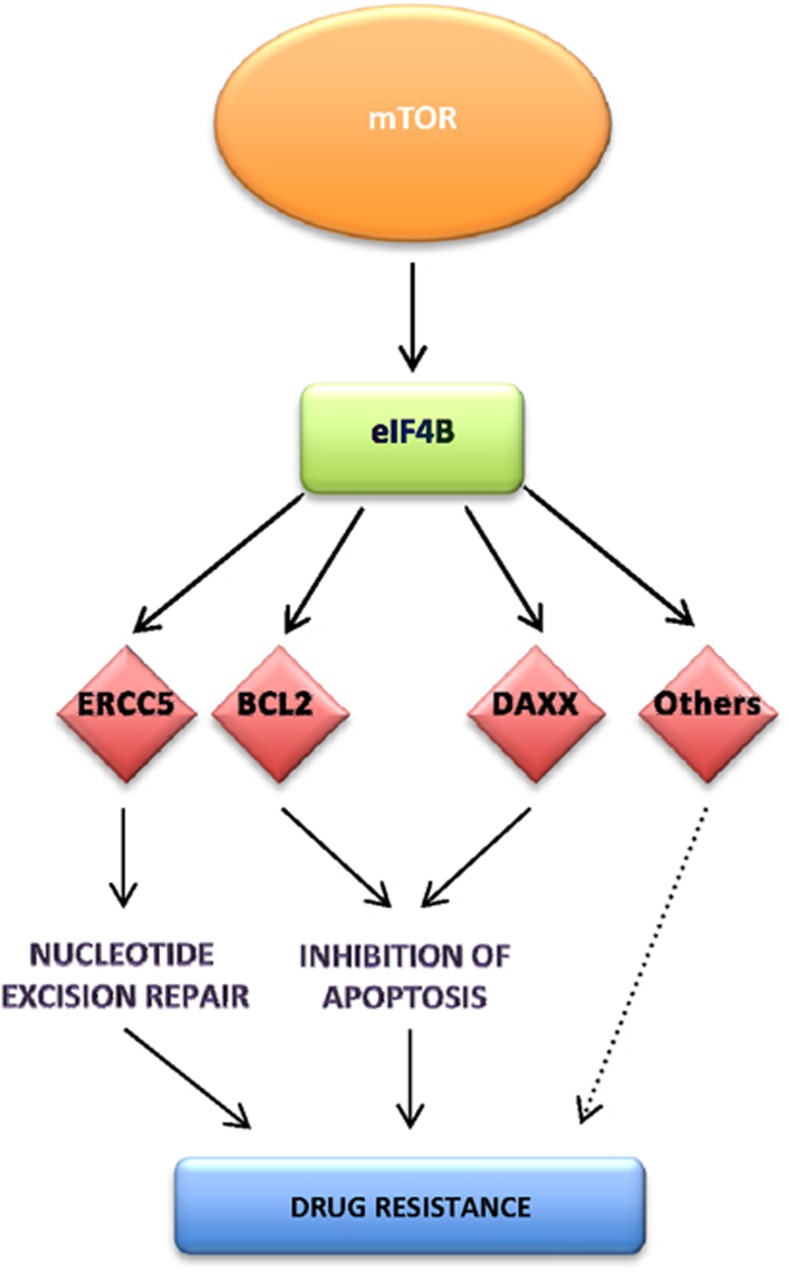
Our data allow us to propose a model whereby aberrant signaling through mTOR leads to an upregulation in eIF4B, which directly drives the translation of a subset of mRNAs that contain structured 5′-UTRs, including those involved in tumorigenesis and chemoresistance, such as DAXX, BCL2 and ERCC5 (Figure 7). Importantly, we identified eIF4B and two of its targets, DAXX and ERCC5, as markers of poor prognosis in DLBCL, suggesting that the increased expression and activation of this protein drives tumorigenesis (Figures 4, 6 and ).
Figure 7.
A schematic diagram to represent the changes in DLBCL that occur as a consequence of eIF4B overexpression. Taken together, the data suggest a model wherein increased signaling through mTOR in DLBCL increases the expression of eIF4B, which drives the translation of mRNAs that contain highly structured 5′-UTRs and lead to the upregulated synthesis of ERCC5, DAXX and BCL2. Increased expression of these proteins negatively impacts upon patient survival, providing three new prognostic markers for this disease: eIF4B, DAXX and ERCC5.
