Abstract
APOBEC1 is a cytidine deaminase that edits messenger RNAs and was the first enzyme in the APOBEC family to be functionally characterized. Under appropriate conditions APOBEC1 also deaminates deoxycytidine in single-stranded DNA (ssDNA). The other ten members of the APOBEC family have not been fully characterized however several have deoxycytidine deaminase activity on ssDNAs. Despite the nucleic acid substrate preferences of different APOBEC proteins, a common feature appears to be their intrinsic ability to bind to RNA as well as to ssDNA. RNA binding to APOBEC proteins together with protein-protein interactions, post-translation modifications and subcellular localization serve as biological modulators controlling the DNA mutagenic activity of these potentially genotoxic proteins.
Keywords: APOBEC, DNA deaminase, mutation, regulation, RNA editing
1. Introduction
The purpose of this review is to familiarize the reader with the proteins in the APOBEC family in order to better appreciate differences in their functional roles as well as to describe cellular and viral control mechanisms that determine APOBEC activities. The review begins with Apolipoprotein B Editing Catalytic subunit 1 (APOBEC1 or A1) because it is the founding member of the family [1]. All family members have in common a zinc-dependent cytidine deaminase domain (ZDD) that is identifiable through its primary amino acid motif and a conserved super-secondary structure.
1.1. Overview of the requirements for apolipoprotein B mRNA editing
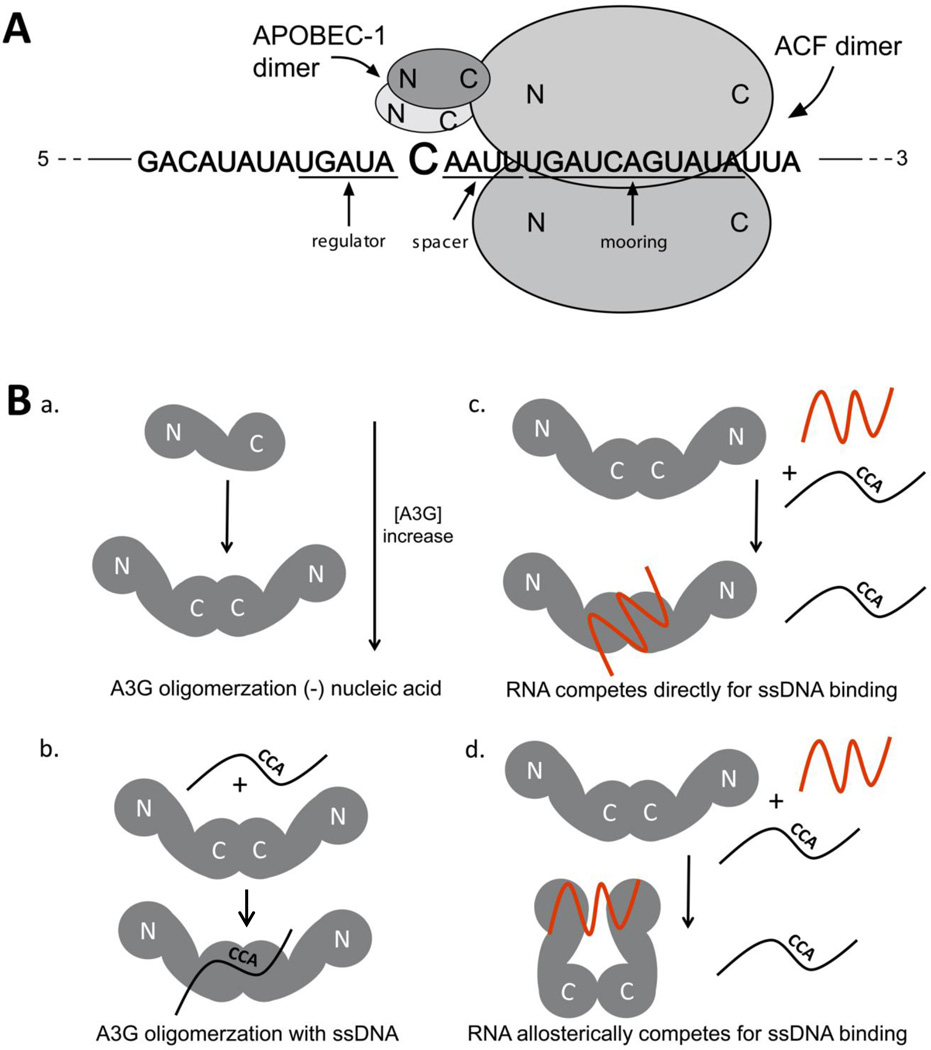
Apolipoprotein B (apoB) mRNA (C to U editing) and the glutamate receptor mRNA (A to I editing) were the first mRNAs discovered to be edited in mammalian cells circa the late 1980s [2]. Human liver and intestine produced a long and short form of the apolipoprotein B (ApoB) protein and the discovery of apoB mRNA editing resulted from research to determine the molecular basis for this polymorphism. Sequencing revealed a single nucleotide difference between mRNA and the genomically encoded sequence which was attributed to post-transcriptional RNA editing [3, 4]. The cis-acting sequences required for editing site recognition flanking the cytidine to be edited, in particular the 11 nt ‘mooring sequence’ (Figure 1A), had already been completely defined [2] before A1 was discovered [5]. However A1 is a low-affinity RNA-binding protein [6–8] and its ability to edit mRNA could only be realized in cells or cell extracts if they contained the RNA-binding protein APOBEC1 Complementation Factor (ACF) [9–11]. A1 dimers [7, 12] and RNA-bridged dimers of ACF [13] make up the minimal composition of the 27S editosome [14, 15] (Figure 1A).
Figure 1. Models for A1 and A3G complexes with nucleic acids.
A. The tripartite apoB mRNA editing motif consisting of the mooring sequence, and spacer 3’ of the C to be edited and the enhancer element 5’ of the edited C is shown with a cartoon of an editosome assembled upon it. An A1 C-terminal dimer is positioned for site-selective C to U editing by virtue of its association with ACF dimers that are shown bound to the mooring sequence. For the purposes of presentation only one A1 dimer is shown bound to one ACF through C-terminal to N-terminal (respectively) interactions (although the precise stoichiometry is unknown). Most of the three RNA recognition motifs comprising greater than the N-terminal half of ACF are required for optimal A1 binding. ACF dimerization requires only the N-terminal half of ACF.
B. a. An A3G monomer is shown containing an N- and C-terminal ZDD (label as ‘N’ and ‘C’). Nucleic acid deficient A3G forms a heterogeneous mixture of oligomers consisting mostly of dimers in solution and an A3G concentration-dependent small population of monomers and tetramers. b. A3G dimers to bind ssDNA substrates (black line with the CCA editing site) and must form at minimum a tetramer for enzymatic activity. c. RNA (red line) competes for ssDNA binding by displacing ssDNA and binding at the same site or d. RNA competes for ssDNA binding by binding at a distal site and causing an allosteric change in A3G conformation, preventing ssDNA binding.
1.2. Site-specific editing
The primary editing site at nt 6666 in apoB mRNA is a CAA glutamine codon that is deaminated to a UAA premature stop codon. Unedited and edited mRNAs coexist at varying ratios in editing-competent cells because tissue-specific and metabolically regulated differences in editing efficiency and because edited apoB mRNA is stabilized through the ability of ACF to blunt nonsense codon mediated mRNA degradation [16]. The specificity of this editing event is apparent in that apoB mRNA contains 3,315 cytidines of which 375 are in the correct reading frame and 100 are CAA. The mooring sequence is necessary and sufficient in determining whether a 5’ located cytidine is a candidate for editing [17–21]. However, editing activity itself is determined by the expression and nuclear retention of A1 and ACF [14, 22, 23] (see Sections 3.1 and 4) and has a temporal and spatial ‘window of opportunity’ for editosome assembly and function that occurs subsequent to pre-mRNA splicing but prior to mRNA nuclear export [20, 24].
1.2.1. RNA substrates
An open question in A1 research has been how much of the transcriptome is edited? Within apoB mRNA the cytidine at nt 6802 also is edited (converting an ACA threonine codon to an AUA isoleucine codon) [17]. Editing at C6802 is associated with editing at C6666 and therefore is unlikely to impact the biology of ApoB protein. The other example is the mRNA encoding the tumor suppressor neurofibromin which was evaluated for mooring sequences that might support editing because of the occurrence of truncated proteins in NF1 tumor tissues. NF1 mRNA contained three mooring sequence-like motifs of which one supported mooring sequence dependent C to U editing at nt 2914 [19, 25]. Editing of cytidines at C6666 and C6802 of apoB mRNA and C2914 in NF1 mRNA were mooring sequence-dependent; suggesting that the mooring sequence itself might be predictive of other mRNAs that could support C to U editing and therefore may be of utility in computational analyses of the transcriptome. Computational modeling using a weighted matrix which considered the editing efficiencies of all natural and experimental editing sites predicted that mooring sequences existed in multiple mRNAs although no new editing events were identified [26]. Transcriptome-wide, comparative RNA sequencing revealed mooring sequence-dependent editing of cytidines to uridines within the 3’ UTRs of 32 mRNAs [21]. Further deep sequencing studies and the application of advanced computational prediction of editing sites [27, 28] are likely to reveal abundant and biologically significant C to U post-transcriptional editing events as have been revealed for A to I pre-mRNA editing [29].
In the absence of A1, apoB mRNA is not edited and therefore A1 is the sole family member capable of recognizing and using apoB mRNA as a substrate [30, 31]. In tissues where A1 is expressed, deaminase activity on RNA is highly regulated through A1’s: (i) interactions with ACF, (ii) shuttling between its storage form in the cytoplasm and the assembly of editosomes in the nucleus (Sections 3.1 and 4) and (iii) regulation of A1 protein expression and abundance [1]. Experimental overexpression of A1 in rat hepatoma cells resulted in promiscuous editing of cytidines in apoB mRNA as far as 50 nt 5’ of the mooring sequence [20] and hyperediting activity on mRNAs that otherwise were not edited [32, 33]. Overexpression of A1 was associated with oncogenesis [34], which has been inferred to be due to its RNA editing capability. Recently, A1 was shown to edit ssDNA when overexpressed in an E. coli mutator assay [35, 36]. This raises the question of whether deregulation of A1 expression in mammalian cells could also lead to genomic DNA mutation and cancer.
