Summary
Mutations within BRCA1 predispose carriers to a high risk of breast and ovarian cancers. BRCA1 functions to maintain genomic stability through the assembly of multiple protein complexes involved in DNA repair, cell-cycle arrest, and transcriptional regulation. Here, we report the identification of a DNA damage-induced BRCA1 protein complex containing BCLAF1 and other key components of the mRNA-splicing machinery. In response to DNA damage, this complex regulates pre-mRNA splicing of a number of genes involved in DNA damage signaling and repair, thereby promoting the stability of these transcripts/proteins. Further, we show that abrogation of this complex results in sensitivity to DNA damage, defective DNA repair, and genomic instability. Interestingly, mutations in a number of proteins found within this complex have been identified in numerous cancer types. These data suggest that regulation of splicing by the BRCA1-mRNA splicing complex plays an important role in the cellular response to DNA damage.
Graphical Abstract
Highlights
-
•
BRCA1 forms a complex with mRNA splicing proteins following DNA damage
-
•
The BRCA1-mRNA splicing complex promotes the stability of genes involved in the DDR
-
•
BRCA1-dependent splicing of DDR genes maintains their protein expression
-
•
The BRCA1-mRNA splicing complex is required for DNA repair and genomic stability
BRCA1 functions to maintain genomic integrity by forming multiple protein complexes with different functions. Here, Savage et al. report the identification of a BRCA1 protein complex that promotes the mRNA splicing and expression of genes required for maintaining genomic stability.
Introduction
The DNA damage response (DDR) pathway has evolved to protect cells from both endogenous and exogenous sources of DNA damage and ultimately to prevent tumorigenic transformation. One of the key players in the DDR pathway is BRCA1. Heterozygous mutations within BRCA1 predispose carriers to a high risk of breast and ovarian cancer (Savage and Harkin, 2009). BRCA1 functions to maintain genomic stability and plays key roles in cell-cycle checkpoint activation, homologous recombination (HR)-mediated DNA double-strand break (DSB) repair, and transcriptional regulation (Savage and Harkin, 2009). BRCA1 broadly functions as a scaffolding protein, facilitating the assembly of multiple and distinct multiprotein complexes, with various functions within the DDR. The formation and function of these complexes are thought to be regulated by phosphorylation of BRCA1 by the ATM, ATR, and Chk2 kinases in response to DNA damage, and a number of BRCA1 functions have been attributed to these phosphorylation events. For example, DNA damage-induced phosphorylation of BRCA1 serine-1423 and serine-1524 by ATM or ATR is required for resistance to ionizing radiation (IR) and G1/S and G2/M-phase arrest, respectively, whereas phosphorylation of serine-1387 is specifically required for intra-S phase arrest (Cortez et al., 1999). However, despite the broad functions associated with different BRCA1 phosphorylation sites, the mechanistic role(s) of specific DNA damage-induced BRCA1 phosphorylation events remains largely unknown.
Here we identify a DNA damage-induced BRCA1 binding protein, BCLAF1, which mediates the formation of a BRCA1-mRNA splicing complex following DNA damage. We show that through this interaction with BCLAF1, BRCA1, which is constitutively bound to a subset of genes, recruits the mRNA splicing machinery, resulting in enhanced pre-mRNA splicing of BRCA1/BCLAF1 target genes, thereby promoting transcript stability and protein expression. Intriguingly, many of the genes/proteins regulated by the BRCA1/BCLAF1 complex are involved in the DDR, and depletion of BRCA1, BCLAF1, and other members of the BRCA1-mRNA splicing complex (U2AF65) results in sensitivity to DNA damage and defective DNA repair.
Results
Identification of BCLAF1 as a BRCA1pSer1423-Interacting Protein
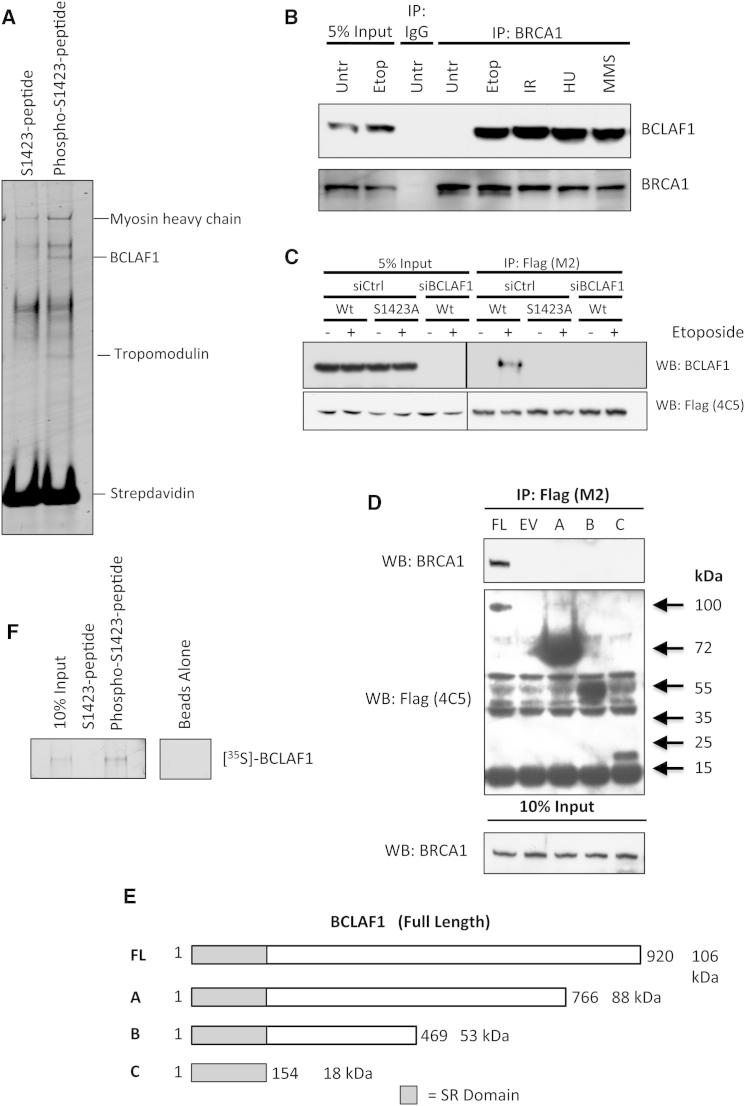
To examine how DNA damage-induced BRCA1 phosphorylation events might mechanistically regulate associated BRCA1 functions, we performed phosphopeptide pull-down assays followed by LC-MS/MS, to identify phospho-BRCA1-interacting proteins. Using this approach, we identified BCLAF1 as a BRCA1 phosphoserine-1423 (pSer-1423)-interacting protein (Figure 1A and see Figure S1A available online). Coimmunoprecipitation confirmed that BCLAF1 interacts with BRCA1 in response to DNA alkylation (MMS), stalled replication forks (HU), and DNA double-strand breaks (IR and etoposide) and is not mediated indirectly through RNA/DNA bridging (Figures 1B, S1B, and S1C). This suggests that this complex forms as part of a general DDR mechanism and is likely a reflection of BRCA1Ser-1423 being a substrate of both ATM and ATR, which are activated in response to DSBs or DNA single-strand breaks/stalled replication forks, respectively. In keeping with this, substitution of BRCA1Ser-1423 with alanine abrogated the damage-induced interaction between BRCA1 and BCLAF1, confirming BCLAF1 as a BRCA1pSer-1423 interacting protein (Figure 1C). Coimmunoprecipitation experiments with a series of Flag-tagged BCLAF1 truncated proteins (harvested from etoposide treated cells) revealed that deletion of the C-terminal region of BCLAF1 abolishes its ability to interact with BRCA1 (Figures 1D and 1E). The C terminus of BCLAF1 contains no defined domains, though it is positively charged under physiological conditions (pI = ∼9.5), suggesting that the interaction between BRCA1pSer-1423 and the BCLAF1 C terminus may occur directly. Consistent with this, in vitro-translated BCLAF1 bound strongly to the phosphorylated Ser1423-BRCA1 peptide, but not its nonphosphorylated counterpart (Figures 1F and S1D).
Figure 1.
BCLAF1 Interacts with Phosphorylated BRCA1 following DNA Damage
(A) Colloidal Coomassie-stained gel of peptide pull-down assays carried out from 293T cell nuclear extracts with phosphorylated BRCA1-S1423 peptide and its nonphosphorylated counterpart. The indicated phosphopeptide-interacting band was identified as BCLAF1 by LC-MS/MS.
(B) Coimmunoprecipitation assay demonstrating an interaction between BRCA1 and BCLAF1 in 293T cells treated with etoposide (1 μM, 16 hr), IR (2 Gy, 1 hr), MMS (200 μM, 6 hr), and HU (5 μM, 3 hr).
(C) Coimmunoprecipitation assay demonstrating DNA damage-induced interaction of BCLAF1 with ectopic Flag-BRCA1 is abrogated by BRCA1-S1423A phosphosite substitution. A Flag-BRCA1 IP was also carried out from cells depleted of BCLAF1 to confirm the specificity of the BCLAF1 antibody.
