Abstract
Ring NTPases represent a large and diverse group of proteins that couple their nucleotide hydrolysis activity to a mechanical task involving force generation and some type of transport process in the cell. Because of their shape, these enzymes often operate as gates that separate distinct cellular compartments to control and regulate the passage of chemical species across them. In this manner, ions and small molecules are moved across membranes, biopolymer substrates are segregated between cells or moved into confined spaces, double-stranded nucleic acids are separated into single strands to provide access to the genetic information, and polypeptides are unfolded and processed for recycling. Here we review the recent advances in the characterization of these motors using single-molecule manipulation and detection approaches. We describe the various mechanisms by which ring motors convert chemical energy to mechanical force or torque and coordinate the activities of individual subunits that constitute the ring. We also examine how single-molecule studies have contributed to a better understanding of the structural elements involved in motor-substrate interaction, mechanochemical coupling, and intersubunit coordination. Finally, we discuss how these molecular motors tailor their operation—often through regulation by other cofactors—to suit their unique biological functions.
Introduction
Cells are nonequilibrium systems that constantly consume and dissipate energy to maintain or undergo transitions among steady states. Most essential cellular processes are unidirectional and are carried out by nanometer-scale machine-like devices that operate as molecular motors, coupling a downhill chemical reaction to a mechanical task (1–3). The chemical free energy may come from substrate binding, bond hydrolysis, or electrochemical gradients. These microscopic engines differ from their macroscopic counterparts in that they operate at energies only marginally higher than the energy of the thermal bath, and thus, thermal fluctuations play an important role in their operation. How these molecular machines carry out their cellular functions is of great interest to biologists and biophysicists.
Many molecular motors display ring-shaped architectures and consist of multiple subunits. In these ring motors, each individual subunit can couple its mechanical function to the energy source and one central question is to determine how the activities of these subunits are coordinated during the operation of the ring. A major fraction of ring motors belong to the Additional Strand Conserved E (ASCE) superfamily of NTPases, utilizing the energy from nucleotide binding and hydrolysis to catalyze a diversity of cellular activities, including ATP synthesis, DNA translocation, duplex unwinding, and protein unfolding (4,5) (Fig. 1).
Figure 1.
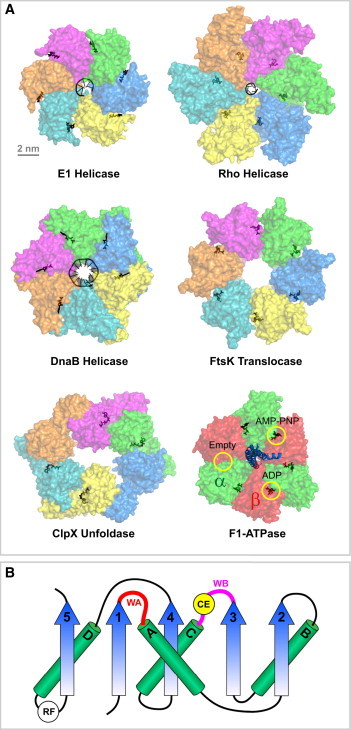
The architecture of ring-shaped motors from the ASCE superfamily of NTPases. (A) A crystal structure gallery of representative ASCE ring motors. The bovine papillomavirus replicative helicase E1 with a ssDNA substrate (PDB:2GXA) (7); The E. coli transcription termination factor Rho helicase with ssRNA (PDB: 3ICE) (8); The Bacillus stearothermophilus replicative helicase DnaB with ssDNA (PDB: 4ESV) (9); The motor domain of the Pseudomonas aeruginosa dsDNA translocase FtsK (PDB: 2IUU) (94); The E. coli protein unfoldase and polypeptide translocase ClpX (PDB: 3HWS) (110); The bovine mitochondrial F1-ATPase with the three catalytic sites encircled (α-subunits shown in green, β in red, and γ in blue) (PDB: 1BMF) (6). In all structures, nucleotides and their analogs are bound at the interface between adjacent subunits (black). Note that with the exception of the F1-ATPase, each ring consists of identical subunits that are colored differently for clarity. (B) Diagram of the core ASCE fold (modified from Lyubimov et al. (28)). WA, Walker A motif; WB, Walker B motif; CE, catalytic glutamate; RF, arginine finger. Note that the positions of CE and RF may vary in different motors and only the most common locations are shown.