1.2.2. ssDNA substrates
A1 has no known mammalian DNA substrate but it has DNA deaminase activity sufficient to induce reversion mutations when overexpressed in E. coli that have been placed under selection pressure [35, 36]. In addition, A1 expressed in neurons may have a protective function against herpes simplex virus that involves ssDNA deamination of the viral genome [37]. The sequence requirements for A1 deamination were lax but deamination occurred within transcribed or otherwise single-stranded regions of DNA and the dC that was deaminated had a nearest neighbor preference of a 5’ T [35].
Unlike RNA editing, ssDNA editing did not require a cofactor and this appears to be true for the other APOBEC family members (Section 2). Many in the field have asked whether A1 RNA editing is a curious exception or will RNA substrates be identified for the other APOBEC family members? This question is made all the more interesting by the fact that many of the APOBEC family members bind RNA as well as ssDNA and in the RNA-bound state, were not active as ssDNA deaminases (discussed below). In this regard, A1 from various species differed in their capacity for site-specific RNA editing [38, 39]. Reptilian A1, that lacks a C-terminal dimerization and regulatory domain only deaminated ssDNA [40]. In contrast, the yeast APOBEC homolog known as CDD1 site-selectively edited apoB mRNA when this mRNA was expressed in yeast [41, 42]. CDD1 is a 15.5 kDa protein representing little more than the deaminase catalytic domain and yet it had deaminase activity on RNA, ssDNA and free cytosine [43]. It is therefore unclear what structural features of the APOBEC proteins determine their range of substrates.
2. The Eleven proteins in the APOBEC family
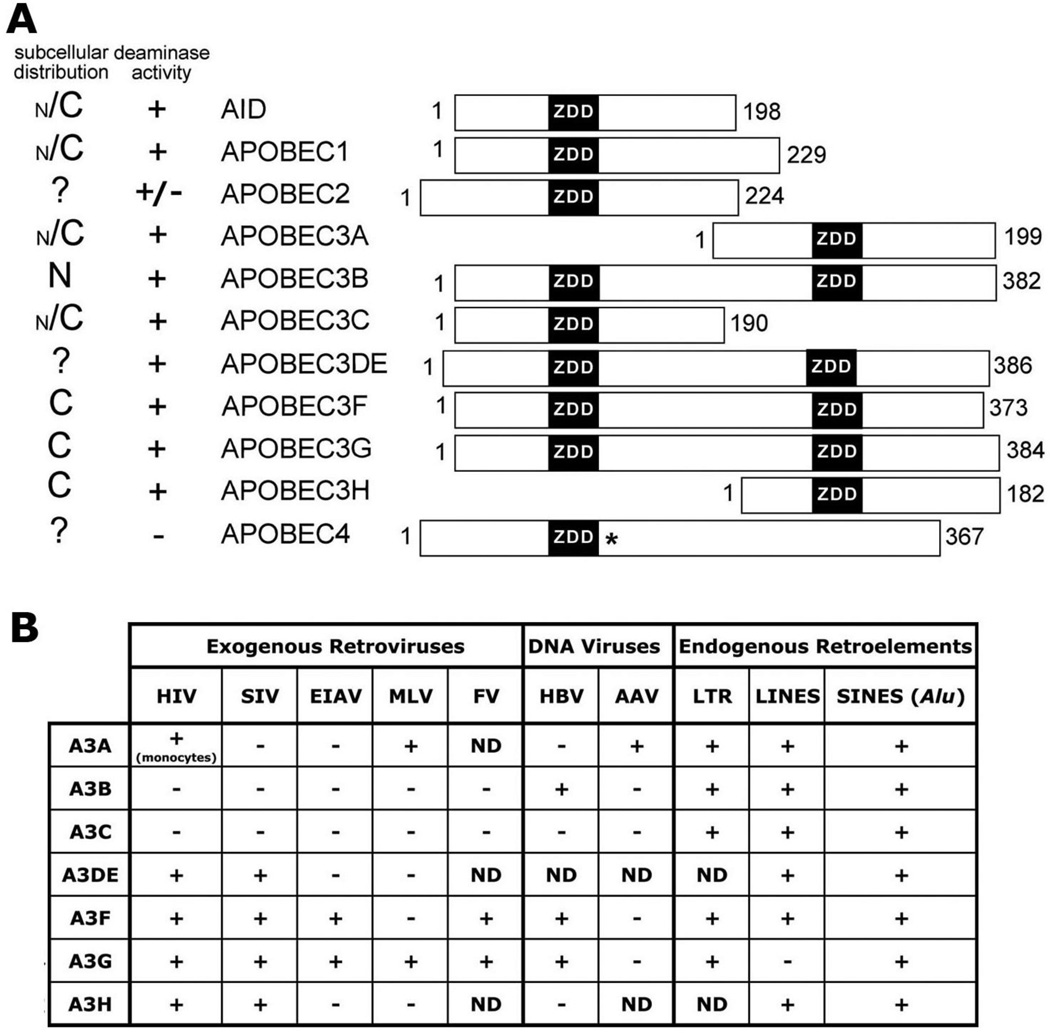
The APOBEC family of proteins are readily identifiable in amino acid similarity searches through the occurrence of the zinc-dependent cytidine or deoxycytidine deaminase domain (ZDD) (H/C)-x-E-x25–30P-C-x-x-C (Figure 2A). Zinc is coordinated through the underlined H or C residues in the context of a super secondary structure consisting of five antiparallel beta strands forming a beta sheet that is supported through two alpha helices. Deaminase activity of this ZDD involves hydrolytic removal of the exocyclic amine from cytidine (C) or deoxycytidine (dC) to form uridine (U) or deoxyuridine (dU) [44].
Figure 2. APOBEC family localization and activity.
A. Bar diagrams of human APOBEC proteins and their relative alignments according to exon junctions. The ZDD motifs (black) and number of amino acids for each protein are indicated. The (*) next to the ZDD for APOBEC4 indicates that it is divergent from the consensus ZDD. The relative subcellular distribution is on the left with N for nuclear and C for cytoplasmic, and ‘?’ if localization is unknown. The size of N or C indicates relative distribution. The deaminase activity is indicated by a + or − and the +/− for APOBEC2 indicates mixed results depending on the system (see text). B. A chart of A3 proteins’ activity on different mobile genetic elements, with + for active and − for inactive and ND when activity has been not determined. The (monocytes) under the A3A HIV activity indicates that it has only been shown to be active in monocyte derived cells. The information in A and B was compiled from references within the text.
2.1. Activation Induced Deaminase, AID
Upon encountering a foreign antigen, immunoglobulin genes within germinal center B cells undergo several rounds of modifications allowing antibodies to be expressed with strong antigen-binding affinity and different effector functions [45, 46]. Activation induced cytidine deaminase (AID) was discovered more than a decade ago using subtractive hybridization by comparing the transcripts from resting and activated murine B cells that underwent immunoglobulin gene diversification [47]. AID’s deaminase activity [35] is responsible for dU mutations that are converted through low fidelity excision repair to a variety of point mutations or cause DNA strand breaks. This mutagenic activity leads to Somatic Hypermutation (SHM) and immunoglobulin gene recombination events known as Class Switch Recombination (CSR) and Gene Conversion (GC) [48–52].
The human AID gene maps to chromosome 12p13 and encodes a 198 amino acid protein containing a single ZDD [53] (Figure 2A). AID gene mutations have been identified in patients with a rare immunodeficiency known as Hyper IgM sydrome, HIGM (characterized by high serum levels of IgM and lack of other immunoglobulin isotypes (IgG, IgE, IgA)) [54]. This phenotype also was observed in AID knockout mice [47, 55] which demonstrated that AID was required for immunoglobulin diversification. On the other hand AID expression has been implicated in several pathologies including Non-Hodgkin B cell lymphomas and solid tumors [56]. Increased expression of alternative splice variants of AID has been reported in patients with chronic lymphocytic leukemia (CLL) [57–59]. Alternatively spliced transcripts lacking C-terminal residues or the entire exon that contains the nuclear export signal have been observed in cancer cells. Although this suggested that AID hyperactivity led to genomic instability, the role of AID deaminase activity remains to be confirmed [60, 61]. The formation of lung micro-adenomas and massive T cell lymphomas in transgenic mice ubiquitously expressing AID confirmed the hypothesis that deregulated expression of AID can lead to malignant transformation [62]. In fact, up-regulation of AID expression through the NFΚB signaling pathway in response to hepatitis C infection of hepatocytes also was associated with genomic mutations, however these mice did not develop cancers [63]. This suggested that an additional control system buffering the effects of endogenous AID expression in B cells may exist.
In support of this possibility, constitutive expression of AID in mice that was restricted to B cells was not associated with developmental defects, excessive DNA mutations or tumors even though a large amount of AID protein was evident [64]. Why AID expressed in germinal centers normally only targets variable and switch regions of immunoglobulin genes for mutagenesis is unknown. Part of the answer might be that B cell-specific regulation inhibits the expression of AID through Ca2+/calmodulin mediated inhibition of the E2A protein that is required for AID gene transcription [65, 66]. Accumulating evidence indicates that AID selectivity for its ssDNA substrates may be regulated through AID interactions with cis-acting elements in the Ig loci or by binding to SHM- and CSR-specific protein cofactors [49, 67]. The data clearly show that AID and its ssDNA deaminase activity are essential for a responsive B cell immune system but it is also clear that AID must be highly regulated to control its oncogenic potential.
2.2. APOBEC2
APOBEC2 (A2) was discovered by a search of mouse and human EST databases for genes homologous to A1. Human A2 is located on chromosome 6 and expressed in cardiac tissue and skeletal muscle [35, 68]. Before any function was ascribed to A2, its crystal structure became the first and only full length APOBEC family member to be solved to date [69]. Although this structure has been used for homology modeling of other family members, A2 seems to be functionally quite different. While the other APOBECs are clearly capable of deaminase activity on ssDNA in the absence of cofactors, A2 was non-mutagenic in yeast or bacteria based mutator assays [35, 70]. However, a recent paper suggested that A2 DNA mutagenic activity targeted specific tumor suppressor genes and that A2 overexpression in mice resulted in liver and lung tumors [68]. The potential of A2 activity on DNA is of interest considering that A2 is essential for muscle development [71]. A2 may have a specific and unique function in muscle and heart tissue. Whether A2 acts alone or has a cofactor is not known. The data suggest that A2 may be far less prone to off target activity given its weak interaction with RNAs and lack of autonomous deaminase activity in bacterial or yeast based systems [70, 71].