(D) Mapping of the BRCA1-interacting region within BCLAF1. Coimmunoprecipitation experiments were carried out from etoposide-treated cells transfected with the Flag-BCLAF1 truncation mutant constructs depicted in (E).
(E) Schematic diagram of BCLAF1 truncation constructs used for BRCA1 coimmunoprecipitation experiments in (D).
(F) Peptide pull-down assays carried out with [35S] in vitro-translated BCLAF1, indicating that BCLAF1 interacts directly and specifically with the phosphorylated BRCA1-S1423 peptide and not its unphosphorylated counterpart. See also Figure S1.
BCLAF1 Promotes Resistance to DNA Damage and Is Required for Efficient DNA Repair and Maintenance of Genomic Stability
BCLAF1 was first identified as a Bcl2-associated transcription factor that promotes apoptosis. Further studies found that BCLAF1 binds the TP53 promoter in response to Adriamycin treatment, where it is required for PKC-delta-mediated TP53 transcription and apoptosis (Liu et al., 2007). However, we and others have been unable to demonstrate a role for BCLAF1 in the regulation of TP53 expression following DNA damage (McPherson et al., 2009). Additionally, BCLAF1 null (−/−) mice do not appear to have an altered apoptotic response; rather they exhibit immunodevelopment defects and die within 24–48 hr after birth due to gross lung malformation (McPherson et al., 2009).
BRCA1 mediates resistance to DNA-damaging agents, and phosphorylation of BRCA1Ser-1423 has also been linked with this function (Cortez et al., 1999). Therefore, to evaluate if BCLAF1 may also play a role in this process, we assessed the effect of BCLAF1 depletion on cellular survival following DNA damage. Interestingly, BCLAF1 depletion resulted in sensitization to both IR and etoposide to an equivalent level as BRCA1 depletion (Figures 2A, 2B, and S2A). To determine whether BRCA1 and BCLAF1 promote resistance to DNA damage through a common pathway, we examined the effect of BCLAF1 depletion on cellular survival following DNA damage in the BRCA1-deficient MDA-MB-436 breast cancer cell line, which we stably transfected with an empty vector (EV) or a BRCA1 expression vector (Elstrodt et al., 2006). Surprisingly, BCLAF1 depletion did not sensitize the BRCA1-deficient MDA-MB-436-EV cells and only sensitized these cells when reconstituted with ectopic BRCA1, suggesting that BCLAF1 and BRCA1 may be epistatic, at least in mediating resistance to IR-induced DSBs (Figures S2C and S2D).
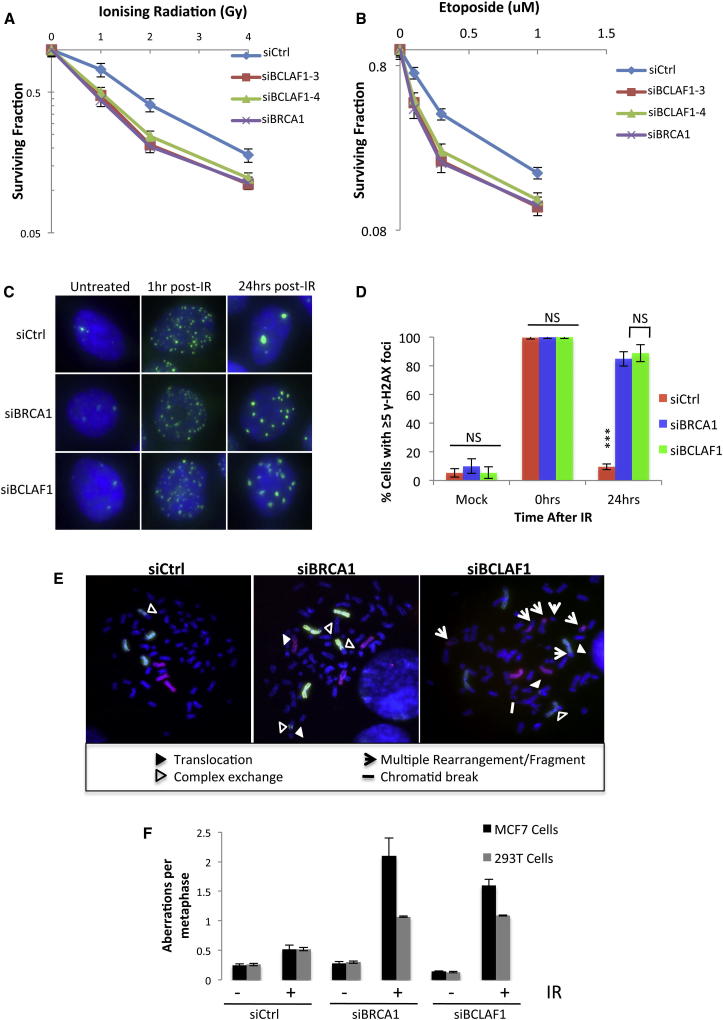
Figure 2.
BRCA1/BCLAF1 Mediates Resistance to DNA Damage and Is Required for Efficient DNA Repair and Maintenance of Genomic Stability
(A and B) Clonogenic survival assays demonstrating that depletion of BRCA1 or BCLAF1 (two independent siRNAs) induces sensitivity to ionizing radiation (IR) and etoposide in (MCF7) cells. Mean surviving fraction of three independent experiments is plotted ± SEM.
(C) Representative immunofluorescent staining of γ-H2AX marked DNA damage in untreated 293T cells depleted of either BRCA1 or BCLAF1 and 1 and 24 hr following 2Gy IR.
(D) Quantification of three independent experiments described above (≥200 cells were scored/experiment). Mean fraction of cells containing ≥5 γ-H2AX foci is plotted ± SEM. Significant differences in the fraction of cells containing ≥5 γ-H2AX foci were assessed using Student’s two-tailed t test and are indicated by ∗∗∗p < 0.001.
(E) Representative metaphase spreads of control (siCtrl) and BRCA1- or BCLAF1-depleted 293T cells either untreated or 24 hr following 2Gy IR. FISH-mediated whole chromosome painting (chromosome 1, green; chromosome 2, red) was used to identify complex chromosome aberrations.
(F) Quantification of total chromosome aberrations in control, BRCA1, and BCLAF1 depleted 293T and MCF7 cells 24 hr after mock irradiation or irradiation with 2 Gy IR. Graphs represent the mean number of chromosome aberrations/metaphase from three independent experiments ± SEM (≥200 metaphases scored/experiment). See also Figure S2.
The direct role of BRCA1 in DNA DSB repair is thought to contribute strongly to its ability to promote cellular survival following DNA damage. To evaluate if BCLAF1 may also play a role in DNA repair, we assessed DNA repair kinetics in both BRCA1- and BCLAF1-depleted cells 0 and 24 hr following DNA damage. Surprisingly, like BRCA1-depleted cells, BCLAF1-depleted cells also exhibited a significant defect in their ability to resolve γ-H2AX-marked DNA breaks 24 hr after IR treatment (Figures 2C, 2D, S2D, and S2E). Moreover, depletion of BCLAF1 using an shRNA also resulted in sensitization to IR and defective DNA repair, which was rescued by ectopic expression of shRNA resistant BCLAF1 (Figures S2F–S2H). Additionally, IR-treated cells depleted of BRCA1 or BCLAF1 displayed a marked increase in chromosome aberrations in comparison to control cells, indicating that loss of BCLAF1 results in increased genomic instability following DNA damage (Figures 2E and 2F). BRCA1Ser-1423 phosphorylation has also been linked with G1/S and G2/M checkpoint function. However, we did not observe any checkpoint defects in BCLAF1-depleted cells, suggesting that the BRCA1pSer-1423-dependent interaction with BCLAF1 does not play a role in DNA damage-induced cell-cycle arrest (data not shown). BRCA1 plays a direct role in HR-mediated DSB repair, during which it localizes to DNA break sites. Given the dramatic DNA repair defect observed in BCLAF1-depleted cells, we examined BCLAF1 cellular localization following DNA damage. Unlike BRCA1, we found that BCLAF1 was excluded from DNA break sites induced by laser microirradiation (Figure S2I).
Similar findings were recently reported for BCLAF1 and THRAP3, a protein sharing 48% identity with BCLAF1 and which has also been identified as a DNA damage-induced ATM/ATR phosphorylation substrate (Beli et al., 2012). BCLAF1 and THRAP3 associate within a complex containing a number of mRNA processing proteins, which promotes the efficient splicing of Cyclin-D1 pre-mRNAs, functioning to generate stable postspliced Cyclin-D1 transcripts (Bracken et al., 2008). In support of a role in mRNA splicing/processing, Beli et al. found that exclusion of THRAP3 and associated factors such as BCLAF1 from DNA break sites was concomitant with inhibition of transcription and subsequent loss of mRNA processing at sites of DNA damage mediated by ATM/ATR/DNA-PK (Beli et al., 2012). Taken together, these findings support a role for BCLAF1 in mRNA processing/splicing and suggest that, unlike BRCA1, BCLAF1’s role in DNA repair is likely to be indirect.