Various experimental approaches have been used to investigate the operating mechanisms of molecular motors. High-resolution structures, especially those containing both the nucleotide-bound motor and its mechanical substrate, provide detailed albeit static snapshots of motor operation (6–9). Bulk biochemical methods can offer dynamic information about motor function, but they average the molecular signals over ensembles. This averaging often obscures crucial mechanistic features of the motor such as transient intermediates or parallel reaction pathways. Single-molecule techniques have yielded tremendous insights into various biological processes by enabling researchers to follow the dynamics of individual motor complexes in real time. Quantities describing molecular machines, such as displacement, force, torque, and rate, can be directly measured with single-molecule methods, making them ideally suited for motor studies.
There are two broad classes of single-molecule techniques: fluorescence-based detection (single particle tracking, single-molecule fluorescence resonance energy transfer, DNA curtains, etc.), and force-based manipulation (optical tweezers, magnetic tweezers, atomic force microscopy, flow stretching, etc.). Comprehensive reviews of these methods can be found in the literature (10–19). Recently, high-speed atomic force microscopy in an aqueous environment was added to the single-molecule toolbox (20), and this new methodology has already begun to provide remarkable real-time movies of motor proteins in action (21–23). Moreover, new hybrid instruments that combine single-molecule fluorescence and force microscopy carry great potential for simultaneously observing multiple aspects of motor dynamics (24–26).
The structure and function of ring-shaped molecular motors have been extensively reviewed (5,27–30). We will limit the scope of this review to single-molecule studies on ASCE ring NTPases. In particular, we will cover the following motors in detail:
-
1.
Packaging motors that pump viral genomes into preformed proteinaceous capsids during the assembly of double-stranded DNA (dsDNA) viruses (31,32) (Fig. 2 A);
-
2.
Replicative helicases, ubiquitous enzymes that catalyze strand separation of basepaired nucleic acids at the replication fork (33) (Fig. 2 B);
-
3.
FtsK and SpoIIIE, membrane-bound and septum-localized DNA translocases involved in bacterial chromosome segregation in Escherichia coli and Bacillus subtilis, respectively (34) (Fig. 2 C);
-
4.
F1-ATPase, the soluble portion of the F0F1-ATP synthase that utilizes a trans-membrane proton gradient to produce ATP through the rotation of the central γ-subunit inside a cylinder made of three αβ dimers (35,36) (Fig. 2 D); and
-
5.
ClpX, a bacterial unfoldase that unravels proteins and translocates unfolded polypeptides into the associated peptidase ClpP for degradation (37) (Fig. 2 E).
Figure 2.
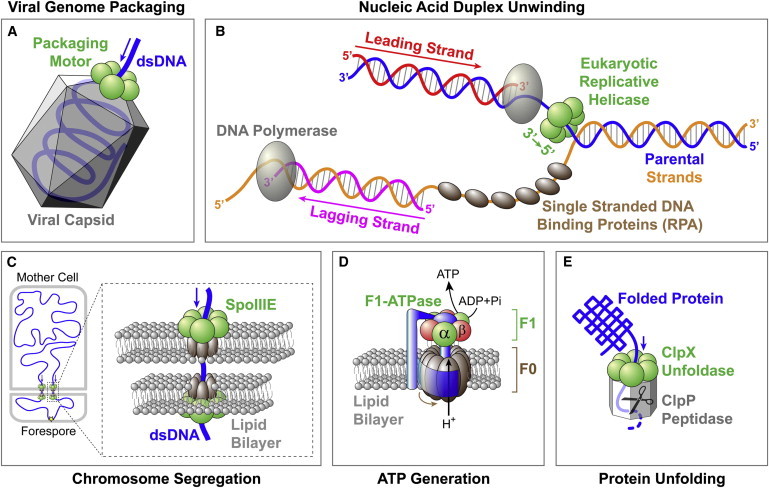
Biological systems that contain ring-shaped molecular motors. (A) Packaging motor translocates the viral genome into a preformed capsid. (B) Eukaryotic replicative helicase unwinds duplex DNA at the replication fork. Note that the bacterial helicase DnaB has a 5′→3′ polarity and translocates along the lagging-strand template. (C) SpoIIIE translocates dsDNA at the bacterial division septum during sporulation. (D) F0F1-ATP synthase generates ATP by using the proton gradient across the membrane. (E) ClpXP unfolds and degrades proteins.