2.3. APOBEC3
The main function of APOBEC3 (A3) proteins (clustered on human chromosome 22) is as sentinels in innate immunity to mobile genetic elements (i.e. endogenous retroelements and exogenous viruses) (Figure 2B). Many forms of A3 genes are found in mammals (Figure 2A): a single A3 gene in rodents, cats, pigs, and sheep, two in cows, three in dogs and horses, and seven in primates [72]. A3B, A3DE, A3F, and A3G differ from A1, AID, A3A, A3C and A3H in that they contain two deaminase domains instead of one within a single polypeptide (Figure 2A) [44, 73]. Deaminase activity on particular mobile genetic elements varies greatly between these homologs (Figure 2B). The goal of this section is evaluate these differences.
The most studied A3 family member is A3G. The function of A3G as an antiviral host factor was discovered in 2002 through cDNA transfer experiments designed to identify a host cell suppressor of the HIV-1 accessory protein known as the viral infectivity factor (Vif) [74]. Vif binds to A3G and induces its destruction via the ubiquitination and proteasome degradation pathway [75]. Viruses deficient in Vif had low infectivity if they were produced in cell lines known as ‘nonpermissive’ (express A3G) but otherwise exhibited near wild type infectivity levels when produced in ‘permissive’ cell lines [75]. Transfection of permissive cells with A3G was necessary and sufficient for conversion to the nonpermissive phenotype for Vif-deficient HIV-1 infectivity [74]. From these findings it became clear that wild type HIV-1 expressed Vif to overcome the A3G innate immune system.
The primary antiviral mechanism of A3G required that it be encapsidated into HIV viral particles [75]. During reverse transcription A3G hypermutated minus-strand HIV DNA with mutations becoming fixed as G to A mutations upon plus-strand synthesis [76, 77]. A3G also acted on a wide variety of distantly related retroviruses that package genomic RNA. The retroviruses A3G has been reported to affect SIV [78], equine infectious anemia virus (EIAV) [79], murine leukemia virus (MLV) [80], and foamy virus (FV) [81] (Figure 2B). The interspecies transmission of SIV was likely prevented by A3G’s insensitivity to SIV Vif, while MLV and EIAV virions do not have a Vif equivalent to protect the viruses against A3G.
A3F was capable of anti-HIV, SIV, EIAV and FV activity nearly equivalent to that of A3G [78, 79, 81]. However, A3F preferred a dTC sequence context for deaminase targeting opposed to the dCC preference for A3G. This enabled A3F to target DNA sequences for hypermutation other than those mutated by A3G [82]. A3DE also had antiviral effects on HIV infectivity in a Vif-sensitive manner although to a lesser degree than A3G or A3F [80]. A specific haplotype II (in Africans) of A3H had activity similar to A3G while the haplotype I, III and IV were not antiviral [83]. A recent report examined antiviral activities of all human and rhesus macaque A3s stably expressed in a Sup T1 cell line and found that A3DE, A3F, A3G, and A3H (haplotype II) from both species had similar expression patterns in T cells and activity against HIV and SIV [78]. In this study A3A, A3B and A3C were inactive against HIV (Figure 2B) [78]. In a different study, A3A had anti-HIV activity that was linked to a specific A3A protein variant. Deaminase activity in INF-α treated monocytes and monocyte derived macrophages correlated specifically to a variant that was expressed equal to wild type A3A but whose translation initiated with methionine at residue 13 instead of residue 1 [83]. It is unclear whether this variant was the key difference between the studies or if a cofactor specifically expressed in monocytes activated A3A against HIV. A3A was also a potent inhibitor of a parvovirus, AAV-2 (Figure 2B), and its activity on these viruses correlated with protein structural divergence from A3G [84].
A3G, A3F and A3B inhibited hepatitis B virus (HBV) infection (Figure 2B) [85]. HBV is considered a pararetrovirus because it reverse transcribes “pre-genomic” RNA like retroviruses, but unlike retroviruses reverse transcription occurs before viral release and therefore a DNA genome is packaged. Liver cells are the target of HBV and A3G, A3F and A3B expression in human liver was up-regulated by INF-α [85]. However, the low and variable levels of A3s in the liver may have caused low levels of mutations that imparted HBV with some selective advantages. Specifically this mutagenic activity produced a truncation mutation in the HBx gene that was linked to hepatocellular carcinoma [86].
The original A3 targets may have been endogenous retroelements [87, 88]. Endogenous retroelements’ ability to copy themselves into random locations in the genome leads to genomic instability and disease [88]. There are three major classes of retroelements: (i) long terminal repeat (LTR) based endogenous retroviruses, and non-LTR based (ii) autonomous long interspersed nuclear elements (LINEs) and (iii) non-autonomous short interspersed nuclear elements (SINEs). LINEs are autonomous because within their code is everything needed to reverse transcribe and re-insert their sequence into another location within the cell’s genome. Conversely, SINEs are non-autonomous because they must use the machinery encoded in LINEs in trans for reverse transcription and genomic re-insertion [88].
Various retroelement reporter assays revealed that A3G and A3F inhibited LTR-based endogenous retroviruses (i.e. IAP, Mus-D and Ty1) [89, 90]. Hypermutations were detected in these sequences [89, 90] as well as a reduced number of reverse transcripts [89] consistent with the known A3G mechanism on exogenous retroviruses. In contrast, A3G inhibited SINE retrotransposition (i.e. Alu and hY) by sequestering these RNAs as ribonucleoprotein complexes (Section 4) [87, 91]. This mechanism was deaminase-independent consistent with A3G being enzymatically inactivated by cellular RNAs in HMM (Section 3.3 and 4) [75, 87, 91].
The expansion of A3s in primates correlates well with the decreased presence of active retroelements in humans [88]. There are seven A3s in humans and the only currently active retroelements in humans are non-LTR based retroelements (i.e. LINEs and SINEs). Conversely, mice have only one APOBEC3 protein and their genomes contain active LTR-based and non-LTR based retroelements [88, 89]. Moreover, mice carry 50–60 times more active LINE-1 retroelements in their genomes than humans and the proportion of LINE-1 causing disease is 35% greater in mice compared to humans [88]. In this regard all human A3s inhibit LINE-1 and/or Alu retrotransposition, with the one exception that A3G does not inhibit LINE-1 (Figure 2B) [92–97]. A3DE, A3F and A3H were localized to the cytoplasm and were able to block either LTR-based or non-LTR based retroelements in manners similar to A3G [89, 94, 95, 97, 98]. In contrast, A3A, A3B and A3C were capable of nuclear localization and may be able to directly inhibit nuclear reverse transcription of retroelements [92–96]. Overall these multiple fronts of defense highlight the diversity within human A3 genes as a key determinant in combating the genotoxic threat posed by endogenous retroelements. However, as with A1, A2 and AID, up-regulation of nuclear A3A and A3B deaminases may become genotoxic and pose a risk of inducing cancer [85, 99]. This threat along with the variable activities and expression levels on different mobile genetic elements underscores the need to further understand cell type-specific regulation of A3 proteins.
2.4. APOBEC4
APOBEC4 (A4) has no ascribed function and was discovered by a computational homology search that revealed its location on human chromosome 1 and inspection of the ESTs suggested that it is expressed in testes [100]. The A4 sequence is distinctly divergent from other APOBEC genes and contains a significant alteration in the presumptive deaminase domain compared to the consensus ZDD motif (PCx6C instead of PCxxC) [100] (Figure 2A). A recent report showed that A4 was non mutagenic when expressed in yeast and bacteria [70]. As suggested for A2, A4 might require a specific cofactor for mutagenic activity or it may have a deaminase-independent function.
3. Subcellular compartmentalization of APOBEC proteins regulates their activity
3.1. A1 in 27S editosome and 60S pre-editosomal complexes
A1 distribution in both the nucleus and cytoplasm is determined in part by an N-terminal nuclear localization signal and a C-terminal cytoplasmic retention signal (Figure 3A) [101, 102]. Although RNA editing can occur in the cytoplasm when A1 is overexpressed [103], cytoplasmic A1 editing activity is normally suppressed and RNA editing is restricted to the cell nucleus within a temporal and spatial window that occurs subsequent to pre-mRNA splicing and prior to mRNA nuclear export [24, 104].
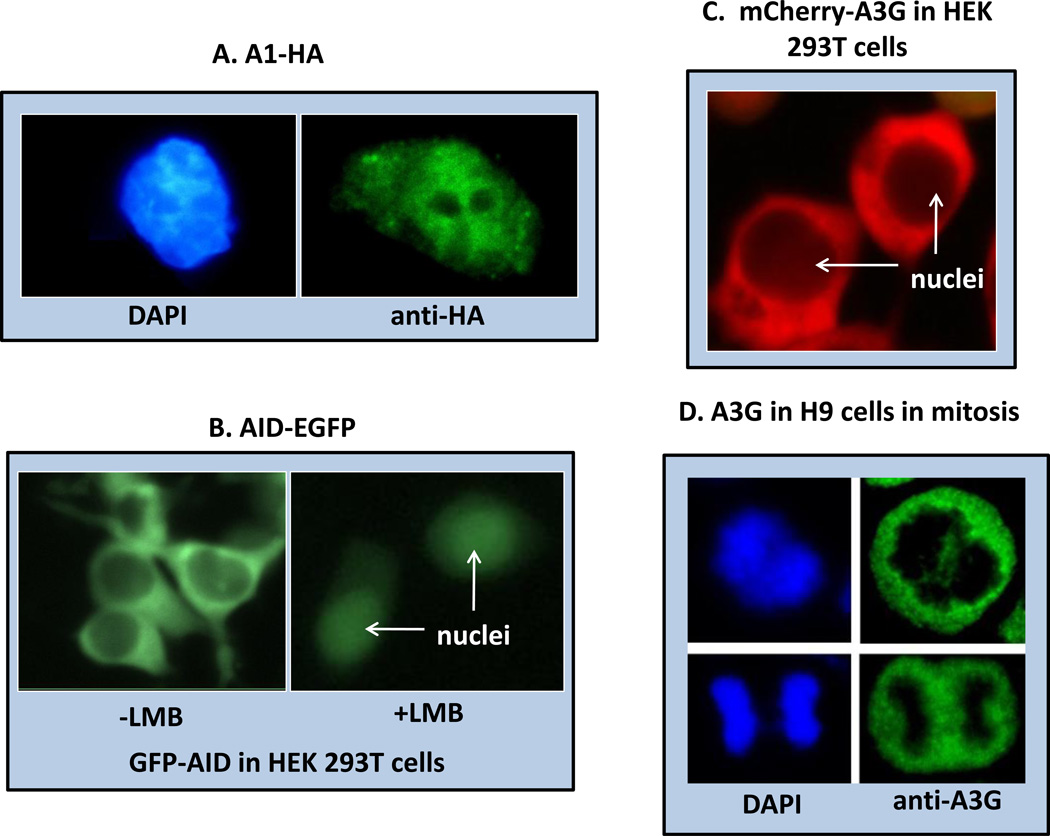
Figure 3. Localization of the APOBEC proteins by in situ fluorescence.