BRCA1/BCLAF1 Interaction Mediates the Formation of a BRCA1-mRNA Splicing Complex, which Drives the Splicing of a Subset of Genes following DNA Damage
BCLAF1 contains a Serine-Arginine (SR) rich region within its N terminus, which is consistent with a role in pre-mRNA processing and/or splicing (Cáceres et al., 1997). In addition, as mentioned above, BCLAF1 has been shown to form part of an mRNA splicing/processing complex required for the production of stable spliced Cyclin-D1 transcripts (Bracken et al., 2008). BCLAF1 has also been copurified with the core splicing machinery, within both spliced and unspliced human mRNP complexes, further suggesting a role in mRNA splicing (Merz et al., 2007). Indeed, when examining its subcellular localization, BCLAF1 was localized to pannuclear speckles, in both unperturbed and DNA damage-treated cells, a pattern consistent with interchromatin granule clusters formed by proteins, such as U2AF65, involved in pre-mRNA processing and splicing (Cáceres et al., 1997) (Figure S2J).
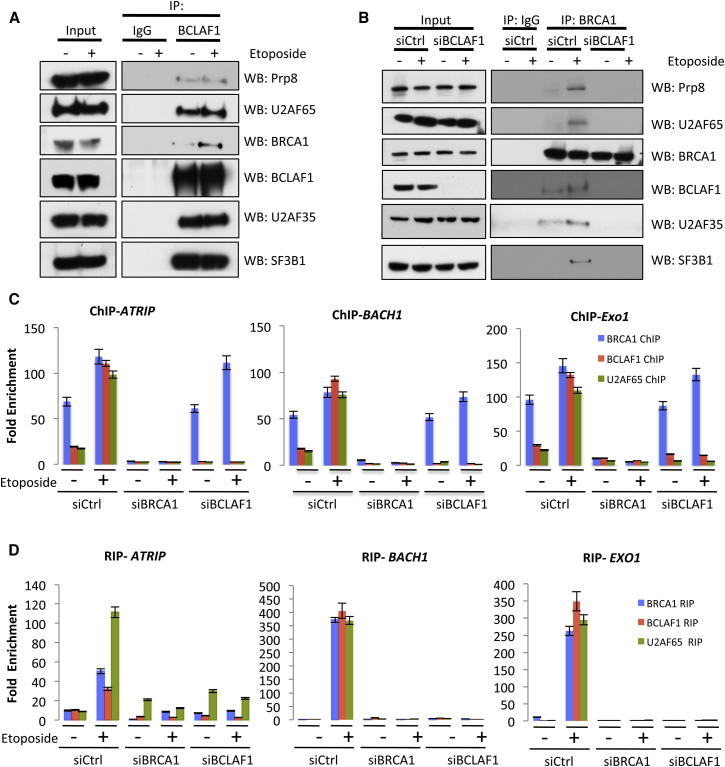
Given this previously identified role for BCLAF1 in pre-mRNA splicing and interaction with a number of core splicing machinery proteins, we examined the ability of BCLAF1 and BRCA1 to interact with known components of the BCLAF1 interacting spliceosome (Merz et al., 2007). Coimmunoprecipitation confirmed that BCLAF1 constitutively interacts with a number of these core mRNA splicing proteins such as Prp8, U2AF65, U2AF35, and SF3B1, independently of DNA damage (Figure 3A). In contrast, BRCA1 coprecipitated with Prp8, U2AF65, U2AF35, and SF3B1 only in response to DNA damage (Figure 3B). Furthermore, depletion of BCLAF1 resulted in abrogation of the damage-induced interaction between BRCA1 and these proteins, suggesting that BCLAF1 mediates the interaction between phosphorylated BRCA1 and core components of the spliceosome in response to DNA damage (Figure 3B).
Figure 3.
BRCA1/BCLAF1 Forms an mRNA Splicing Complex which Is Recruited to Target Gene Promoters and Transcripts following DNA Damage
(A) Coimmunoprecipitation assays demonstrating that BCLAF1 interacts with the spliceosome proteins Prp8, U2AF65, U2AF35, and SF3B1 in both the presence and absence of DNA damage.
(B) Coimmunoprecipitation assays demonstrating DNA damage-induced interaction between BRCA1 and the spliceosome proteins Prp8, U2AF65 U2AF35, and SF3B1 in response to DNA damage. Additionally, depletion of BCLAF1 results in abrogation of DNA damage-induced interaction between BRCA1 and these proteins.
(C) BRCA1, BCLAF1, and U2AF65 ChIP-qPCRs demonstrating constitutive binding of BRCA1 to ATRIP, BACH1, and EXO1 promoters irrespective of DNA damage in control (siCtrl) cells. The ChIPs also demonstrate that BCLAF1 and U2AF65 are recruited to these promoters only in etoposide-treated cells and that depletion of BRCA1 or BCLAF1 results in loss of DNA damage-induced BCLAF1 and U2AF65 recruitment, respectively. Graphs represent the mean fold enrichment quantified from three independent experiments ± SEM.
(D) BRCA1, BCLAF1, and U2AF65 RIP-qRT-PCRs demonstrating that BRCA1, BCLAF1, and U2AF65 only bind to ATRIP, BACH1, and EXO1 mRNAs in response to DNA damage. In addition, depletion of BCLAF1 results in loss of BRCA1 and U2AF65 mRNA binding to all three transcripts. Graphs represent the mean fold enrichment quantified from three independent experiments ± SEM. See also Figure S3.
We have previously demonstrated that BRCA1 is bound to a large subset of gene promoters throughout the genome, though it does not regulate the transcription of the majority of these genes in unperturbed cells (Gorski et al., 2011). However, in response to various stresses, such as DNA damage, expression of many of these genes is regulated in a BRCA1-dependent manner. BRCA1Ser-1423 phosphorylation is thought to occur at DNA break sites, where it colocalizes with active ATM/ATR. In contrast, we and others have observed that BCLAF1, which only interacts with BRCA1pSer-1423, is excluded from DNA break sites (Beli et al., 2012). Therefore, to examine whether BRCA1Ser-1423 phosphorylation is restricted to DNA break sites, or may occur on BRCA1 bound to chromatin more globally, we performed high-resolution confocal microscopy with BRCA1pSer-1423 antibodies on cells following extraction of non-chromatin-bound proteins. As expected, this revealed that BRCA1pSer-1423 is concentrated at DNA break sites (marked by γ-H2AX) but also revealed the presence of chromatin bound BRCA1pSer-1423 throughout the nucleus, which was abolished by depletion of BRCA1 using specific siRNAs. This suggests that BRCA1pSer-1423 exists in distinct chromatin bound complexes at different loci within the nucleus, which are likely to have distinct functions following DNA damage (Figure S2K).
It is well accepted that mRNA splicing occurs cotranscriptionally through the sequential recruitment of spliceosomal proteins to the chromatin/mRNA template of actively transcribed genes and is required to promote transcript maturation and stability. Therefore, we hypothesized that BRCA1, constitutively bound at gene promoters, may regulate their expression following DNA damage, through the recruitment of BCLAF1 and the cotranscriptional spliceosome, thereby promoting mRNA splicing and transcript production/stability.
In order to test this model and identify BRCA1/BCLAF1 target genes, we performed chromatin immunoprecipitation array hybridization (ChIP-chip) with BCLAF1 antibodies in unperturbed and etoposide-treated cells in the presence and absence of BRCA1 (data not shown). This strategy identified 675 genomic regions bound by BCLAF1 in response to etoposide treatment (Figure S3A). Interestingly, 610 of these regions, which mapped to 782 genes, were not bound by BCLAF1 in BRCA1-depleted cells, suggesting that, as hypothesized, BRCA1 may recruit BCLAF1 and the associated spliceosome to genetic promoter regions in order to promote cotranscriptional splicing of target genes following DNA damage (Figure S3A). Ingenuity Pathway Analysis of BRCA1/BCLAF1 regulated promoters/genes revealed that the top network involving these genes was the DNA Replication, Recombination and Repair, and Cancer network (p = 1 × 10−43). In keeping with this, many of these genes, such as ATRIP, BACH1, and EXO1, are involved in the DDR pathway, suggesting that BRCA1/BCLAF1-mediated splicing of a large subset of DDR genes may regulate cellular survival/DNA repair in response to DNA damage.