The data indicate that the increase in eIF4B expression occurs at the level of translation (Supplementary Figure 3 and Figure 5) and, in agreement with previous data,38 that this is downstream of mTOR signaling (Figures 5 and 7). It has been suggested that although signaling through mTORC1 is required for the selective upregulation of terminal oligopyrimidine tract containing mRNAs, this pathway does not directly control the translation of mRNAs with long and/or structured 5′-UTRs.38 However, we show that in DLBCL, upregulation of eIF4B levels by the mTOR pathway increases the translation of selected mRNAs that contain structured 5′-UTRs (Figures 4, 5 and ). Our data are supported by studies, which show that silencing of eIF4B in HeLa cells reduces the translation of selected mRNAs that contain structured 5′-UTRs.35
We demonstrate that high DAXX expression correlated with poor patient survival (Figure 6). The cellular role of DAXX is complex, with opposing effects being attributed for DAXX in tumor necrosis factor and FAS signaling to apoptosis.42, 43, 44 In addition, mutations that inactivate DAXX function are associated with prolonged survival of patients with pancreatic cancer.45
It is well established that increased expression of BCL2 is associated with DLBCL and it has been shown previously that high expression of BCL2 is associated with poor survival,46, 47 particularly for germinal center B-cell-like such as DLBCL.47 The data presented here suggest that increased translation of BCL2 also contributes to the elevated levels of this protein in DLBCL (Figures 1). Similarly, we show that ERCC5 expression is increased in DLBCL (Figures 1). ERCC5 is an essential component of the nucleotide excision repair pathway that is used to repair damaged DNA following exposure to therapeutic agents such as cisplatin.48 In addition, it has been shown recently that eIF4B is present as part of a gene expression signature of acquired resistance to cisplatin in patients with gastric cancer.49 Inhibition of these DNA repair pathways, as part of combination therapy with mTOR inhibitors, could therefore provide alternative treatment options for patients with DLBCL that respond poorly to conventional therapy.48 In this regard, a recent trial has been established to evaluate mTOR inhibitors for the treatment of patients with relapsed or refractory DLBCL.50
In summary, our data show that increased eIF4B expression makes a significant contribution to the development of tumorigenesis in DLBCL. Targeting the proteins that are translationally upregulated and which have not been previously identified as associated with this disease, for example, DAXX or ERCC5, in addition to direct methods to reduce eIF4B expression or activity, could provide new therapeutic options, particularly for patients who do not respond to conventional treatment.
Acknowledgments
This work was funded by grants from the LLR (formerly LRF; EH, KH, LCC and PG) and MRC program funding (AEW, MB and MM). TS was funded by HSH Prince Albert II of Monaco and the Government of the Principality of Monaco. AEW holds a BBSRC Professorial Fellowship and MB holds a MRC Senior Research Fellowship. RVS was funded by the BBSRC.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Stoneley M, Willis AE. Aberrant regulation of translation initiation in tumorigenesis. Curr Mol Med. 2003;3:597–603. doi: 10.2174/1566524033479474. [DOI] [PubMed] [Google Scholar]
- Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, et al. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol Cell. 2007;28:501–512. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Le Quesne JP, Spriggs KA, Bushell M, Willis AE. Dysregulation of protein synthesis and disease. J Pathol. 2010;220:140–151. doi: 10.1002/path.2627. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell M, Wood W, Carpenter G, Pain VM, Morley SJ, Clemens MJ. Disruption of the interaction of mammalian protein synthesis eukaryotic initiation factor 4B with the poly(A)-binding protein by caspase- and viral protease-mediated cleavages. J Biol Chem. 2001;276:23922–23928. doi: 10.1074/jbc.M100384200. [DOI] [PubMed] [Google Scholar]
- Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, et al. mRNA helicases: the tacticians of translational control. Nat Rev Mol Cell Biol. 2011;12:235–245. doi: 10.1038/nrm3083. [DOI] [PubMed] [Google Scholar]
- Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N. eIF4E—from translation to transformation. Oncogene. 2004;23:3172–3179. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- Averous J, Proud CG. When translation meets transformation: the mTOR story. Oncogene. 2006;25:6423–6435. doi: 10.1038/sj.onc.1209887. [DOI] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Bea S, Zettl A, Wright G, Salaverria I, Jehn P, Moreno V, et al. Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction. Blood. 2005;106:3183–3190. doi: 10.1182/blood-2005-04-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JZ, Sanger WG, Greiner TC, Staudt LM, Weisenburger DD, Pickering DL, et al. The t(14;18) defines a unique subset of diffuse large B-cell lymphoma with a germinal center B-cell gene expression profile. Blood. 2002;99:2285–2290. doi: 10.1182/blood.v99.7.2285. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Staudt LM. Gene expression profiling of diffuse large B-cell lymphoma. Leuk Lymphoma. 2003;44 (Suppl 3:S41–S47. doi: 10.1080/10428190310001623775. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- Savage KJ, Monti S, Kutok JL, Cattoretti G, Neuberg D, De Leval L, et al. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood. 2003;102:3871–3879. doi: 10.1182/blood-2003-06-1841. [DOI] [PubMed] [Google Scholar]
- Friedberg JW. New strategies in diffuse large B-cell lymphoma: Translating findings from gene expression analyses into clinical practice. Clin Cancer Res. 2011;17:6112–6117. doi: 10.1158/1078-0432.CCR-11-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhaeusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Li JJ, Bickel PJ, Biggin MD.System wide analyses have underestimated protein abundances and transcriptional importance in mammalsarXiv.org eprint archive, arXiv:1212.0587v6. [DOI] [PMC free article] [PubMed]
- Clemens MJ, Bushell M, Morley SJ. Degradation of eukaryotic polypeptide chain initiation factor (eIF) 4G in response to induction of apoptosis in human lymphoma cell lines. Oncogene. 1998;17:2921–2931. doi: 10.1038/sj.onc.1202227. [DOI] [PubMed] [Google Scholar]
- West MJ, Stoneley M, Willis AE. Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene. 1998;17:769–780. doi: 10.1038/sj.onc.1201990. [DOI] [PubMed] [Google Scholar]
- Johannes G, Carter MS, Eisen MB, Brown PO, Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc Natl Acad Sci USA. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvilleur E, Bauer M, Goldschneider D, Mergui X, de la Motte A, Benard J, et al. p73alpha isoforms drive opposite transcriptional and post-transcriptional regulation of MYCN expression in neuroblastoma cells. Nucleic Acids Res. 2008;36:4222–4232. doi: 10.1093/nar/gkn394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics. 2006;22:2825–2827. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- Obermann EC, Went P, Zimpfer A, Tzankov A, Wild PJ, Stoehr R, et al. Expression of minichromosome maintenance protein 2 as a marker for proliferation and prognosis in diffuse large B-cell lymphoma: a tissue microarray and clinico-pathological analysis. BMC Cancer. 2005;5:162. doi: 10.1186/1471-2407-5-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmele W, Stegner HE. [Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue] Pathologe. 1987;8:138–140. [PubMed] [Google Scholar]
- Therneau TM, Li H. Computing the Cox model for case cohort designs. Lifetime Data Anal. 1999;5:99–112. doi: 10.1023/a:1009691327335. [DOI] [PubMed] [Google Scholar]
- Lorenz WA, Clote P. Computing the partition function for kinetically trapped RNA secondary structures. PLoS One. 2011;6:e16178. doi: 10.1371/journal.pone.0016178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MH, Hermans J, Parker J, Krol AD, Kluin-Nelemans JC, Haak HL, et al. Clinical significance of bcl2 and p53 protein expression in diffuse large B-cell lymphoma: a population-based study. J Clin Oncol. 1996;14:2131–2138. doi: 10.1200/JCO.1996.14.7.2131. [DOI] [PubMed] [Google Scholar]