A. APOBEC-1 (A1) with a C-terminal HA tag was expressed by transfection in McArdle rat hepatoma cells. Immunocytochemical localization of the HA tag in fixed cells shows that A1 is distributed throughout the cytoplasm and nucleus (DAPI) but is not localized in the nucleolus. A1 shuttles between the cytoplasm and nucleus. B. Activation Induced Deaminase (AID) as a C-terminal GFP chimeric protein was expressed in transfected human embryonic kidney cells 293T and visualized in live cells. AID appears predominantly in the cytoplasm (left panel, four transfected cells shown) but its rapid shuttling activity can be demonstrated by inhibiting its CRM1-dependent nuclear export with Leptomycin B (LMB) (right panel, two transfected cells shown). C. A3G as an N-terminal mCherry chimeric protein was expressed in transfected HEK293T cells and visualized in live cells. A3G is restricted to the cytoplasm because it has no nuclear localization signal but does have a cytoplasmic retention signal. D. Natively expressed A3G in synchronized H9 cells after fixing and staining with anti-A3G polyclonal antibody. DAPI stained mitotic chromosomes (left top, anaphase; left bottom, telophase) are segregated in the mitotic cells from cytoplasmic A3G. Original magnification was 40X.
Glycerol gradient velocity sedimentation of nuclear extracts revealed A1 and ACF co-sedimenting with editing activity as 27S complexes [14, 105]. A1 and ACF in cytoplasmic extracts co-sedimented as 60S complexes whose RNA editing activity could be activated in vitro and converted to 27S complexes by incubating 60S complexes at 30 °C [105] or by treating them with elevated monovalent salt concentrations [15, 106]. Other than A1, ACF and RNA, the molecular composition of 27S and 60S complexes is unknown. It is also not known whether the interaction of A1 with ACF in inactive 60S complexes is different than that in 27S complexes which are editing competent.
The proportion of apoB mRNA edited in tissues is determined by shuttling of A1 and ACF from cytoplasmic 60S complexes into the nucleus where they are recovered as 27S complexes [14, 16, 101, 107]. This process is regulated tissuespecifically, during development and in response to metabolic regulation (reviewed in [108]). Though different from A1, ACF contains nuclear localization and nuclear export signals [16, 107] and therefore it is possible that A1 and ACF may shuttle independently. ACF shuttling occurred in the absence of A1 but the converse situation is not known. Nuclear retention of A1 and ACF was strongly influenced by ACF phosphorylation that in turn was regulated by insulin and ethanol through protein kinase C [14, 22, 109, 110]. The distribution of A1 and ACF in cytoplasmic 60S or nuclear 27S complexes was regulated through the abundance of ACF and determined in part by leptin inhibition of ACF gene transcription [109] and through the expression of ACF variants (due to alternative pre-mRNA splicing) that have different affinities for A1 and different levels of nuclear retention [43, 111, 112]. These findings underscore the importance of subcellular localization of A1 and its organization as low or high molecular mass complexes as key regulatory determinants for A1 mRNA editing activity.
3.2. AID Cytoplasmic Retention
Controlling the abundance of AID in different cellular compartments is one of the major regulatory mechanisms restricting its contact with the genomic material [113–115]. Despite its function in the nucleus, AID is predominantly localized in the cytoplasm, even when it is overexpressed (Figure 3B) [116]. This pattern is produced by the action of an active transport system [117]. When the export is blocked with a CRM-1 specific inhibitor, Leptomycin B (LMB), the nucleo-cytoplasmic shuttling feature of AID becomes more obvious with the majority of AID sequestered in the nucleus [118, 119] (Figure 3B).
While findings from different groups confirmed the existence of a CRM-1 dependent nuclear export signal (NES) at the C-terminus of AID, there is no consensus about the exact location and nature (classical bipartite or conformational NLS) of the N-terminal nuclear localization signal (NLS) [117, 119]. AID has been shown to interact with several importin alpha isoforms (impα1, impα3, and impα5) [117], suggesting that the generic impα/β import pathway may be mediating AID nuclear entry. CTNNBL1 (catenin, beta-like 1), an NLS-binding protein containing architectural homology and binding affinity to impα5 but not to imp β, was also associated with AID when AID was overexpressed in HEK293T cells [120]. For this reason a role for CTNNBL1 in nuclear localization of AID as either an adapter protein mediating AID-impα5 interaction or AID association with spliceosome components has been proposed [120].
A cytoplasmic retention signal may contribute to the steady state cytoplasmic localization of AID [117]. Under conditions rendering both NLS and NES inactive (combination of oxidative stress, N-terminal tagging of AID, mutagenesis or LMB treatment), AID remained in the cytoplasm, rather than as predicted from AID’s size, passively diffusing into the nucleus and displaying a homogenous cellular distribution [117]. Mutational analysis revealed a retention signal overlapping with the NES but distinguishable from it with the specific amino acid substitutions (D188A, L198S) [117].
Protein factors have been shown to stabilize or destabilize AID in various cellular fractions [121, 122]. Heat-shock protein 90 bound to the N-terminal region of AID and stabilized cytoplasmic AID by shielding it against E3 ubiquitin ligase mediated proteosomal degradation [121]. Moreover, analysis of the half-life of different AID mutants demonstrated that, compared to wild type AID, nuclear export deficient AID was 3-fold less stable whereas nuclear import impaired AID had a 3-fold longer half-life suggesting that cytoplasmic AID is more stable [122].
3.3. APOBEC3 cytoplasmic ribonucleoprotein complexes
A3G is strongly retained in the cytoplasm of interphase cells and remains excluded from chromosomes during mitosis (Figure 3C and D). This localization is due to a cytoplasmic retention signal between amino acids 113–128 [123]. Interestingly this region is also crucial for Vif, Gag and RNA interactions, but A3G binding to RNA was not required for cytoplasmic retention [123–125]. The N-terminal half of A3G expressed in a quail cell line was not restricted to the cytoplasm (in contrast to what was observed in mammalian cells) suggesting that there may be a mammalian specific factor involved in the cytoplasmic retention of A3G [126].
A3G is present in two distinct cytoplasmic forms: low molecular mass (LMM) and RNA-bound, high molecular mass (HMM) complexes. Cytokines IL-2, IL-15, and IL-7 stimulated the formation of HMM [127] but poly(I:C) and TNF-α increased A3G expression as LMM [128]; suggesting that these various forms of A3G may have functional significance. Vif also has been suggested to facilitate HMM formation [129]. On the other hand, HMM formation was not a common feature among all A3s. A3DE, A3F and A3H all have similar antiviral profiles to A3G and share in the ability to form HMM complexes and/or associate with Alu RNA [97]. On the other hand, A3A in INF-α activated PBMCs did not have to be treated with RNase to activate its deaminase activity, suggesting that it is not inhibited by RNA in HMM complexes [83].
Immunocytochemistry revealed a homogeneous cytoplasmic distribution of A3G along with concentrated foci determined to be P-bodies and stress granules (Figure 3) [87, 98, 130]. These cytoplasmic aggregates are composed of various cellular RNAs and their associated proteins and function as RNA and ribonucleoprotein degradation or recycling centers. A3G promoted dissociation of miRNA-targeted mRNA from P bodies, thus allowing for translation of these mRNAs [131] and the interaction of A3G with Ago1 and Ago2 (proteins associated with the RNA interference pathway) [132] adds credence to the idea that A3G may function as an RNAi regulator in P bodies. Collectively, these data support a model whereby the oligomeric state and/or localization of A3 proteins is influenced by both cellular and viral factors which, in turn, may be a manifestation of how cells regulate the functionality of these proteins.
4. RNA binding to APOBEC as a cellular regulator of ssDNA deaminase activity
Although there may be several different HMM complexes with which A3G associates, they all share a common characteristic that RNA binding to A3G inhibits ssDNA deaminase activity [75, 133]. Both A1 and A3G were retained as large cytoplasmic complexes that were catalytically inactive. The catalytic activity of A3G that was sequestered in ribonucleoprotein complexes could be restored in vitro by RNase digestion of HMM [134]. Most of the interactions of A3G with other proteins within these complexes were through RNA bridging as RNase digestion reduced megaDalton size A3G ribonucleoprotein complexes to dimers and mononers. Interestingly, A3G-RNA complexes in viral particles were inactive until RNaseH activity of reverse transcriptase degraded the RNA of the DNA-RNA hybrid [135]. Although A3G has the ability to bind RNAs nonselectively, several studies have shown that A3G (and A3F) can select for cellular and viral RNAs that become incorporated with it into viral particles [136–140]. A3G:7SL and A3G:viral RNA interactions may facilitate the association of A3G and the nucleocapsid (NC) region of HIV-1 Gag; a requirement for A3G viral packaging [141–143]. RNAs of diverse sequence and propensity for secondary structure, and as short as 25 nt bound A3G in vitro and prevented the formation of A3G:ssDNA complexes necessary for catalytic activity [144]. A3C also was incorporated into HIV particles and although it did not associate with 7SL RNA or HIV NC, it did interact with the matrix (MA) region of HIV Gag in what may be a 5.8S ribosomal RNA-dependent manner [145, 146]. Furthermore, AID is bound to mRNA in the cytoplasm of cells [147] and in vitro studies have shown AID to have very little catalytic activity unless pre-treated with RNase [148].