To validate ChIP-chip-identified target genes, we tested BRCA1, BCLAF1, and U2AF65 (a BCLAF1 interacting spliceosome assembly factor) binding to the promoter regions of these genes and an additional three DDR genes not identified from the ChIP-chip screen (CHEK2, BRCA2, and ATM) as negative controls. This confirmed that BRCA1 constitutively associates with the promoter regions of ATRIP, BACH1, and EXO1, in both unperturbed and etoposide-treated cells (Figures 3C and S3B–S3D). In contrast, BCLAF1 and U2AF65 binding to these regions was significantly induced upon DNA damage. As BCLAF1 only interacts with BRCA1 in response to DNA damage, we tested whether BRCA1 is required for BCLAF1 recruitment to these genes. Indeed, BRCA1 depletion resulted in loss of damage-induced BCLAF1 and U2AF65 recruitment to DNA (Figures 3C and S3B–S3D). Concurrently, we also found that BRCA1 and BCLAF1 depletion resulted in loss of recruitment of U2AF65 to these promoter regions, suggesting that BRCA1-dependent recruitment of BCLAF1 mediates binding of the core splicing machinery to these genes. Importantly, we did not observe any significant binding of BRCA1, BCLAF1, or U2AF65 to the promoter regions of the negative control genes, CHEK2, BRCA2, and ATM (Figure S3E). We also found that the splicing proteins U2AF35 and SF3B1, which also interact with BRCA1 following DNA damage through interaction with BCLAF1, are recruited to these promoters following DNA damage in a BRCA1- and BCLAF1-dependent manner (Figure S3F).
Consistent with a role in cotranscriptional splicing, we also observed significant enrichment of BRCA1, BCLAF1, and U2AF65 on ATRIP, BACH1, and EXO1 mRNA transcripts in response to DNA damage that was not evident in unperturbed cells (Figures 3D and S3G). Moreover, depletion of BRCA1 resulted in loss of BCLAF1 and U2AF65 association with these transcripts (Figures 3D and S3G). Intriguingly, depletion of BCLAF1 also resulted in loss of damage-induced BRCA1 binding, suggesting that BRCA1 does not interact directly with these mRNA transcripts but is likely associated with target transcripts through interactions with BCLAF1 and the associated mRNA binding spliceosome (Figures 3D and S3G). Additionally, we did not observe any enrichment of BRCA1, BCLAF1, or U2AF65 with CHEK2, BRCA2, or ATM transcripts (Figure S3H).
We also assessed binding of BRCA1, BCLAF1, and U2AF65 to a number of regions along the ATRIP, BACH1, and EXO1 genes using ChIP-qRT-PCR. This revealed that BRCA1 constitutively binds to exonic and exon/intron boundary regions (but not intronic regions) within these genes, albeit with reduced binding associated with progression toward the 3′ end of these genes (Figures S4A–S4C). This is consistent with a role in mRNA processing/splicing, where reduced binding, with progression along genes, is associated with reduced transcript tethering concurrent with transcriptional termination. In contrast, BCLAF1 and U2AF65 were enriched at BRCA1-bound regions only following DNA damage. In support of this, examination of publically available BRCA1 ChIP-seq data, derived from the normal-like breast cell line MCF10A (GEO accession number GSE40591 [Barrett et al., 2013]), also revealed BRCA1 binding across ATRIP, BACH1, and EXO1 exons (Figures S4D and S4E). Moreover, a genome-wide analysis of all BRCA1 binding peaks within this data set revealed enrichment of BRCA1 binding peaks within gene promoters (and 5′ UTRs) as well as coding exons but not introns, again supporting a role for BRCA1 in mRNA splicing (Figure S4F).
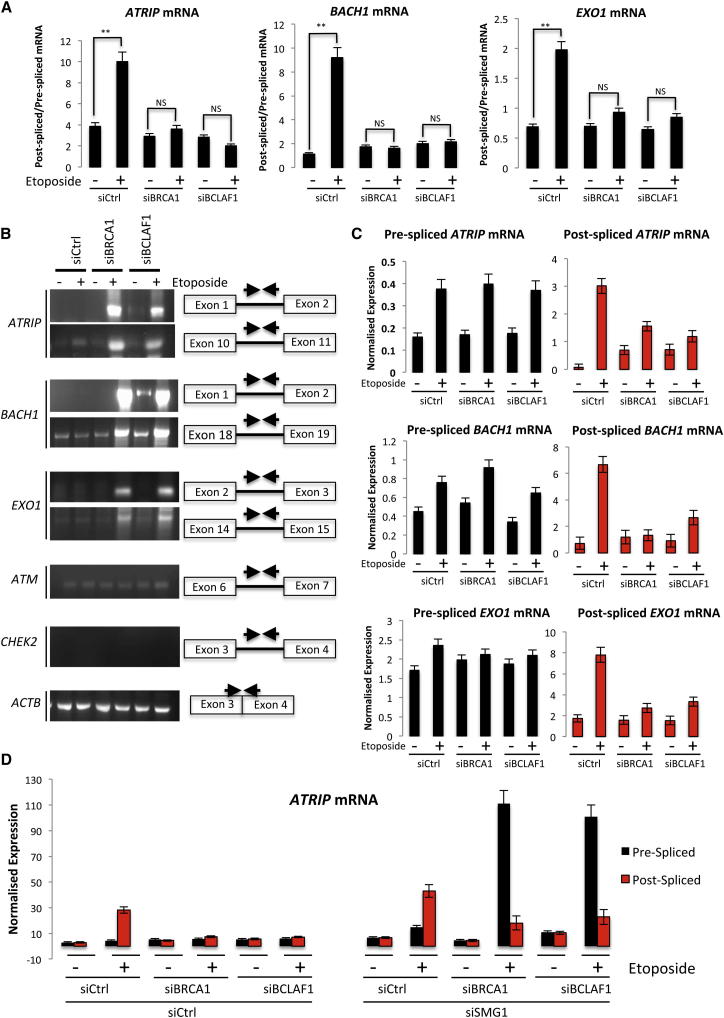
Consistent with this, we found that the mRNA splicing of ATRIP, BACH1, and EXO1 transcripts was significantly upregulated in response to DNA damage in both a BRCA1- and a BCLAF1-dependent manner using two independent siRNAs (Figures 4A and S5A). Additionally, saturating RT-PCR analysis with intron-targeted primers revealed the presence of introns in ATRIP, BACH1, and EXO1 transcripts following DNA damage in BRCA1- and BCLAF1-depleted cells, but not control cells, confirming that splicing of these transcripts following DNA damage requires BRCA1 and BCLAF1 (Figure 4B). In response to DNA damage, transcription of ATRIP, BACH1, and EXO1 is upregulated. However, BRCA1 or BCLAF1 depletion does not affect the transcription of these genes as indicated by similar levels of pre-mRNA production, in the absence or presence of DNA damage (Figure 4C). Similarly, RNA Pol II loading and activity on these gene promoters is unaffected by BRCA1 or BCLAF1 depletion (Figure S5B). In contrast, we observed a marked reduction in the production of postspliced ATRIP, BACH1, and EXO1 transcripts following depletion of BRCA1 or BCLAF1 (Figure 4C). Additionally, the increased ratio of postspliced/prespliced mRNA observed in BRCA1/BCLAF1-depleted cells following DNA damage is not due to increased decay of prespliced transcripts in these cells (Figure S5C). Instead, mRNA decay experiments revealed reduced levels of postspliced ATRIP, BACH1, and EXO1 transcripts, in comparison to prespliced transcripts, in BRCA1/BCLAF1-depleted cells following inhibition of transcription, which is consistent with a role for BRCA1/BCLAF1 in the cotranscriptional splicing of these genes (Figure S5C). Importantly, we did not observe changes in ATRIP, BACH1, or EXO1 splice variant expression following DNA damage, suggesting that DNA damage induced BRCA1/BCLAF1-mediated splicing of these genes does not facilitate alternative splicing (data not shown).
Figure 4.
The BRCA1/BCLAF mRNA Splicing Complex Promotes the Splicing and Stability of ATRIP, BACH1, and EXO1 Transcripts following DNA Damage
(A) Ratio of postspliced to prespliced ATRIP, BACH1, and EXO1 mRNAs in control (siCtrl) and BRCA1- or BCLAF1-depleted cells mock treated or treated with etoposide. mRNA levels were assessed by qRT-PCR using exon 9-exon 10 (post-spliced-ATRIP) and exon 9-intron 9 (pre-spliced-ATRIP), exon 15-exon 16 (post-spliced-BACH1) and exon 15-intron 15 (pre-spliced- BACH1), and exon 1-exon 2 (post-spliced-EXO1) and exon 1-intron 1 (pre-spliced- EXO1) primers and normalized to ACTB mRNA. Graphs represent the mean ratios of postspliced/prespliced mRNA from three independent experiments ± SEM. Significance of changes in splicing ratios was assessed using Student’s two-tailed t test with significant changes indicated by ∗∗p < 0.01.
(B) Semiquantitative PCR analysis of a cDNA generated from DNase-treated RNA, collected from control (siCtrl) and BRCA1- or BCLAF1-depleted cells, mock treated or treated with Etoposide. Primers targeting two independent intronic regions within ATRIP, BACH1, and EXO1 and a single intronic region within ATM and CHEK2 (control genes) were used for semiquantitative PCR analysis. Exon-spanning primers targeted against ACTB were used as a loading control.
(C) Normalized expression of prespliced and postspliced mRNAs evaluated in (A).