- Pichon X, Wilson LA, Stoneley M, Bastide A, King HA, Somers J, et al. RNA binding protein/RNA element interactions and the control of translation. Curr Protein Pept Sci. 2012;13:294–304. doi: 10.2174/138920312801619475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EH, Zhang F, Warringer J, Sunnerhagen P, Hinnebusch AG. Depletion of eIF4G from yeast cells narrows the range of translational efficiencies genome-wide. BMC Genomics. 2011;12:68. doi: 10.1186/1471-2164-12-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagden SP, Willis AE. The biological and therapeutic relevance of mRNA translation in cancer. Nat Rev Clin Oncol. 2011;8:280–291. doi: 10.1038/nrclinonc.2011.16. [DOI] [PubMed] [Google Scholar]
- Rogers GW, Jr, Richter NJ, Lima WF, Merrick WC. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem. 2001;276:30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- Shahbazian D, Parsyan A, Petroulakis E, Topisirovic I, Martineau Y, Gibbs BF, et al. Control of cell survival and proliferation by mammalian eukaryotic initiation factor 4B. Mol Cell Biol. 2010;30:1478–1485. doi: 10.1128/MCB.01218-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebestyen A, Sticz TB, Mark A, Hajdu M, Timar B, Nemes K, et al. Activity and complexes of mTOR in diffuse large B-cell lymphomas—a tissue microarray study. Mod Pathol. 2012;25:1623–1628. doi: 10.1038/modpathol.2012.141. [DOI] [PubMed] [Google Scholar]
- Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 2012;485:109–113. doi: 10.1038/nature11083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto Y, Bashir MM, Givol I, Cossman J, Jaffe E, Croce CM. DNA rearrangements in human follicular lymphoma can involve the 5' or the 3' region of the bcl-2 gene. Proc Natl Acad Sci USA. 1987;84:1329–1331. doi: 10.1073/pnas.84.5.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggins AG, Pepper CJ. The role of Bcl-2 family proteins in chronic lymphocytic leukaemia. Leuk Res. 2010;34:837–842. doi: 10.1016/j.leukres.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Barrans SL, Carter I, Owen RG, Davies FE, Patmore RD, Haynes AP, et al. Germinal center phenotype and bcl-2 expression combined with the International Prognostic Index improves patient risk stratification in diffuse large B-cell lymphoma. Blood. 2002;99:1136–1143. doi: 10.1182/blood.v99.4.1136. [DOI] [PubMed] [Google Scholar]
- Perlman R, Schiemann WP, Brooks MW, Lodish HF, Weinberg RA. TGF-beta-induced apoptosis is mediated by the adapter protein Daxx that facilitates JNK activation. Nat Cell Biol. 2001;3:708–714. doi: 10.1038/35087019. [DOI] [PubMed] [Google Scholar]
- Yang X, Khosravi-Far R, Chang HY, Baltimore D. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell. 1997;89:1067–1076. doi: 10.1016/s0092-8674(00)80294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LY, Chen JD. Daxx silencing sensitizes cells to multiple apoptotic pathways. Mol Cell Biol. 2003;23:7108–7121. doi: 10.1128/MCB.23.20.7108-7121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer MH, Hermans J, Wijburg E, Philippo K, Geelen E, van Krieken JH, et al. Clinical relevance of BCL2, BCL6, and MYC rearrangements in diffuse large B-cell lymphoma. Blood. 1998;92:3152–3162. [PubMed] [Google Scholar]
- Dunleavy K, Wilson WH. Differential role of BCL2 in molecular subtypes of diffuse large B-cell lymphoma. Clin Cancer Res. 2011;17:7505–7507. doi: 10.1158/1078-0432.CCR-11-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord CJ, Ashworth A. Bringing DNA repair in tumors into focus. Clin Cancer Res. 2009;15:3241–3243. doi: 10.1158/1078-0432.CCR-09-0434. [DOI] [PubMed] [Google Scholar]
- Kim HK, Choi IJ, Kim CG, Kim HS, Oshima A, Michalowski A, et al. A gene expression signature of acquired chemoresistance to cisplatin and fluorouracil combination chemotherapy in gastric cancer patients. PLoS One. 2011;6:e16694. doi: 10.1371/journal.pone.0016694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witzens-Harig M, Memmer ML, Dreyling M, Hess G. A phase I/II trial to evaluate the safety, feasibility and activity of salvage therapy consisting of the mTOR inhibitor Temsirolimus added to standard therapy of Rituximab and DHAP for the treatment of patients with relapsed or refractory diffuse large cell B-Cell lymphoma - the STORM trial. BMC Cancer. 2013;13:308. doi: 10.1186/1471-2407-13-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.