RNA binding is emerging as a general means of inactivating ssDNA deaminase activity in the APOBEC family; however the mechanism whereby RNA inhibits deaminase activity is unknown. For A3G, deaminase activity resides in the C-terminal ZDD while RNA binding has been suggested to occur through an interaction with the N-terminal ZDD [149, 150]. RNA binding may regulate deaminase activity allosterically by binding to the N-terminus of A3G and inducing a conformational change in A3G so it can no longer bind ssDNA, or competitively by directly displacing ssDNA from the C-terminal catalytic domain (Figure 1B) [144]. This model is not directly applicable to enzymes with a single ZDD although RNA inhibition of the deaminase activity of these APOBEC proteins also may be because RNA and ssDNA interactions with the enzyme are mutually exclusive.
5. Concluding Remarks
Other than A2 and A4, binding to an appropriate nucleic acid by APOBEC proteins results in deamination. When limited in frequency and targeted to specific genes, this mutagenic activity can be beneficial to organisms as well as to viral pathogens. There are however clear indications that APOBEC mutagenic activities and nucleic acid binding capabilities can be genotoxic for retroviruses and are used as such in host cell defense. Excessive APOBEC activity and/or off-target mutations within the cellular genome can be genotoxic and oncogenic. Thus cells have multiple mechanisms that regulate the expression of APOBEC proteins, control their enzymatic activity and restrict their access to DNA or RNA substrates.
Use of RNA as a substrate is considered unique to A1. Even though RNA is not commonly used as a substrate, the majority of APOBEC family members bind to both RNA and ssDNA. An emerging theme is that RNA binding to APOBEC enzymes and the resultant homo-multimerization of these enzymes inhibits their ssDNA binding and deaminase activities. Typically RNA binding sequesters APOBEC as large ribonucleoprotein aggregates that are compartmentalized within the cell cytoplasm or within viral particles. Ribonucleoprotein complexes form rapidly after APOBEC translation and are typically not RNA sequence-specific. Thus the availability of free and active APOBEC deaminases is anticipated to be limited under most circumstances. While this may diminish deaminase-dependent antiviral activity, APOBEC binding to retroviral RNAs may impair their ability to reverse transcribe or translate and thereby provide deaminase-independent antiviral activity for the host cell. Inhibition of deaminase activity is reversible and involves release of APOBEC from its interaction with RNA. The mechanism whereby some cell types maintain APOBEC in an RNA-depleted and active state is not known.
The identification of RNA editing substrates has largely depended on prior knowledge of protein isoforms. Proof that an APOBEC family member can edit these RNAs has relied on the identification of a cell type or cell extract that expressed an appropriate complementation factor. Although RNA editing by AID has been suggested since the discovery of the enzyme’s role in SHM and CSR, no edited transcript has been found and the AID RNA editing hypothesis has fallen out of favor. Future discoveries may reveal a broader RNA editing phenotype for this family but current research is focused on APOBEC ssDNA deaminase activities where RNA plays a role as a regulatory cofactor.
Acknowledgements
The authors thank Jenny ML Smith for critical reading of this manuscript. Data from the authors’ laboratory that have been described in this manuscript were obtained with the support of Public Health Services Grants NIDDK DK43739, NIAID AI54369, AI095007, AI58789, NINDS NS067671, an Air Force Office of Scientific Research Grant F49620 and a Bill and Melinda Gates foundation Grand Challenges in Exploration grant awarded to HCS. Additional support was from Developmental Center for AIDS Research grant (NIAID P30 078498) awarded to Steve Dewhurst, a Public Health Services Grant NIAID AI076085 awarded to Joseph E. Wedekind and an institutional Ruth L. Kirschstein National Research Service Award Public Health Services T32 Grant GM068411 awarded to Robert Bambara and Lynn Maquat.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smith HC. The APOBEC1 Paradigm for Mammalian Cytidine Deaminases that Edit DNA and RNA. In: Grosjean H, editor. Austin. Chapter 15. Landes BioScience; 2009. pp. 181–202. [Google Scholar]
- 2.Smith HC, Gott JM, Hanson MR. A guide to RNA editing. RNA. 1997;3:1105–1123. [PMC free article] [PubMed] [Google Scholar]
- 3.Powell LM, Wallis SC, Pease RJ, Edwards YH, Knott TJ, Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein- B48 in intestine. Cell. 1987;50:831–840. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- 4.Chen SH, Habib G, Yang CY, Gu ZW, Lee BR, Weng SA, et al. Apolipoprotein B-48 is the product of a messenger RNA with an organ- specific in-frame stop codon. Science. 1987;238:363–366. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- 5.Teng B, Burant CF, Davidson NO. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–1819. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- 6.Anant S, MacGinnitie AJ, Davidson NO. apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, is a novel RNA-binding protein. J Biol Chem. 1995;270:14762–14767. [PubMed] [Google Scholar]
- 7.Navaratnam N, Fujino T, Bayliss J, Jarmuz A, How A, Richardson N, et al. Escherichia coli cytidine deaminase provides a molecular model for ApoB RNA editing and a mechanism for RNA substrate recognition. J Mol Biol. 1998;275:695–714. doi: 10.1006/jmbi.1997.1506. [DOI] [PubMed] [Google Scholar]
- 8.Navaratnam N, Bhattacharya S, Fujino T, Patel D, Jarmuz AL, Scott J. Evolutionary origins of apoB mRNA editing: catalysis by a cytidine deaminase that has acquired a novel RNA-binding motif at its active site. Cell. 1995;81:187–195. doi: 10.1016/0092-8674(95)90328-3. [DOI] [PubMed] [Google Scholar]
- 9.Mehta A, Kinter MT, Sherman NE, Driscoll DM. Molecular cloning of apobec-1 complementation factor, a novel RNA- binding protein involved in the editing of apolipoprotein B mRNA. Mol Cell Biol. 2000;20:1846–1854. doi: 10.1128/mcb.20.5.1846-1854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mehta A, Driscoll DM. Identification of Domains in APOBEC-1 Complementation Factor Required for RNA Binding and Apolipoprotein B mRNA editing. RNA. 2002;8:69–82. doi: 10.1017/s1355838202015649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanc V, Henderson JO, Kennedy S, Davidson NO. Mutagenesis of apobec-1 complementation factor reveals distinct domains that modulate RNA binding, protein-protein interaction with apobec-1, and complementation of C to U RNA-editing activity. J Biol Chem. 2001;276:46386–46393. doi: 10.1074/jbc.M107654200. [DOI] [PubMed] [Google Scholar]
- 12.Lau PPH-J, Zhu HA, Baldini C, Charnsangavej L, Chan Dimeric structure of a human apo B mRNA editing protein and cloning and chromosomal localization of its gene. Proc Natl Acad Sci USA. 1994;91:8522–8526. doi: 10.1073/pnas.91.18.8522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galloway CA, Kumar A, Krucinska J, Smith HC. APOBEC-1 complementation factor (ACF) forms RNA-dependent multimers. Biochem Biophys Res Commun. 2010;398:38–43. doi: 10.1016/j.bbrc.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sowden MP, Ballatori N, de Mesy Jensen KL, Hamilton Reed L, Smith HC. The editosome for cytidine to uridine mRNA editing has a native complexity of 27S: identification of intracellular domains containing active and inactive editing factors. J Cell Science. 2002;115:1027–1039. doi: 10.1242/jcs.115.5.1027. [DOI] [PubMed] [Google Scholar]
- 15.Smith HC. Analysis of protein complexes assembled on apolipoprotein B mRNA for mooring sequence-dependent RNA editing. Methods. 1998;15:27–39. doi: 10.1006/meth.1998.0603. [DOI] [PubMed] [Google Scholar]
- 16.Chester A, Somasekaram A, Tzimina M, Jarmuz A, Gisbourne J, O'Keefe R, et al. The apolipoprotein B mRNA editing complex performs a multifunctional cycle and suppresses nonsense-mediated decay. Embo J. 2003;22:3971–3982. doi: 10.1093/emboj/cdg369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Navaratnam ND, Patel RR, Shah JC, Greeve LM, Powell TJ, Knott J, Scott An additional editing site is present in apolipoprotein B mRNA. Nucleic Acids Res. 1991;19:1741–1744. doi: 10.1093/nar/19.8.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Backus JW, Schock D, Smith HC. Only cytidines 5' of the apolipoprotein B mRNA mooring sequence are edited. Biochim Biophys Acta. 1994;1219:1–14. doi: 10.1016/0167-4781(94)90240-2. [DOI] [PubMed] [Google Scholar]
- 19.Skuse GR, Cappione AJ, Sowden M, Metheny LJ, Smith HC. The neurofibromatosis type I messenger RNA undergoes base-modification RNA editing. Nucleic Acids Res. 1996;24:478–485. doi: 10.1093/nar/24.3.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sowden M, Hamm JK, Spinelli S, Smith HC. Determinants involved in regulating the proportion of edited apolipoprotein B RNAs. RNA. 1996;2:274–288. [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenberg BR, Hamilton CE, Mwangi MM, Dewell S, Papavasiliou FN. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3' UTRs. Nat Struct Mol Biol. 2011;18:230–236. doi: 10.1038/nsmb.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehmann DM, Galloway CA, Macelrevey C, Sowden MP, Wedekind JE, Smith HC. Functional characterization of APOBEC-1 complementation factor phosphorylation sites. Biochim Biophys Acta. 2007;1773:408–418. doi: 10.1016/j.bbamcr.2006.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehmann DM, Galloway CA, Sowden MP, Smith HC. Metabolic regulation of apoB mRNA editing is associated with phosphorylation of APOBEC-1 complementation factor. Nucleic Acids Research. 2006;34:3299–3308. doi: 10.1093/nar/gkl417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sowden MP, Smith HC. Commitment of apolipoprotein B RNA to the splicing pathway regulates cytidine-to-uridine editing-site utilization. Biochem J. 2001;359:697–705. doi: 10.1042/0264-6021:3590697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukhopadhyay D, Anant S, Lee RM, Kennedy S, Viskochil D, Davidson NO. C-->U editing of neurofibromatosis 1 mRNA occurs in tumors that express both the type II transcript and apobec-1, the catalytic subunit of the apolipoprotein B mRNA-editing enzyme. Am J Hum Genet. 2002;70:38–50. doi: 10.1086/337952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith HCS, Mark P, Kefang Xie, Wedekind Joseph, E. In: Structure and Function of Mammalian Cytidine Deaminases that Mediate Expressed Sequence Diversification. Grosjean H, editor. NY: Springer-Verlag; 2005. pp. 1610–1145. [Google Scholar]