(D) Expression levels of postspliced and prespliced ATRIP mRNAs in control (siCtrl) and BRCA1- or BCLAF1-depleted cells, transfected with control siRNA (siCtrl) or depleted of SMG1 (siSMG1), a key regulator of the non-sense-mediated decay pathway. Normalized expression levels were quantified as in (A). Graphs represent the mean normalized expression from three independent experiments ± SEM. See also Figures S4 and S5.
As previously mentioned, mRNA splicing is required to maintain transcript stability, as unspliced transcripts are rapidly degraded through the non-sense-mediated decay (NMD) pathway. This likely explains why we did not observe increased levels of pre-spliced ATRIP, BACH1, or EXO1 mRNAs after DNA damage in BRCA1- and BCLAF1-depleted cells. Consistent with this, siRNA-mediated depletion of SMG1, a key player in the NMD pathway, led to a marked increase in prespliced ATRIP, BACH1, and EXO1 mRNAs in BRCA1- and BCLAF1-depleted cells following DNA damage (Mendell et al., 2004) (Figures 4D, S5D, and S5E).
BRCA1 Ser-1423 Phosphorylation Is Required for BCLAF1 Recruitment and Target Gene Splicing following DNA Damage
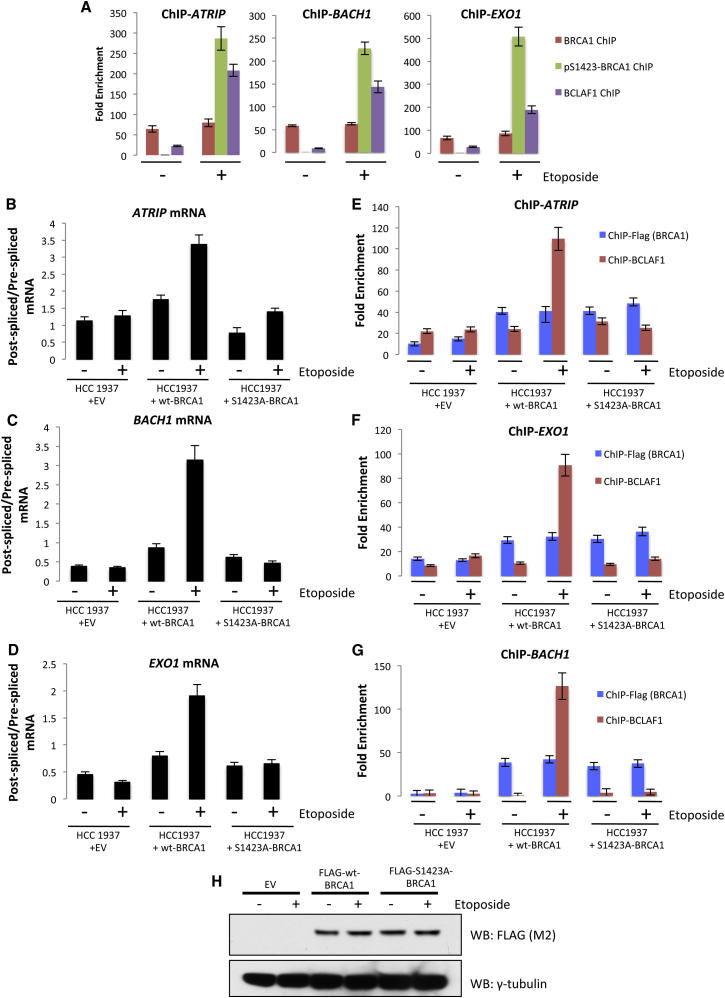
Taken together, our data suggest a model in which phosphorylated BRCA1, bound at a subset of gene promoters following DNA damage, recruits BCLAF1 and associated spliceosomal proteins, thereby facilitating DNA damage-induced mRNA splicing. To confirm that BRCA1, bound at the ATRIP, BACH1, and EXO1 promoters, is indeed phosphorylated at serine-1423 following DNA damage, we performed ChIP-qRT-PCR with BRCA1pSer-1423 antibodies. This revealed marked enrichment of BRCA1pSer-1423 at the ATRIP, BACH1, and EXO1 promoters only following DNA damage (Figure 5A). Also in support of this model, reconstitution of BRCA1 mutant cells (HCC1937) with wild-type BRCA1 restored their ability to upregulate ATRIP, BACH1, and EXO1 mRNA splicing following DNA damage, whereas reconstitution with BRCA1S1423A phosphomutant protein did not (Figures 5B–5D). Additionally, wild-type BRCA1 was able to recruit BCLAF1 to the ATRIP, BACH1, and EXO1 promoter regions following DNA damage, whereas the BRCA1S1423A phospho mutant was not (Figures 5E–5H). Consistent with this, inhibition of ATM and ATR (mediators of BRCA1S1423 phosphorylation) also abrogated DNA damage-induced ATRIP, BACH1, and EXO1 mRNA splicing and recruitment of BCLAF1 and U2AF65 to their promoters (Figures S6A–S6C).
Figure 5.
BRCA1 Ser-1423 Phosphorylation Is Required for BCLAF1 Recruitment and Target Gene Splicing following DNA Damage
(A) BRCA1, pS1423-BRCA1, and BCLAF1 ChIP-qPCRs demonstrating constitutive binding of BRCA1 to ATRIP, BACH1, and EXO1 promoters irrespective of DNA damage and DNA damage-dependent enrichment of pS1423-BRCA1 and BCLAF1 on these promoters. Graphs represent the mean fold enrichment quantified from three independent experiments ± SEM.
(B–D) Ratio of postspliced to prespliced ATRIP, BACH1, and EXO1 mRNA in BRCA1-deficient cells (HCC1937) transfected with empty vector (EV), wild-type Flag-BRCA1 (wt-BRCA1), or S1423A Flag-BRCA1 (S1423A-BRCA1). Cells were mock treated or treated with etoposide. mRNA levels were assessed as described in Figure 4A. Graphs represent the mean ratios of postspliced/prespliced mRNA from three independent experiments ± SEM ATRIP, BACH1, and EXO1 splicing is upregulated in cells expressing wild-type BRCA1 but not S1423A-BRCA1, indicating that phosphorylation of BRCA1 S1423 is required for DNA damage-induced splicing.
(E–G) FLAG and BCLAF1 ChIP-qRT-PCR analysis carried out in BRCA1-deficient cells (HCC1937) transfected with empty vector (EV), wild-type Flag-BRCA1 (wt), or S1423A Flag-BRCA1 (S1423A). Cells were mock treated or treated with etoposide. Graphs represent the mean fold enrichment from three independent experiments ± SEM.
(H) Representative western blots demonstrating wild-type and S1423A Flag-BRCA1 in HCC1937 cells used for splicing assays and ChIPs presented above. See also Figure S6.
BRCA1/BCLAF1-Mediated mRNA Splicing Maintains ATRIP, BACH1, and EXO1 Protein Expression and Resistance to DNA Damage
As BRCA1 and BCLAF1 were required for efficient splicing and stability of spliced ATRIP, BACH1, and EXO1 transcripts following DNA damage, we next assessed the effect of BRCA1 or BCLAF1 depletion on ATRIP, BACH1, and EXO1 protein expression. In keeping with the reduced expression of ATRIP, BACH1, and EXO1 spliced transcripts, we observed substantially reduced expression of all three proteins in BRCA1- and BCLAF1-depleted cells following DNA damage (Figure 6A). However, to our surprise, in control cells we did not observe increased levels of ATRIP, BACH1, and EXO1 protein levels following DNA damage that would be expected, given the increased expression of spliced transcript observed. This suggests that either ATRIP, BACH1, and EXO1 translation is attenuated following DNA damage or or that turnover of these proteins may be increased and that BRCA1/BCLAF1-regulated cotranscriptional splicing serves to maintain the expression of these proteins in response to DNA damage.
Figure 6.
BRCA1/BCLAF1-Mediated mRNA Splicing Is Required for Maintenance of ATRIP, BACH1, and EXO1 Protein Expression
(A) Representative western blots demonstrating that depletion of either BRCA1 or BCLAF1 results in downregulated expression of ATRIP, BACH1, and EXO1 proteins in response to DNA damage.
(B) Representative western blot demonstrating DNA damage-dependent accumulation of ATRIP, BACH1, and EXO1 proteins over time following inhibition of proteosomal mediated protein degradation with MG132 (10 μM). Cells were mock treated or treated with etoposide (1 μM) for 30 min prior to MG132 treatment.
(C) Representative western blots demonstrating DNA damage-dependent increased protein turnover in control and BRCA1- or BCLAF1-depleted cells following inhibition of protein translation with Cyclohexamide (10 μg/mL). Cells were mock treated or treated with etoposide (1 μM) for 30 min prior to Cyclohexamide treatment.
(D) Representative western blots demonstrating BRCA1 and BCLAF depletion in cells used for experiments shown in (C).
(E–G) Quantification of ATRIP, BACH1, and EXO1 protein levels shown in (C). Image densitometry values were normalized to 0 hr and decay curves fitted and used to calculate protein half-lives.