Basic Motor Properties
Velocity
The velocity of a molecular motor, v, is given by v = d × kcat × ε, where d is the step size, kcat is the rate of chemical catalysis, and ε is the coupling coefficient, i.e., the probability that the motor takes a mechanical step per chemical cycle. For NTPases, kcat, and hence v, depends on the nucleotide concentration. The motor velocity also depends on externally applied loads in single-molecule force-manipulation assays, which can shed light on the force-generating mechanism of the motor (discussed later in this article).
The velocity varies greatly among different motors. For dsDNA translocases, the fastest known motor is FtsK, which translocates DNA at ∼5 kbp⋅s−1 at room temperature (38,39), going up to a remarkable ∼17 kbp⋅s−1 at 37°C (40). The bacteriophage φ29 packaging motor has a maximum velocity of ∼120 bp⋅s−1 at room temperature (41,42). By comparison, the bacteriophage λ motor packages DNA at ∼600 bp⋅s−1 (43), whereas the bacteriophage T4 motor can reach 2 kbp⋅s−1 (44). The translocation velocity of viral packaging motors scales roughly with the genome length (19.3 kbp for φ29, 48.5 kbp for λ, and 171 kbp for T4), reflecting similar timeframes for viral genome packaging in vivo. Replicative helicases operate at a wide range of velocities, from ∼440 bp⋅s−1 for the bacteriophage T7 gene product 4 (gp4) (45) to ∼4 bp⋅s−1 for the simian virus 40 helicase large T antigen (SV40 LTag) and the eukaryotic helicase MCM2-7 (46,47). Note that the unwinding rate by standalone helicases might be different from the rate of fork progression, because the velocity of the helicase is thought to be regulated by other replisome components (48–50).
ClpX is the only polypeptide translocase whose velocity has been directly measured in single-molecule experiments thus far (51–53). ClpX translocates unfolded polypeptides at ∼6 nm⋅s−1 in the presence of the associated peptidase ClpP, and at ∼8 nm⋅s−1 in the absence of ClpP (52). Unlike rigid dsDNA, unfolded polypeptides are relatively elastic, with a persistence length (∼0.65 nm) smaller than the ClpX step size (∼1 nm). As a result, the number of amino acids translocated per step by ClpX varies depending on the force the motor exerts on the polypeptide (e.g., 1 nm corresponds to ∼8 amino acids at 4 pN force but only ∼4 amino acids at 13 pN force).
Processivity
Traditionally, the processivity of a motor protein was used as a measure of the motor’s ability to undergo consecutive rounds of substrate movement before it is interrupted by dissociation. Thus, processivity was mathematically defined as kforward/(kforward + koff), where kforward is the rate of motor translocation and koff is the rate of motor dissociation from the substrate. In the context of single-molecule studies, a functionally relevant definition of processivity should include all processes that are off the main reaction pathway such as motor slippage and pausing. Hence, processivity can be rewritten as kforward/(kforward + koff + kslippage + kpausing). A motor’s processivity can be modulated by external force (38,54), nucleotide concentration (55,56), alternative nucleotide fuels (45), or, in general, any variable affecting the rates in the expression above. For instance, at saturating [ATP] and low external force, viral packaging motors and FtsK can move along several kilobasepairs of DNA without slipping or pausing (38,43,55,56). The processivity of these ring motors is adversely affected when the intersubunit coordination is disrupted by mutating a subset of NTPase subunits (57) or binding nonhydrolyzable nucleotide analogs to the ring (56,58).