- 27.Thompson J, Gopal S. Genetic algorithm learning as a robust approach to RNA editing site prediction. BMC Bioinformatics. 2006;7:145. doi: 10.1186/1471-2105-7-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paz-Yaacov N, Levanon EY, Nevo E, Kinar Y, Harmelin A, Jacob-Hirsch J, et al. Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates. Proc Natl Acad Sci U S A. 2010;107:12174–12179. doi: 10.1073/pnas.1006183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farajollahi S, Maas S. Molecular diversity through RNA editing: a balancing act. Trends Genet. 2010;26:221–230. doi: 10.1016/j.tig.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirano K, Young SG, Farese RV, Jr, Ng J, Sande E, Warburton C, et al. Targeted disruption of the mouse apobec-1 gene abolishes apolipoprotein B mRNA editing and eliminates apolipoprotein B48. J Biol Chem. 1996;271:9887–9890. doi: 10.1074/jbc.271.17.9887. [DOI] [PubMed] [Google Scholar]
- 31.Nakamuta M, Chang BHJ, Zsigmond E, Kobayashi K, Lei H, Ishida BY, Oka K, Li E, Chan L. Complete phenotypic characterization of the apobec-1 knockout mice with a wild-type genetic background and a human apolipoprotein B transgenic background, and restoration of apolipoprotein B mRNA editing by somatic gene transfer of Apobec-1. J Biol Chem. 1996;271:25981–25988. doi: 10.1074/jbc.271.42.25981. [DOI] [PubMed] [Google Scholar]
- 32.Yamanaka S, Poksay KS, Driscoll DM, Innerarity TL. Hyperediting of multiple cytidines of apolipoprotein B mRNA by APOBEC-1 requires auxiliary protein(s) but not a mooring sequence motif. J Biol Chem. 1996;271:11506–11510. doi: 10.1074/jbc.271.19.11506. [DOI] [PubMed] [Google Scholar]
- 33.Yamanaka S, Poksay KS, Arnold KS, Innerarity TL. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA- editing enzyme. Genes Dev. 1997;11:321–333. doi: 10.1101/gad.11.3.321. [DOI] [PubMed] [Google Scholar]
- 34.Yamanaka S, Balestra M, Ferrell L, Fan J, Arnold KS, Taylor S, Taylor JM, Innerarity TL. Apolipoprotein B mRNA editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci USA. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 36.Petersen-Mahrt SK, Neuberger MS. In vitro deamination of cytosine to uracil in single-stranded DNA by apolipoprotein B editing complex catalytic subunit 1 (APOBEC1) J Biol Chem. 2003;278:19583–19586. doi: 10.1074/jbc.C300114200. [DOI] [PubMed] [Google Scholar]
- 37.Gee P, Ando Y, Kitayama H, Yamamoto SP, Kanemura Y, Ebina H, et al. APOBEC1-mediated editing and attenuation of HSV-1 DNA implicates an antiviral role in neurons during encephalitis. J Virol. 2011 doi: 10.1128/JVI.05288-11. in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sowden MP, Eagleton MJ, Smith HC. Apolipoprotein B RNA sequence 3' of the mooring sequence and cellular sources of auxiliary factors determine the location and extent of promiscuous editing. Nucleic Acids Research. 1998;26:1644–1652. doi: 10.1093/nar/26.7.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hersberger M, Patarroyo-White S, Qian X, Arnold KS, Rohrer L, Balestra ME, et al. Regulatable liver expression of the rabbit apolipoprotein B mRNA-editing enzyme catalytic polypeptide 1 (APOBEC-1) in mice lacking endogenous APOBEC-1 leads to aberrant hyperediting. Biochem J. 2003;369:255–262. doi: 10.1042/BJ20020694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Severi F, Chicca A, Conticello SG. Analysis of reptilian APOBEC1 suggests that RNA editing may not be its ancestral function. Mol Biol Evol. 2011;28:1125–1129. doi: 10.1093/molbev/msq338. [DOI] [PubMed] [Google Scholar]
- 41.Dance GS, Beemiller P, Yang Y, Mater DV, Mian IS, Smith HC. Identification of the yeast cytidine deaminase CDD1 as an orphan C-->U RNA editase. Nucleic Acids Res. 2001;29:1772–1780. doi: 10.1093/nar/29.8.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie K, Sowden MP, Dance GS, Torelli AT, Smith HC, Wedekind JE. The structure of a yeast RNA-editing deaminase provides insight into the fold and function of activation-induced deaminase and APOBEC-1. Proc Natl Acad Sci U S A. 2004;101:8114–8119. doi: 10.1073/pnas.0400493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith HC. Measuring editing activity and identifying cytidine-to-uridine mRNA editing factors in cells and biochemical isolates. Methods Enzymol. 2007;424:389–416. doi: 10.1016/S0076-6879(07)24018-2. [DOI] [PubMed] [Google Scholar]
- 44.Wedekind JE, Dance GS, Sowden MP, Smith HC. Messenger RNA editing in mammals: new members of the APOBEC family seeking roles in the family business. Trends Genet. 2003;19:207–216. doi: 10.1016/S0168-9525(03)00054-4. [DOI] [PubMed] [Google Scholar]
- 45.Kracker sDA. Insights into the B cell specific process of immunoglobulin class switch recombination. Immunol Lett. 2011;138:97–103. doi: 10.1016/j.imlet.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 46.Di Noia JMNM. Molecular Mechanisms of Antibody Somatic Hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 47.Muramatsu MSV, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific Expression of Activation-induced Cytidine Deaminase (AID), a Novel Member of the RNA-editing Deaminase Family in Germinal Center B Cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 48.Martin ASM. Antibody alterations. Nature. 2001;412:870–871. doi: 10.1038/35091184. [DOI] [PubMed] [Google Scholar]
- 49.Papavasiliou FNSD. Somatic Hypermutation of Immunoglobulin Genes: Merging Mechanisms for Genetic Diversity. Cell. 2002;109:S35–S44. doi: 10.1016/s0092-8674(02)00706-7. [DOI] [PubMed] [Google Scholar]
- 50.Odegard VhSD. Targeting of somatic hypermutation. Nature Rev Immun. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 51.Arakawa HHJ, Buerstedde JM. Requirement of the Activation-Induced Deaminase (AID) Gene for Immunoglobulin Gene Conversion. Science. 2001;295:1301. doi: 10.1126/science.1067308. [DOI] [PubMed] [Google Scholar]
- 52.Fugmann SD, Schatz DG. Immunology. One AID to unite them all. Science. 2002;295:1244–1245. doi: 10.1126/science.1070023. [DOI] [PubMed] [Google Scholar]
- 53.Muto TMM, Taniwaki M, Kinoshita K, Honjo T. Isolation, Tissue Distribution, and Chromosomal Localization of the Human Activation-Induced Cytidine Deaminase (AID) Gene. Genomics. 2000;68:85–88. doi: 10.1006/geno.2000.6268. [DOI] [PubMed] [Google Scholar]
- 54.Revy PMT, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Lagelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-Induced Cytidine Deaminase (AID) Deficiency Causes the Autosomal Recessive Form of the Hyper-IgM Syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 55.Muramatsu MKK, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 56.Okazaki IMKA, Honjo T. Role of AID in Tumorigenesis. Adv Immunol. 2007;94:245–273. doi: 10.1016/S0065-2776(06)94008-5. [DOI] [PubMed] [Google Scholar]
- 57.McCarthy HWW, Barron LL, Cromwell CC, Wang J, Coombes KR, Rangel R, Elenitoba-Johnson KS, Keating MJ, Abruzzo LV. High expression of activation-induced cytidine deaminase (AID) and splice variants is a distinctive feature of poor-prognosis chronic lymphocytic leukemia. Blood. 2003;101:4903–4908. doi: 10.1182/blood-2002-09-2906. [DOI] [PubMed] [Google Scholar]
- 58.Marantidou FDA, Stalika E, Korkolopoulou P, Saetta A, Anagnostopoulos A, Laoutaris N, Stamatopoulos K, Belessi C, Scouras Z, Patsouris E. Activation-induced cytidine deaminase splicing patterns in chronic lymphocytic leukemia. Blood Cells Mol Dis. 2010;44:262–267. doi: 10.1016/j.bcmd.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 59.Albesiano EMB, Damle RN, Allen SL, Rai KR, Chiorazzi N. Activation-induced cytidine deaminase in chronic lymphocytic leukemia B cells: expression as multiple forms in a dynamic, variably sized fraction of the clone. Blood. 2003;102:3333–3339. doi: 10.1182/blood-2003-05-1585. [DOI] [PubMed] [Google Scholar]
- 60.Wu XDJ, Chang SK, Nowakowski GS, Jelinek DF. Alternative splicing regulates activation-induced cytidine deaminase (AID): implications for suppression of AID mutagenic activity in normal and malignant B cells. Blood. 2008;112:4675–4682. doi: 10.1182/blood-2008-03-145995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Maldegem FJR, van Dijk R, Bende RJ, van Noesel CJ. Activation-induced cytidine deaminase splice variants are defective because of the lack of structural support for the catalytic site. J Immunol. 2010;184:2487–2491. doi: 10.4049/jimmunol.0903102. [DOI] [PubMed] [Google Scholar]
- 62.Okazaki IMHH, Kakazu N, Yamada S, Muramatsu M, Kinoshita K, Honjo T. Constitutive expression of AID leads to tumorigenesis. J Exp Med. 2003;197:1173–1181. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Endo Y, Marusawa H, Kinoshita K, Morisawa T, Sakurai T, Okazaki IM, et al. Expression of activation-induced cytidine deaminase in human hepatocytes via NF-kappaB signaling. Oncogene. 2007;26:5587–5595. doi: 10.1038/sj.onc.1210344. [DOI] [PubMed] [Google Scholar]