To examine whether their translation is attenuated following DNA damage, we inhibited protein degradation in control and etoposide-treated cells using the proteasome inhibitor MG132. Cells were treated with etoposide for 30 min prior to MG132 treatment, after which cells were harvested at increasing time points to visualize protein production (Figure 6B). Intriguingly, ATRIP, BACH1, and EXO1 protein levels all increased more rapidly following DNA damage. This is consistent with the increased expression of spliced ATRIP, BACH1, and EXO1 transcripts observed in these cells following DNA damage and suggests that increased turnover of these proteins is responsible for their lack of increased protein expression following DNA damage, rather than decreased translation.
To test this, we inhibited protein translation in control and etoposide-treated cells transfected with scrambled, BRCA1, or BCLAF1 targeted siRNAs using cyclohexamide. Cells were treated with etoposide for 30 min prior to cyclohexamide treatment, after which cells were harvested at increasing time points to visualize protein degradation (Figures 6C and 6D). Additionally, image densitometry was used to determine ATRIP, BACH1, and EXO1 protein half-lives (t1/2) in these cells (Figures 6E–6G). This revealed that the degradation/turnover of all three proteins was increased following DNA damage and that this was unaffected by depletion of BRCA1 or BCLAF1. Taken together, this suggests that BRCA1/BCLAF1-mediated cotranscriptional splicing and transcript stabilization serve to maintain levels of target gene proteins in response to higher rates of protein turnover following DNA damage.
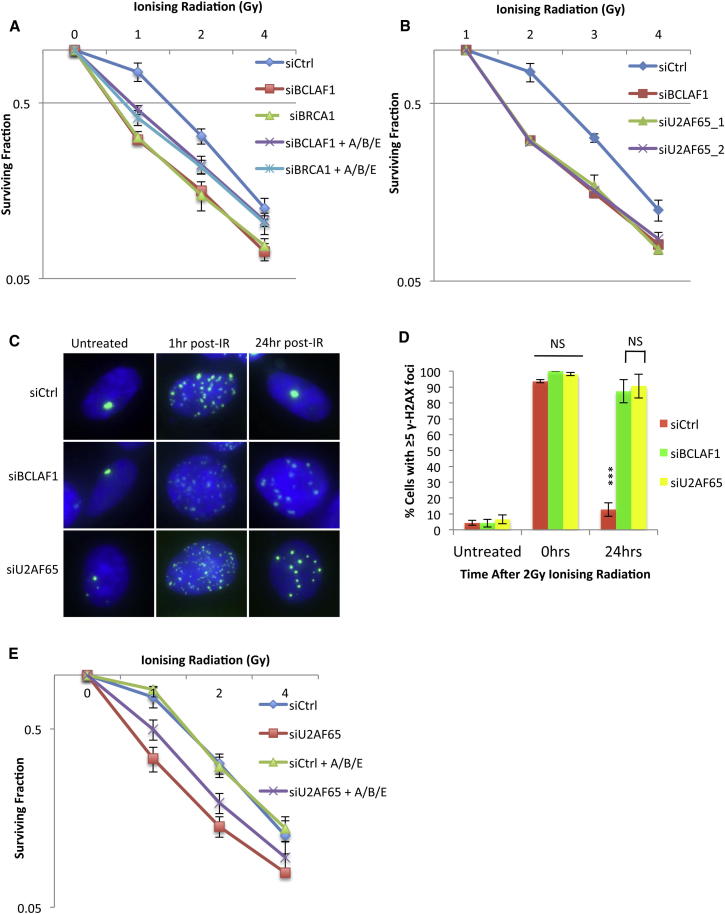
Given that loss of BRCA1/BCLAF1-mediated splicing of ATRIP, BACH1, and EXO1 results in reduced but not absent protein levels following DNA damage, we examined whether reduction of these proteins could contribute to the DNA damage sensitivity observed in BRCA1/BCLAF1-depleted cells. Indeed, reduction but not complete absence of any of these proteins, using titrated siRNA concentrations, resulted in sensitization to IR induced DNA damage (Figures S7A and S7B). In contrast, ectopic expression of ATRIP, BACH1, or EXO1 alone was unable to rescue the sensitivity of BRCA1- or BCLAF1-depleted cells to IR (data not shown). However, the combined ectopic expression of ATRIP, BACH1, and EXO1 was able to partially rescue the IR sensitivity of both BRCA1- and BCLAF1-depleted cells, suggesting that BRCA1/BCLAF1-mediated splicing of these genes is required, at least in part, for BRCA1 and BCLAF1’s ability to mediate resistance to DNA damaging agents (Figures 7A and S7C).
Figure 7.
BRCA1/BCLAF1-Mediated mRNA Splicing of ATRIP, BACH1, and EXO1 Promotes Resistance to DNA Damage and Efficient DNA Repair
(A) Clonogenic survival assays demonstrating that ectopic expression of ATRIP, BACH1, and EXO1 (A/B/E) in BRCA1- or BCLAF1-depleted 293T cells partially rescues their sensitivity to IR. Mean surviving fraction of three independent experiments is plotted ± SEM.
(B) Clonogenic survival assays demonstrating that depletion of BCLAF1 or U2AF65 induces sensitivity to IR in 293T cells. Mean surviving fraction of three independent experiments is plotted ± SEM.
(C) Representative immunofluorescent staining of γ-H2AX marked DNA damage in untreated 293T cells depleted of either BCLAF1 or U2AF65 and 1 and 24 hr following 2Gy IR.
(D. Quantification of three independent experiments described above (≥200 cells were scored/experiment). The mean fraction of cells containing ≥5 γ-H2AX foci is plotted ± SEM. Significant differences in the fraction of cells containing ≥5 γ-H2AX foci were assessed using Student’s two-tailed t test and are indicated by ∗∗∗p < 0.001.
(E) Clonogenic survival assays demonstrating that ectopic expression of ATRIP, BACH1, and EXO1 (A/B/E) in U2AF65-depleted 293T cells partially rescues their sensitivity to IR. Mean surviving fraction of three independent experiments is plotted ± SEM. See also Figure S7.
To further confirm that the defect in DNA repair observed in BCLAF1-depleted cells is due to loss of BRCA1/BCLAF1-mediated mRNA splicing, we tested the role of U2AF65, a splicing factor recruited to target genes by BRCA1/BCLAF1, in mediating sensitivity to DNA damage and DNA repair. Indeed, U2AF65 depletion induced sensitivity to IR and defective DNA repair to a similar extent as BCLAF1 depletion (Figures 7B–7D, S7D, and S7E). Moreover, as is the case for BRCA1- and BCLAF1-depleted cells, ectopic expression of ATRIP, BACH1, and EXO1 partially rescues IR sensitivity in U2AF65-depleted cells (Figures 7E and S7F).
Discussion
This study has identified a function for BRCA1 in the regulation of mRNA splicing in response to DNA damage, through the formation of a DNA damage-induced protein complex with BCLAF1, Prp8, U2AF35/65, SF3B1, and other spliceosome proteins. Consistent with this, a large, as-yet uncharacterized DNA damage-induced BRCA1 complex containing RNA and hnRNP proteins has been previously reported (Chiba and Parvin, 2001).
In addition, our data suggest that BRCA1/BCLAF1-mediated mRNA splicing in response to DNA damage may serve as a mechanism for the processing of key pre-mRNAs required for an efficient DDR and DNA repair. Intriguingly, BRCA1 has been previously reported to inhibit 3′ mRNA polyadenylation and thereby mRNA stability, through BRCA1/BARD1 ubiquitin-dependent degradation of RNA-Pol II (Kleiman et al., 2005). These studies suggest a general role for the BRCA1/BARD1 complex in blocking active transcription on a genome-wide level following DNA damage, presumably to prevent transcription of damaged genes. Despite this, it is accepted that specific genes, for example ATRIP, are actively transcribed in response to DNA damage, suggesting an additional mechanism that facilitates transcription/expression of a subset of genes in the context of a more genome-wide shutdown of transcription. We propose that as part of this mechanism, BRCA1 recruits the mRNA splicing machinery to a subset of promoters of genes required for an efficient DDR (such as ATRIP, BACH1, and EXO1), thereby promoting the cotranscriptional splicing of these genes, positively regulating the stability of their transcripts and subsequent protein expression. Intriguingly, although the BRCA1/BCLAF1 complex promoted the splicing and stability of ATRIP, BACH1, and EXO1 transcripts following DNA damage, we did not observe an increase in the expression of these proteins in control cells, suggesting that following DNA damage the levels of these proteins may also be regulated at the translation and/or protein stability levels. When testing this, we observed no detectable change in the levels of translation of these proteins following DNA damage but instead observed a dramatic increase in the rate of their turnover, suggesting that BRCA1/BCLAF1-mediated cotranscriptional splicing and transcript stabilization may serve as a mechanism through which the levels of these genes/proteins are maintained in response to higher rates of protein turnover following DNA damage. Exactly why the rate of turnover of these proteins is increased following DNA damage will require further investigation. However, it is well accepted that a number of DNA repair proteins are degraded at DNA break sites during the DNA repair process in order to facilitate removal of various proteins at different stages of repair, thereby allowing fine temporal and spatial control of repair processes. For example, KDM4A/JMJD2A is degraded through an RNF8/RNF168-dependent pathway at DNA break sites, thereby facilitating 53BP1 recruitment, loading, and subsequent repair (Mallette et al., 2012). Given that ATRIP, BACH1, and EXO1 are all recruited to DNA break sites following DNA damage, it is possible that these proteins are also degraded at break sites following DNA damage in order to facilitate ordered repair. In this context, BRCA1/BCLAF1-mediated stabilization of spliced transcripts may function in order to maintain levels of these proteins, thereby allowing the efficient repair of all DNA damage.