Directionality
Ring motors often exhibit directionality in the movement relative to their substrate. For single-stranded nucleic acid translocases, the intrinsic asymmetry of the substrate can give rise to the unidirectional translocation, and the structural basis of these motors' directionality has begun to be understood (8,28). Double-stranded nucleic acid translocases and helicases can acquire directionality by tracking only one of the two strands. It is possible that this directionality is dictated by the initial orientation in which the motor is loaded onto the substrate track, using in some cases the ds/ss junction (e.g., the E1 helicase) (59) or specific recognition sequences (e.g., KOPS for FtsK, SRS for SpoIIIE) (60–62).
Interestingly, FtsK and SpoIIIE can reverse directionality during translocation (38–40). One possible model to explain such reversals is that each motor complex consists of two coaxial rings arranged opposite to each other on the DNA. In this model, only one ring is active at any given time; a direction reversal would then require turning off the active ring and switching on the partner ring upon encountering a recognition sequence with a nonpermissive orientation (63). Alternatively, direction can be reversed by a single ring dissociating and then immediately rebinding in the opposite orientation to resume translocation (60). Notably, protein unfoldases like ClpX do not possess an intrinsic translocation polarity and can efficiently move their polypeptide substrates in either direction (64). The polarity of ClpX is dictated by the placement of an 11-aa recognition tag (ssrA) on proteins targeted for degradation (65,66).
Stall force
The stall force of a molecular motor, Fstall, is defined as the force at which the motor velocity drops to zero. Some particularly strong motors, such as viral packaging motors and FtsK, can operate against forces as high as ∼60 pN (39,41,43,44), forces beyond which B-form DNA undergoes an overstretching phase transition (67). We note that the measured stalled forces are often not the thermodynamic stall forces (i.e., the maximum force that the motor can exert given the free energy available for a particular step size), but operational stall forces at which the substrate deforms or the motor slips or pauses indefinitely. In principle, the true thermodynamic stall force can be determined by measuring the force beyond which the motor reaction runs in reverse, as demonstrated by studies in which F1-ATPase was turned into an ATP synthase by mechanically forcing the γ-subunit to rotate in the opposite direction, generating ATP from ADP and Pi (68,69). The mechanochemical reversibility may not be always experimentally attainable due to limits for the maximum force that can be sustained by the motor and/or limits for the maximum [NDP] and [Pi] in the solution.
Step size
The step size of a motor, d, is defined as its net displacement along the substrate per catalytic event. Step sizes of several ASCE ring NTPases were inferred from atomic-resolution structures of the motor cocrystallized with its substrate and bound nucleotides (7–9). Recent advances in single-molecule techniques allow the direct observation of discrete steps for many molecular motors. The step size of the rotary F1-ATPase was measured to be 120° per ATP (70). The φ29 packaging motor was found to translocate DNA in 10-bp bursts, each consisting of four 2.5-bp steps (71). The T7 gp4 helicase unwinds duplex DNA in 2–3 bp bursts, likely with an elementary step size of 1 nt per nucleotide hydrolysis (72). Motor step size can be modulated by external load (73) or other regulatory factors (74).
Although the step size of ultrafast translocases such as FtsK is not directly measurable with current techniques, several alternative strategies can be employed to overcome this limitation:
-
1.
Single-molecule noise spectra were used to infer FtsK’s step size at 12 ± 2 bp (38). Unfortunately, this estimate relies on untested assumptions and must be interpreted with caution.
-
2.
Thermodynamic arguments can be used to estimate an upper bound for a motor’s step size. Because FtsK can exert at least 60 pN of force and the free energy of ATP hydrolysis is ∼100 pN⋅nm in the standard translocation buffer, FtsK’s step size cannot exceed ∼1.7 nm (∼5 bp).
-
3.
Step size can be inferred by measuring the DNA translocation and ATP consumption rates in bulk, which yields an estimate of ∼2 bp per ATP for FtsK (62). However, this ensemble method may lead to an underestimate of the step size if the motor undergoes frequent slipping or futile hydrolysis events.