- 64.Muto TOI, Yamada S, Tanaka Y, Kinoshita K, Muramatsu M, Nagaoka H, Honjo T. Negative regulation of activation-induced cytidine deaminase in B cells. Proc Natl Acad Sci U S A. 2006;103:2752–2757. doi: 10.1073/pnas.0510970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sayegh CEQM, Agata Y, Murre C. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat Immunol. 2003;4:586–593. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- 66.Jannek Hauser NS, Anders Wallenius, Sanna Baradaran, Juha Saarikettu, Thomas Grundstro¨m. B-cell receptor activation inhibits AID expression through calmodulin inhibition of E-proteins. Proc Natl Acad Sci U S A. 2008;105:1267–1272. doi: 10.1073/pnas.0708220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ta VTNH, Catalan N, Durandy A, Fischer A, Imai K, Nonoyama S, Tashiro J, Ikegawa M, Ito S, Kinoshita K, Muramatsu M, Honjo T. AID mutant analyses indicate requirement for class-switch-specific cofactors. Nat Immunol. 2003;4:843–848. doi: 10.1038/ni964. [DOI] [PubMed] [Google Scholar]
- 68.Okuyama S, Marusawa H, Matsumoto T, Ueda Y, Matsumoto Y, Endo Y, et al. Excessive activity of apolipoprotein B mRNA editing enzyme catalytic polypeptide 2 (APOBEC2) contributes to liver and lung tumorigenesis. Int J Cancer. 2011 doi: 10.1002/ijc.26114. in press. [DOI] [PubMed] [Google Scholar]
- 69.Prochnow C, Bransteitter R, Klein MG, Goodman MF, Chen XS. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445:447–451. doi: 10.1038/nature05492. [DOI] [PubMed] [Google Scholar]
- 70.Lada AG, Krick CF, Kozmin SG, Mayorov VI, Karpova TS, Rogozin IB, et al. Mutator effects and mutation signatures of editing deaminases produced in bacteria and yeast. Biochemistry (Mosc) 2011;76:131–146. doi: 10.1134/s0006297911010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sato Y, Probst HC, Tatsumi R, Ikeuchi Y, Neuberger MS, Rada C. Deficiency in APOBEC2 leads to a shift in muscle fiber type, diminished body mass, and myopathy. J Biol Chem. 2010;285:7111–7118. doi: 10.1074/jbc.M109.052977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.OhAinle M, Kerns JA, Malik HS, Emerman M. Adaptive evolution and antiviral activity of the conserved mammalian cytidine deaminase APOBEC3H. J Virol. 2006;80:3853–3862. doi: 10.1128/JVI.80.8.3853-3862.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, et al. An Anthropoid-Specific Locus of Orphan C to U RNA-Editing Enzymes on Chromosome 22. Genomics. 2002;79:285–296. doi: 10.1006/geno.2002.6718. [DOI] [PubMed] [Google Scholar]
- 74.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 75.Smith HC. APOBEC3G: a double agent in defense. Trends Biochem Sci. 2011;36:239–244. doi: 10.1016/j.tibs.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 77.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hultquist JF, Lengyel JA, Refsland EW, Larue RS, Lackey L, Brown WL, et al. Human and Rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H Demonstrate a Conserved Capacity to Restrict Vif-deficient HIV-1. J Virol. 2011 doi: 10.1128/JVI.05238-11. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zielonka J, Bravo IG, Marino D, Conrad E, Perkovic M, Battenberg M, et al. Restriction of equine infectious anemia virus by equine APOBEC3 cytidine deaminases. J Virol. 2009;83:7547–7559. doi: 10.1128/JVI.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dang Y, Wang X, Esselman WJ, Zheng YH. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J Virol. 2006;80:10522–10533. doi: 10.1128/JVI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Delebecque F, Suspene R, Calattini S, Casartelli N, Saib A, Froment A, et al. Restriction of foamy viruses by APOBEC cytidine deaminases. J Virol. 2006;80:605–614. doi: 10.1128/JVI.80.2.605-614.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liddament MT, Brown WL, Schumacher AJ, Harris RS. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr Biol. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 83.Thielen BK, McNevin JP, McElrath MJ, Hunt BV, Klein KC, Lingappa JR. Innate immune signaling induces high levels of TC-specific deaminase activity in primary monocyte-derived cells through expression of APOBEC3A isoforms. J Biol Chem. 2010;285:27753–27766. doi: 10.1074/jbc.M110.102822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bulliard Y, Narvaiza I, Bertero A, Peddi S, Rohrig UF, Ortiz M, et al. Structure-function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities. J Virol. 2011;85:1765–1776. doi: 10.1128/JVI.01651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bonvin M, Achermann F, Greeve I, Stroka D, Keogh A, Inderbitzin D, et al. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology. 2006;43:1364–1374. doi: 10.1002/hep.21187. [DOI] [PubMed] [Google Scholar]
- 86.Xu R, Zhang X, Zhang W, Fang Y, Zheng S, Yu XF. Association of human APOBEC3 cytidine deaminases with the generation of hepatitis virus B x antigen mutants and hepatocellular carcinoma. Hepatology. 2007;46:1810–1820. doi: 10.1002/hep.21893. [DOI] [PubMed] [Google Scholar]
- 87.Chiu YL, Witkowska HE, Hall SC, Santiago M, Soros VB, Esnault C, et al. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103:15588–15593. doi: 10.1073/pnas.0604524103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schumann GG. APOBEC3 proteins: major players in intracellular defence against LINE-1-mediated retrotransposition. Biochem Soc Trans. 2007;35:637–642. doi: 10.1042/BST0350637. [DOI] [PubMed] [Google Scholar]
- 89.Esnault C, Millet J, Schwartz O, Heidmann T. Dual inhibitory effects of APOBEC family proteins on retrotransposition of mammalian endogenous retroviruses. Nucleic Acids Res. 2006;34:1522–1531. doi: 10.1093/nar/gkl054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schumacher AJ, Nissley DV, Harris RS. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc Natl Acad Sci U S A. 2005;102:9854–9859. doi: 10.1073/pnas.0501694102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hulme AE, Bogerd HP, Cullen BR, Moran JV. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene. 2007;390:199–205. doi: 10.1016/j.gene.2006.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bogerd HP, Wiegand HL, Doehle BP, Lueders KK, Cullen BR. APOBEC3A and APOBEC3B are potent inhibitors of LTR-retrotransposon function in human cells. Nucleic Acids Res. 2006;34:89–95. doi: 10.1093/nar/gkj416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, et al. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 94.Muckenfuss H, Hamdorf M, Held U, Perkovic M, Lower J, Cichutek K, et al. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J Biol Chem. 2006;281:22161–22172. doi: 10.1074/jbc.M601716200. [DOI] [PubMed] [Google Scholar]
- 95.Stenglein MD, Harris RS. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J Biol Chem. 2006;281:16837–16841. doi: 10.1074/jbc.M602367200. [DOI] [PubMed] [Google Scholar]
- 96.Kinomoto M, Kanno T, Shimura M, Ishizaka Y, Kojima A, Kurata T, et al. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 2007;35:2955–2964. doi: 10.1093/nar/gkm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tan L, Sarkis PT, Wang T, Tian C, Yu XF. Sole copy of Z2-type human cytidine deaminase APOBEC3H has inhibitory activity against retrotransposons and HIV-1. Faseb J. 2009;23:279–287. doi: 10.1096/fj.07-088781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wichroski MJ, Robb GB, Rana TM. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2006;2:e41. doi: 10.1371/journal.ppat.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Landry S, Narvaiza I, Linfesty DC, Weitzman MD. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 2011;12:444–450. doi: 10.1038/embor.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rogozin IB, Basu MK, Jordan IK, Pavlov YI, Koonin EV. APOBEC4, a new member of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases predicted by computational analysis. Cell Cycle. 2005;4:1281–1285. doi: 10.4161/cc.4.9.1994. [DOI] [PubMed] [Google Scholar]
- 101.Yang YMP, Sowden Y, Yang HC, Smith Intracellular Trafficking Determinants in APOBEC-1, the Catalytic Subunit for Cytidine to Uridine Editing of Apolipoprotein B mRNA. Exp Cell Res. 2001;267:153–164. doi: 10.1006/excr.2001.5255. [DOI] [PubMed] [Google Scholar]
- 102.Yang Y, Smith HC. Multiple protein domains determine the cell type-specific nuclear distribution of the catalytic subunit required for apolipoprotein B mRNA editing. Proc Natl Acad Sci U S A. 1997;94:13075–13080. doi: 10.1073/pnas.94.24.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang Y, Sowden MP, Smith HC. Induction of cytidine to uridine editing on cytoplasmic apolipoprotein B mRNA by overexpressing APOBEC-1. J Biol Chem. 2000;275:22663–22669. doi: 10.1074/jbc.M910406199. [DOI] [PubMed] [Google Scholar]
- 104.Lau PPW, Xiong H-J, Zhu S-H, Chen L, Chan Apo B mRNA editing is an intranuclear event that occurs posttranscriptionally coincident with splicing and polyadenylation. J Biol Chem. 1991;266:20550–20554. [PubMed] [Google Scholar]
- 105.Smith HC, Kuo SR, Backus JW, Harris SG, Sparks CE, Sparks JD. In vitro apolipoprotein B mRNA editing: identification of a 27S editing complex. Proc Natl Acad Sci U S A. 1991;88:1489–1493. doi: 10.1073/pnas.88.4.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang Y, Kovalski K, Smith HC. Partial characterization of the auxiliary factors involved in apolipoprotein B mRNA editing through APOBEC-1 affinity chromatography. J Biol Chem. 1997;272:27700–27706. doi: 10.1074/jbc.272.44.27700. [DOI] [PubMed] [Google Scholar]
- 107.Blanc V, Kennedy SM, Davidson NO. A novel nuclear localization signal in the auxiliary domain of apobec-1 complementation factor (ACF) regulates nucleo-cytoplasmic import and shuttling. J Biol Chem. 2003;278:21148–21204. doi: 10.1074/jbc.M302951200. [DOI] [PubMed] [Google Scholar]
- 108.Smith HC, Wedekind JE, Kefang X, Sowden MP. Mammalian C to U Editing. Topics in Current Biol. 2005;12:365–400. [Google Scholar]