An intriguing finding of our study is that following DNA damage, BRCA1 and BCLAF1 interact on target gene promoters, but not at DNA break sites where BRCA1pSer-1423 is also localized but BCLAF1 is excluded. However, it has been previously reported that exclusion of BCLAF1 and its homologous binding partner THRAP3 from DNA break sites occurs in parallel with inhibition of transcription and subsequent loss of mRNA processing at sites of DNA damage, which is mediated by ATM/ATR/DNA-PK localized at these sites (Beli et al., 2012). This suggests that after DNA damage BRCA1 plays distinct roles that require BRCA1 containing complexes at different loci: one at DNA break sites, where it is directly involved in repair (and transcription/mRNA processing is directly inhibited and BCLAF1 excluded by active ATM, ATR and/or DNA-PK), and another at transcriptionally active regions, near the promoters of DDR factors.
Surprisingly, given that the BRCA1/BCLAF1 complex regulates a relatively large pool of genes following DNA damage, we found that ectopic expression of ATRIP, BACH1, and EXO1 could partially rescue the DNA damage sensitivity phenotype of BRCA1-, BCLAF1-, and U2AF65-depleted cells, suggesting that the BRCA1/BCLAF1-mediated splicing of these genes is important for their ability to mediate resistance to DNA damage. This finding may be a reflection of the highly important role of these three proteins in the DDR, in which all play important and cooperative roles in HR-mediated DSB repair. Nevertheless, the finding that ectopic expression of these three BRCA1/BCLAF1 regulated genes only partially rescues the DNA damage sensitivity of BRCA1-, BCLAF1-, and U2AF65-depleted cells highlights that the regulation of other genes by the BRCA1/BCLAF1 complex may also be important for BRCA1/BCLAF1-mediated DNA damage resistance.
Additionally, although we have not observed BRCA1/BCLAF1-mediated alternative splicing in the genes examined in this study, it is possible that increased cotranscriptional splicing mediated by BRCA1/BCLAF1 may affect the inclusion or skipping of differential exons of regulated genes in response to DNA damage. For example, camptothecin induces increased MDM2 cotranscriptional splicing, resulting in exon skipping and production of alternate MDM2 splice variants (Dutertre et al., 2010). Additionally, UV-induced DNA damage has also been shown to affect cotranscriptional alternative splicing of a subset of UV-responsive genes through inhibition of RNA Pol II elongation during transcription of these genes (Muñoz et al., 2009).
Given the multiple DDR processes within which BRCA1 plays a role, in comparison to BCLAF1, another surprising finding of this study was that BCLAF1 depletion was unable to sensitize BRCA1 mutant cells to IR, as measured by clonogenic survival assays, suggesting that BRCA1 and BCLAF1 function in an epistatic manner within the same pathway. However, clonogenic survival assays only demonstrate cellular survival following a specific genotoxic insult and do not allow the functional separation of different pathways that contribute to cell death. Therefore, although demonstrating that the pathway in which BRCA1 and BCLAF1 cooperate is required for cellular survival following IR, it is unlikely that all of BRCA1’s tumor-suppressive function is mediated through its role in mRNA splicing with BCLAF1 alone.
Taken together, our data suggest that the BRCA1/BCLAF1 mRNA splicing complex may act as a tumor suppressor complex, functioning to promote the splicing and stability of genes required for DNA repair and maintenance of genomic stability. In support of this, a number of SNPs within BCLAF1 have been associated with increased risk of non-Hodgkin’s lymphoma and a number of microRNAs encoded by the oncogenic Kaposi’s sarcoma-associated herpes virus (KSHV) target BCLAF1, resulting in BCLAF1 depletion and sensitivity to DNA damaging agents (Kelly et al., 2010; Ziegelbauer et al., 2009). Additionally, a number of recent studies have found a high incidence of mutually exclusive somatic mutations within members of the BRCA1/BCLAF1 mRNA splicing complex, including BCLAF1, U2AF65, U2AF35, SRSF2, SF3A1, SF3B1, and PRPF40B within various cancer types including myelodysplasia, chronic and acute myeloid leukemias, breast cancers, and lung cancers (Ding et al., 2012; Forbes et al., 2008; Merz et al., 2007; Nik-Zainal et al., 2012; Quesada et al., 2012; Yoshida et al., 2011).
Finally, given BRCA1’s role in predisposition to breast and ovarian cancer, it may also be prudent to investigate the status of these genes in familial cancers not linked to BRCA1/2 mutation. Indeed, mutations within Prp22, a Prp8-interacting protein involved in pre-mRNA splicing, were previously found to cosegregate with breast and ovarian cancer (Friedman et al., 1995).
Experimental Procedures
Cell Lines
Cell lines were obtained from the following sources: London Research Institute, London, for 293T; and ECACC for MCF7 cells. All cell lines were verified by STR profiling (ATCC-LGC Standards, Middlesex, UK).
siRNAs
siRNAs were obtained from QIAGEN. See Supplemental Information for full sequences.
Peptide Pull-Down Assays
Peptide pull-down assays were carried out as previously described (Stucki et al., 2005) using the following biotinylated peptides BRCA1-S1423: AVLEGHGSGPSNSYP; BRCA1-phospho-S1423: AVLEGHGpSGPSNSYP (Genscript).
mRNA Splicing Analysis
Introns/exons were chosen for assessment in splicing assays based on their inclusion in expressed transcripts and their suitability for optimal qRT-PCR primer design. Postspliced to prespliced ATRIP, BACH1 and EXO1 mRNA levels were quantified using qRT-PCR and normalized to ACTB mRNA levels. The ratio of spliced to postspliced to prespliced mRNA was then calculated by dividing the normalized expression levels of postspliced mRNAs by the normalized levels of prespliced mRNAs. See Supplemental Information for a detailed description of the methods and primer sequences used.
Clonogenic Survival Assays
Clonogenic survival assays were performed as previously described (Franken et al., 2006).
Plasmids
Flag-tagged BRCA1 construct Fl4-BRCA1 was a kind gift from Professor Richard Baer, Columbia University, New York, USA. Myc-DDK tagged ATRIP (RC223562), BACH1 (RC224085), and EXO1 (RC200547) plasmids were purchased from Origene. Plasmids were transfected with Genejuice (Merck) as per the manufacturer’s instructions.
Coimmunoprecipitations
Coimmunoprecipitations were carried out as previously described using (BRCA1; Ab1, Millipore) or BCLAF1 (BTF-608A, Bethyl Laboratories) antibodies.
Western Blotting
Western blotting was performed using the invitrogen Novex system and the following antibodies: BRCA1 (D9, Santa-Cruz), BCLAF1 (BTF 608A, Bethyl Labs), U2AF65 (MC3 or H300, Santa-Cruz), Prp8 (E5, Santa-Cruz), BACH1 (4578, Cell Signaling), ATRIP (H300, Santa-Cruz), Exo1 (N18, Santa-Cruz), γ-tubulin (GTU-88, Sigma), SF3B1 (A300-996A, Bethyl Laboratories), and U2AF35 (A307-079A, Bethyl Laboratories).
Chromatin Immunoprecipitations and ChIP-Chip
Chromatin immunoprecipitations (ChIP) and ChIP-chip were carried out as previously described (Gorski et al., 2011) using the NimbleGen Human 3 × 720k RefSeq promoter array. See Supplemental Experimental Procedures for detailed protocol. Immunoprecipitated DNA was quantified by qPCR and expressed as fold enrichment over input compared to enrichment of a nonspecific negative control region. Additionally, to ensure specificity of ChIP antibodies, all ChIP experiments were accompanied by two negative control ChIPs performed with a nonspecific isotype-matched IgG and an anti-HA tag antibody. Nonspecific binding of these antibodies to DNA regions was quantified by qPCR. ChIP with these antibodies never revealed specific enrichment of any of the DNA regions quantified (data not shown).
RNA Immunoprecipitations
See Supplemental Experimental Procedures for detailed RNA immunoprecipitation (RIP) protocol. Immunoprecipitated RNA was quantified by qRT-PCR and expressed as fold enrichment over input compared to enrichment of a negative control region within the ACTB 5′ UTR. Additionally, to ensure specificity of RIP antibodies, all RIP experiments were accompanied by two negative control RIPs performed with a nonspecific isotype-matched IgG and an anti-HA tag antibody. RIP with these antibodies never revealed specific enrichment of any of the mRNA regions quantified (data not shown).