Mechanochemical Coupling
The term “mechanochemistry” represents the mechanism by which a molecular motor couples chemical energy to mechanical force and displacement. Below we discuss the common experimental strategies used in single-molecule studies to dissect the mechanochemical coupling of molecular motors.
Force-velocity dependence
Like chemical products, force can also be considered a product of the reaction. Thus, an opposing external force can function as an inhibitor of the reaction, whereas an assisting force can function as an activator. Under conditions where the translocation step (by definition associated with a force-dependent rate constant) becomes rate-limiting for the reaction, the motor velocity becomes force-sensitive. For a mechanochemical process with a single force-dependent step per cycle, the force-velocity relationship follows a Boltzmann equation:
Here v0 is the velocity at zero force, kBT is the thermal energy, A is a constant describing the degree to which the force-dependent transition is rate-limiting at zero force (if A ≪ 1, the force-independent transitions are rate-limiting; if A ≥ 1, the force-dependent step is rate-limiting), and δ is the distance from the ground state to the transition state (75). The value δ can be thought of as the distance the motor must move along the translocation coordinate before committing to a step. By fitting force-velocity data to the Boltzmann equation, δ was estimated at ∼0.1 nm for the φ29 packaging motor (56) and at ∼0.7 nm for ClpX (51)— corresponding to ∼13% and 70% of the respective motor’s step size.
Interestingly, some motors display a complicated force-velocity dependence that cannot be explained by a single force-dependent step. For example, FtsK exhibits a velocity plateau at small forces, with increasing velocities at large assisting forces and decreasing velocities at large opposing forces (38); the velocity of the bacteriophage λ packaging motor is force-sensitive at low forces but becomes force-insensitive at high forces (43). To rationalize these observations, one must consider mechanochemical models with multiple force-generating transitions or multiple kinetic routes.
Michaelis-Menten kinetics
A motor’s force-velocity dependence at various concentrations of nucleotide and hydrolysis products provides valuable insight into its force-generating mechanism. The reaction pathway of an NTPase typically includes NTP binding, hydrolysis, product release, and mechanical movement that is coupled to certain chemical steps. When the overall reaction is largely irreversible, the average NTPase velocity follows the Michaelis-Menten equation,
where Vmax is the maximum velocity at saturating [NTP] and Km is the nucleotide concentration at which the velocity is half of Vmax. As shown below, a general chemical cycle can be divided into distinct kinetic blocks—regions of contiguous reversible kinetic transitions separated by irreversible kinetic transitions:
| (1) |
It can be shown that Km is a function of all of the kinetic rates in the cycle, Vmax is a function of all of the rates except the NTP docking and undocking rates, and Vmax/Km is a function of only the rates within the block that contains NTP docking (76). By quantifying how the application of mechanical force affects Vmax, Km, and Vmax/Km, one can determine whether the force-dependent transition is ATP docking, within the same kinetic block as ATP docking, or in a separate kinetic block (Table 1). This analysis can also be used to determine which portion of the cycle is affected by other perturbations such as a point mutation.
Table 1.
Dependence of Michaelis-Menten parameters on perturbations such as an external force applied to the motor (see also Eq. 1 in text)
| Location of the affected kinetic transition | Vmax | Km | Vmax/Km |
|---|---|---|---|
| ATP docking (k±1) | Same | Modified | Modified |
| Connected to docking (k±2, … ki, k±(j+1), … k±n) | Modified | Modified | Modified |
| Separate kinetic block (k±(i+1), … kj) | Modified | Modified | Same |
A general chemical cycle is separated into different kinetic blocks by irreversible transitions (ki and kj). How Vmax, Km, and Vmax/Km change with force depends on where the modified kinetic transition is located within the cycle.