- 109.Galloway CA, Sowden MP, Smith HC. Increasing the yield of soluble recombinant protein expressed in E. coli by induction during late log phase. Biotechniques. 2003;34:524–526. 8. doi: 10.2144/03343st04. [DOI] [PubMed] [Google Scholar]
- 110.Lehmann DM, Galloway CA, Sowden MP, Smith HC. Metabolic regulation of apoB mRNA editing is associated with phosphorylation of APOBEC-1 complementation factor. Nucleic Acids Res. 2006;34:3299–3308. doi: 10.1093/nar/gkl417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dance GSC, Sowden MP, Cartegni L, Cooper E, Krainer AR, Smith HC. Two proteins essential for apolipoprotein B mRNA editing are expressed from a single gene through alternative splicing. J Biol Chem. 2002;277:12703–12709. doi: 10.1074/jbc.M111337200. [DOI] [PubMed] [Google Scholar]
- 112.Sowden MP, Lehmann DM, Lin X, Smith CO, Smith HC. Identification of Novel Alternative Splice Variants of APOBEC-1 Complementation Factor with Different Capacities to Support ApoB mRNA Editing. J Biol Chem. 2004;278:197–206. doi: 10.1074/jbc.M307920200. [DOI] [PubMed] [Google Scholar]
- 113.Patenaude AM, Di Noia JM. The mechanisms regulating the subcellular localization of AID. Nucleus. 2010;1:325–331. doi: 10.4161/nucl.1.4.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Stavnezer Complex regulation and function of activation- induced cytidine deaminase. Trends in Immunology. 2011;32:194–201. doi: 10.1016/j.it.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Basu UFA, Alt FW. Post-translational regulation of activation-induced cytidine deaminase. Phil Trans R Soc B. 2009;364:667–673. doi: 10.1098/rstb.2008.0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rada C, Jarvis JM, Milstein C. AID-GFP chimeric protein increases hypermutation of Ig genes with no evidence of nuclear localization. Proc Natl Acad Sci U S A. 2002;99:7003–7008. doi: 10.1073/pnas.092160999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Patenaude AMOA, Hu Y, Campo VH, Kavli B, Buschiazzo A, Di Noia JM. Active nuclear import and cytoplasmic retention of activation-induced deaminase. Nat Struct Molec Biol. 2009;16:517–527. doi: 10.1038/nsmb.1598. [DOI] [PubMed] [Google Scholar]
- 118.McBride KM, Barreto VM, Ramiro AR, Stavropoulos P, Nussenzweig MC. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation induced deaminase. Journal of Experimental Medicine. 2004;199:1235–1244. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ito S. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci USA. 2004;101:1975–1980. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ganesh KAS, Taylor B, Simpson P, Rada C, Neuberger M. CTNNBL1 is a novel nuclear localization sequence-binding protein that recognizes RNA-splicing factors CDC5L and Prp31. J Biol Chem. 2011;286:17091–17102. doi: 10.1074/jbc.M110.208769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Orthwein APA, Affar EB, Lamarre AYJ, Di Noia JM. Regulation of activation-induced deaminase stability and antibody gene diversification by Hsp90. J Exp Med. 2010;207:2751–2765. doi: 10.1084/jem.20101321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Said Aoufouchi AF. 1 Carole Zober, 1 Orietta D ’ Orlando, 2, Sandra Weller J-CW, 1 and Claude-Agn è s Reynaud 1. Proteasomal degradation restricts the nuclear lifespan of AID. J Exp Med. 2008;205:1357–1368. doi: 10.1084/jem.20070950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bennett RP, Presnyak V, Wedekind JE, Smith HC. Nuclear Exclusion of the HIV-1 host defense factor APOBEC3G requires a novel cytoplasmic retention signal and is not dependent on RNA binding. J Biol Chem. 2008;283:7320–7327. doi: 10.1074/jbc.M708567200. [DOI] [PubMed] [Google Scholar]
- 124.Huthoff H, Autore F, Gallois-Montbrun S, Fraternali F, Malim MH. RNA-dependent oligomerization of APOBEC3G is required for restriction of HIV-1. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000330. e1000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huthoff H, Malim MH. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J Virol. 2007;81:3807–3815. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Henry M, Guetard D, Suspene R, Rusniok C, Wain-Hobson S, Vartanian JP. Genetic editing of HBV DNA by monodomain human APOBEC3 cytidine deaminases and the recombinant nature of APOBEC3G. PLoS One. 2009;4:e4277. doi: 10.1371/journal.pone.0004277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stopak KS, Chiu YL, Kropp J, Grant RM, Greene WC. Distinct patterns of cytokine regulation of APOBEC3G expression and activity in primary lymphocytes, macrophages, and dendritic cells. J Biol Chem. 2007;282:3539–3546. doi: 10.1074/jbc.M610138200. [DOI] [PubMed] [Google Scholar]
- 128.Kreisberg JF, Yonemoto W, Greene WC. Endogenous factors enhance HIV infection of tissue naive CD4 T cells by stimulating high molecular mass APOBEC3G complex formation. J Exp Med. 2006;203:865–870. doi: 10.1084/jem.20051856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Goila-Gaur R, Khan MA, Miyagi E, Kao S, Opi S, Takeuchi H, et al. HIV-1 Vif promotes the formation of high molecular mass APOBEC3G complexes. Virology. 2008;372:136–146. doi: 10.1016/j.virol.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kozak SL, Marin M, Rose KM, Bystrom C, Kabat D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J Biol Chem. 2006;281:29105–29119. doi: 10.1074/jbc.M601901200. [DOI] [PubMed] [Google Scholar]
- 131.Huang J, Liang Z, Yang B, Tian H, Ma J, Zhang H. Derepression of microRNA-mediated protein translation inhibition by apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) and its family members. J Biol Chem. 2007;282:33632–33640. doi: 10.1074/jbc.M705116200. [DOI] [PubMed] [Google Scholar]
- 132.Gallois-Montbrun S, Kramer B, Swanson CM, Byers H, Lynham S, Ward M, et al. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J Virol. 2007;81:2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.McDougall WM, Smith HC. Direct evidence that RNA inhibits APOBEC3G ssDNA cytidine deaminase activity. Biochem Biophys Res Commun. 2011 doi: 10.1016/j.bbrc.2011.08.009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wedekind JE, Gillilan R, Janda A, Krucinska J, Salter JD, Bennett RP, et al. Nanostructures of APOBEC3G support a hierarchical assembly model of high molecular mass ribonucleoprotein particles from dimeric subunits. J Biol Chem. 2006;281:38122–38126. doi: 10.1074/jbc.C600253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Soros VB, Yonemoto W, Greene WC. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, subsequently activated by RNase H. PLoS Pathog. 2007;3:e15. doi: 10.1371/journal.ppat.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Khan MA, Goila-Gaur R, Opi S, Miyagi E, Takeuchi H, Kao S, et al. Analysis of the contribution of cellular and viral RNA to the packaging of APOBEC3G into HIV-1 virions. Retrovirology. 2007;4:48. doi: 10.1186/1742-4690-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Svarovskaia ES, Xu H, Mbisa JL, Barr R, Gorelick RJ, Ono A, et al. Human APOBEC3G is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J Biol Chem. 2004;270:35822–35828. doi: 10.1074/jbc.M405761200. [DOI] [PubMed] [Google Scholar]
- 138.Zhang W, Du J, Yu K, Wang T, Yong X, Yu XF. Association of potent human antiviral cytidine deaminases with 7SL RNA and viral RNP in HIV-1 virions. J Virol. 2010;84:12903–12913. doi: 10.1128/JVI.01632-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang T, Tian C, Zhang W, Luo K, Sarkis PT, Yu L, et al. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J Virol. 2007;81:13112–13124. doi: 10.1128/JVI.00892-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang TTC, Zhang W, Sarkis PT, Yu XF. Interaction with 7SL RNA but not with HIV-1 genomic RNA or P bodies is required for APOBEC3F virion packaging. J Molec Biol. 2008;375:1098–1112. doi: 10.1016/j.jmb.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 141.Alce TM, Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J Biol Chem. 2004;4:97–107. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- 142.Cen S, Guo F, Niu M, Saadatmand J, Deflassieux J, Kleiman L. The interaction between HIV-1 Gag and APOBEC3G. J Biol Chem. 2004;279:33177–33184. doi: 10.1074/jbc.M402062200. [DOI] [PubMed] [Google Scholar]
- 143.Luo K, Liu B, Xiao Z, Yu Y, Yu X, Gorelick R, et al. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J Virol. 2004;78:11841–11852. doi: 10.1128/JVI.78.21.11841-11852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.McDougall WM, Smith HC. Direct Evidence that RNA Inhibits APOBEC3G ssDNACytidine Deaminase Activity. Biophys Biochem Res Commun. 2011 doi: 10.1016/j.bbrc.2011.08.009. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang T, Zhang W, Tian C, Liu B, Yu Y, Ding L, et al. Distinct viral determinants for the packaging of human cytidine deaminases APOBEC3G and APOBEC3C. Virology. 2008;377:71–79. doi: 10.1016/j.virol.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Stauch BHH, Perkovic M, Weisel M, Kopietz F, Cichutek K, Münk C, Schneider G. Model structure of APOBEC3C reveals a binding pocket modulating ribonucleic acid interaction required for encapsidation. Proc Natl Acad Sci U S A. 2009;106:12079–12084. doi: 10.1073/pnas.0900979106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nonaka TDT, Toyoshima T, Muramatsu M, Honjo T, Kinoshita K. Carboxy-terminal domain of AID required for its mRNA complex formation in vivo. Proc Natl Acad Sci U S A. 2009;106:2747–2751. doi: 10.1073/pnas.0812957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A. 2003;100:4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Iwatani Y, Takeuchi H, Strebel K, Levin JG. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J Virol. 2006;80:5992–6002. doi: 10.1128/JVI.02680-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Navarro F, Bollman B, Chen H, Konig R, Yu Q, Chiles K, et al. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]