Immunofluorescent Cytochemistry
Immunofluorescent cytochemistry was carried out as previously described (Paul et al., 2011) using anti γ-H2AX, (05–636, Millipore), BCLAF1 (BTF 608A, Bethyl Labs), and U2AF65 (MC3, Santa Cruz) antibodies.
Ionising Radiation and Etoposide Treatment
Irradiations were carried out using an X-RAD 225 X-ray generator (Precision X-ray Inc. Branford, CT, USA) at a dose rate of 0.591 Gy.min−1. Unless otherwise stated, etoposide treatments were using 1 μM for 16 hr.
Metaphase Spreads and Chromosomal Aberration Analysis
Metaphase spreads and chromosome 1 and 2 FISH were carried out as previously described (Manti et al., 2006) using XCP1 and XCP2 whole-chromosome paint probes (MetaSystems, Zeiss, Germany).
Acknowledgments
This work was supported by grants from Cancer Focus Northern Ireland (K.I.S), Cancer Research UK (C538/A8132 to K.I.S., J.J.G., E.M.B., and D.P.H.), the Research and Development Office Northern Ireland (G.W.I.), the UK Medical Research Council (G0700754 to S.S.M. and D.J.M.), and the Leukaemia and Lymphoma Research of the United Kingdom (A.P. and J.B.). We would also like to thank Professor Steve Jackson (University of Cambridge) for his valuable input and discussion.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Contributor Information
Kienan I. Savage, Email: k.savage@qub.ac.uk.
D. Paul Harkin, Email: d.harkin@qub.ac.uk.
Accession Numbers
BCLAF1 ChIP-chip data can be found at the Gene Expression Omnibus under accession number GSE47016.
Supplemental Information
References
- Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beli P., Lukashchuk N., Wagner S.A., Weinert B.T., Olsen J.V., Baskcomb L., Mann M., Jackson S.P., Choudhary C. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol. Cell. 2012;46:212–225. doi: 10.1016/j.molcel.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken C.P., Wall S.J., Barré B., Panov K.I., Ajuh P.M., Perkins N.D. Regulation of cyclin D1 RNA stability by SNIP1. Cancer Res. 2008;68:7621–7628. doi: 10.1158/0008-5472.CAN-08-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres J.F., Misteli T., Screaton G.R., Spector D.L., Krainer A.R. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba N., Parvin J.D. Redistribution of BRCA1 among four different protein complexes following replication blockage. J. Biol. Chem. 2001;276:38549–38554. doi: 10.1074/jbc.M105227200. [DOI] [PubMed] [Google Scholar]
- Cortez D., Wang Y., Qin J., Elledge S.J. Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- Ding L., Ley T.J., Larson D.E., Miller C.A., Koboldt D.C., Welch J.S., Ritchey J.K., Young M.A., Lamprecht T., McLellan M.D. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutertre M., Sanchez G., De Cian M.C., Barbier J., Dardenne E., Gratadou L., Dujardin G., Le Jossic-Corcos C., Corcos L., Auboeuf D. Cotranscriptional exon skipping in the genotoxic stress response. Nat. Struct. Mol. Biol. 2010;17:1358–1366. doi: 10.1038/nsmb.1912. [DOI] [PubMed] [Google Scholar]
- Elstrodt F., Hollestelle A., Nagel J.H., Gorin M., Wasielewski M., van den Ouweland A., Merajver S.D., Ethier S.P., Schutte M. BRCA1 mutation analysis of 41 human breast cancer cell lines reveals three new deleterious mutants. Cancer Res. 2006;66:41–45. doi: 10.1158/0008-5472.CAN-05-2853. [DOI] [PubMed] [Google Scholar]
- Forbes S.A., Bhamra G., Bamford S., Dawson E., Kok C., Clements J., Menzies A., Teague J.W., Futreal P.A., Stratton M.R. The catalogue of somatic mutations in cancer (COSMIC) Curr. Protoc. Hum. Genet. 2008;Chapter 10 doi: 10.1002/0471142905.hg1011s57. Unit 10 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken N.A., Rodermond H.M., Stap J., Haveman J., van Bree C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006;1:2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- Friedman L.S., Ostermeyer E.A., Lynch E.D., Welcsh P., Szabo C.I., Meza J.E., Anderson L.A., Dowd P., Lee M.K., Rowell S.E. 22 genes from chromosome 17q21: cloning, sequencing, and characterization of mutations in breast cancer families and tumors. Genomics. 1995;25:256–263. doi: 10.1016/0888-7543(95)80133-7. [DOI] [PubMed] [Google Scholar]
- Gorski J.J., Savage K.I., Mulligan J.M., McDade S.S., Blayney J.K., Ge Z., Harkin D.P. Profiling of the BRCA1 transcriptome through microarray and ChIP-chip analysis. Nucleic Acids Res. 2011;39:9536–9548. doi: 10.1093/nar/gkr679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J.L., Novak A.J., Fredericksen Z.S., Liebow M., Ansell S.M., Dogan A., Wang A.H., Witzig T.E., Call T.G., Kay N.E. Germline variation in apoptosis pathway genes and risk of non-Hodgkin’s lymphoma. Cancer Epidemiol. Biomarkers Prev. 2010;19:2847–2858. doi: 10.1158/1055-9965.EPI-10-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman F.E., Wu-Baer F., Fonseca D., Kaneko S., Baer R., Manley J.L. BRCA1/BARD1 inhibition of mRNA 3′ processing involves targeted degradation of RNA polymerase II. Genes Dev. 2005;19:1227–1237. doi: 10.1101/gad.1309505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Lu Z.G., Miki Y., Yoshida K. Protein kinase C delta induces transcription of the TP53 tumor suppressor gene by controlling death-promoting factor Btf in the apoptotic response to DNA damage. Mol. Cell. Biol. 2007;27:8480–8491. doi: 10.1128/MCB.01126-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallette F.A., Mattiroli F., Cui G., Young L.C., Hendzel M.J., Mer G., Sixma T.K., Richard S. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 2012;31:1865–1878. doi: 10.1038/emboj.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manti L., Durante M., Grossi G., Ortenzia O., Pugliese M., Scampoli P., Gialanella G. Measurements of metaphase and interphase chromosome aberrations transmitted through early cell replication rounds in human lymphocytes exposed to low-LET protons and high-LET 12C ions. Mutat. Res. 2006;596:151–165. doi: 10.1016/j.mrfmmm.2005.12.010. [DOI] [PubMed] [Google Scholar]
- McPherson J.P., Sarras H., Lemmers B., Tamblyn L., Migon E., Matysiak-Zablocki E., Hakem A., Azami S.A., Cardoso R., Fish J. Essential role for Bclaf1 in lung development and immune system function. Cell Death Differ. 2009;16:331–339. doi: 10.1038/cdd.2008.167. [DOI] [PubMed] [Google Scholar]
- Mendell J.T., Sharifi N.A., Meyers J.L., Martinez-Murillo F., Dietz H.C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- Merz C., Urlaub H., Will C.L., Lührmann R. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. RNA. 2007;13:116–128. doi: 10.1261/rna.336807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz M.J., Pérez Santangelo M.S., Paronetto M.P., de la Mata M., Pelisch F., Boireau S., Glover-Cutter K., Ben-Dov C., Blaustein M., Lozano J.J. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137:708–720. doi: 10.1016/j.cell.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Nik-Zainal S., Alexandrov L.B., Wedge D.C., Van Loo P., Greenman C.D., Raine K., Jones D., Hinton J., Marshall J., Stebbings L.A., Breast Cancer Working Group of the International Cancer Genome Consortium Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul I., Savage K.I., Blayney J.K., Lamers E., Gately K., Kerr K., Sheaff M., Arthur K., Richard D.J., Hamilton P.W. PARP inhibition induces BAX/BAK-independent synthetic lethality of BRCA1-deficient non-small cell lung cancer. J. Pathol. 2011;224:564–574. doi: 10.1002/path.2925. [DOI] [PubMed] [Google Scholar]
- Quesada V., Conde L., Villamor N., Ordóñez G.R., Jares P., Bassaganyas L., Ramsay A.J., Beà S., Pinyol M., Martínez-Trillos A. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 2012;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- Savage K.I., Harkin D.P. BRCA1 and BRCA2: role in the DNA damage response, cancer formation and treatment. In: Shiloh Y., Khanna K.K., editors. The DNA Damage Response: Implications on Cancer Formation and Treatment. Springer; New York: 2009. pp. 415–443. [Google Scholar]
- Stucki M., Clapperton J.A., Mohammad D., Yaffe M.B., Smerdon S.J., Jackson S.P. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., Sato Y., Sato-Otsubo A., Kon A., Nagasaki M. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- Ziegelbauer J.M., Sullivan C.S., Ganem D. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat. Genet. 2009;41:130–134. doi: 10.1038/ng.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.