This analysis was applied to study the mechanochemical coupling in the φ29 packaging motor and the ClpX unfoldase (53,56). In both cases, Vmax and Km decrease with increasing force, yet the ratio Vmax/Km remains constant. As a result, in conditions where ATP binding is rate-limiting ([ATP] < Km), the velocity (v ≈ (Vmax/Km) × [ATP]) does not depend on force. These observations rule out a scheme in which ATP docking drives translocation. The observation that Vmax/Km is independent of force also suggests that the force-generating chemical transition is not within the same kinetic block as ATP docking, which includes the irreversible ATP tight-binding step and the reversible ADP release step (53,56,58,71). Thus, force generation is coupled to either ATP hydrolysis or Pi release. It is unlikely that nucleotide hydrolysis results in a significant free energy drop along the reaction coordinate (77), whereas the release of phosphate involves a large change in free energy and is essentially irreversible because high concentrations of Pi have no measurable effect on velocity (53,56). Therefore, Pi release is the most likely candidate to power substrate translocation in the φ29 ATPase gp16 and ClpX. In contrast, F1-ATPase executes two power strokes in each hydrolysis cycle: ATP binding drives the initial 80° rotation of each 120° step and Pi release drives the remaining 40° rotation (77–79).
Passive and active unwinding mechanisms of helicases
Helicases are enzymes that separate double-stranded nucleic acids into single strands and play crucial roles during all stages of nucleic acid metabolism (80–82). The mechanism by which a helicase unwinds a duplex is classified as “passive” or “active” (82–84). Passive helicases rectify thermally driven fluctuations of the nucleic acid duplex by translocating and binding to the newly exposed single strand. In contrast, active helicases destabilize the duplex near the junction and shift the close↔open duplex equilibrium. The activeness of a helicase can be quantified by the amount of destabilization energy it contributes per basepair unwound, ΔGd (85). ΔGd = 0 for a purely passive helicase and ΔGd = 3.4 kBT (G–C base-pairing energy) for an optimally active helicase. Optical tweezers and magnetic tweezers were used to analyze how unwinding velocity depends on external force and substrate sequence (G–C versus A–T/U). Activeness varies widely among different helicases, from the mostly passive phage T4 gp41 (ΔGd = 0.05 kBT) (86), the partially active phage T7 gp4 (ΔGd = 1–2 kBT) (87) and E. coli DnaB (ΔGd = 0.5–1.6 kBT) (54), to the highly active non-ring-shaped helicases such as hepatitis C virus NS3 (ΔGd = 2.5 kBT) (88). However, caution should be exercised when interpreting these varied numbers as the reflection of distinct unwinding mechanisms, because they may also be due to differences in the modeling approaches and experimental geometries used in different studies (54,81,89).
Intersubunit Coordination
The coordination between the catalytic cycles of individual subunits is a key feature in the operation of ring NTPases. Whereas each ring subunit is often capable of hydrolyzing NTP and undergoing the associated mechanical transition, the functionality of the ring depends on the manner in which the activities of individual subunits are orchestrated. Structural and bulk biochemical studies have suggested various models of intersubunit coordination for ring motors. In the sequential model, subunits bind and hydrolyze nucleotides in a well-defined order, most likely ordinally around the ring (7,8,90–95). In the concerted model, all subunits simultaneously bind and hydrolyze nucleotides and release products, such that they are always at the same nucleotide state (96). In the stochastic model, each subunit undergoes its own catalytic cycle independently of the others (97). Single-molecule studies, as presented in detail below, have provided fresh insights into the existing models and revealed novel coordination schemes.
F1-ATPase
Rotation of the γ-subunit was first observed via an optical microscope by attaching an actin filament to the γ-rotor (98). At sufficiently low ATP concentrations, the rotation was found to proceed in 120° steps (70) (Fig. 3 A), consistent with the threefold symmetry of the (αβ)3 hexamer. Using a 40-nm gold bead that generates a much lower hydrodynamic drag than the actin filament, the 120° step was further resolved into 80° and 40° substeps (79). The duration of the dwells preceding the 80° substeps is inversely proportional to [ATP] (79), indicating that ATP binding occurs during these dwells.
Figure 3.
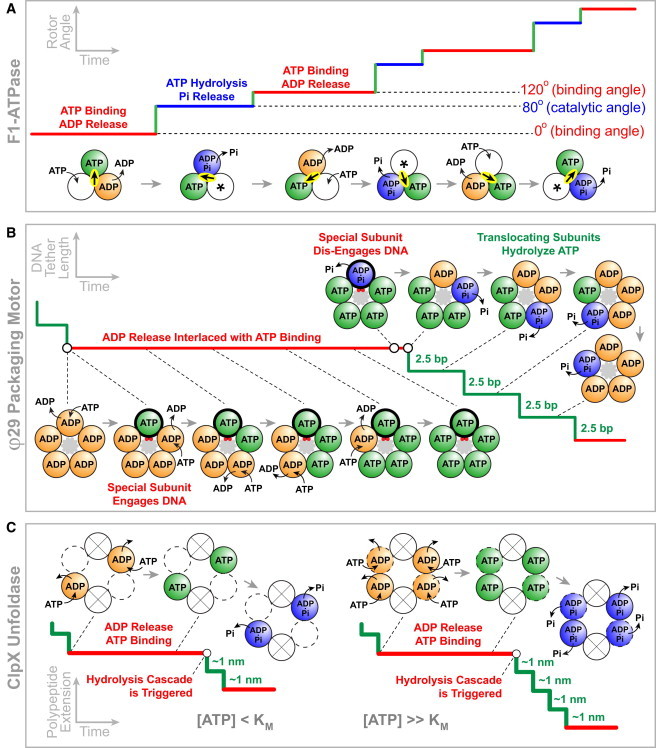
Models of mechanochemical coupling and intersubunit coordination for three ASCE ring ATPases. (A) F1-ATPase model adapted from Adachi et al. (78). (Solid line) The γ-subunit rotation angle versus time. (Circles) The three catalytic sites. (Asterisk) Sites accessible to nucleotides from solution. (Highlighted arrow) The γ-subunit orientation. (B) φ29 gp16 model adapted from Chistol et al. (58). (Solid line) Motor position on DNA versus time. (Circles) The five catalytic sites. (Gray-shaded area) DNA. The special subunit (thick outline) contacts a pair of DNA backbone phosphates (red circles) during the dwell phase. (C) ClpX model adapted from Sen et al. (53) and Stinson et al. (112). (Solid line) Motor position on the unfolded polypeptide versus time. (Circles) The six catalytic sites: (solid outline) two high-affinity catalytic sites that bind nucleotides at any [ATP]; (dashed outline) two low-affinity catalytic sites that bind nucleotides only at high [ATP]; (crossed circles) the two ATPase sites that remain empty due to ring distortion.
Hydrolysis of ATP likely occurs in the dwell preceding the 40° substeps, because this dwell is prolonged in a hydrolysis-deficient mutant (β-E190D) or in the presence of ATP-γ-S in the solution (99). Furthermore, experiments with Cy3-ATP, a fluorescent ATP analog that is presumably hydrolyzed slowly, allowed the simultaneous observation of rotation and ATP binding. It suggested that the ATP that was bound when the γ-subunit is at 0° is hydrolyzed when the γ-subunit points at 200° (100). Thus, the dwells before the 80° substep and the 40° substep are termed the “ATP-binding dwell” and the “catalytic dwell”, respectively (Fig. 3A).
The addition of Pi to the reaction mixture specifically prolongs the catalytic dwell, and very high [Pi] induces frequent reversals of 40° substeps (78). These results strongly indicate that Pi release drives the 40° rotation. However, the exact timing of Pi release cannot be determined unambiguously: it may occur either immediately after hydrolysis during the same catalytic dwell (scenario 1), or during the next catalytic dwell after 120° rotation (scenario 2). A later study showed that there is an open nucleotide-binding site (asterisk in Fig. 3 A) freely accessible to ATP in the solution and that free Pi from the solution competes with the nucleotide binding, inconsistent with scenario 2 in which product Pi is retained in this site and thus supporting scenario 1 (101).
Finally, experiments monitoring the binding and release of Cy3-ATP (78), together with experiments monitoring the site occupancy (number of nucleotides bound to the three catalytic sites) (101), suggested that ADP release occurs during the 80° substep from 240° to 320° and possibly helps drive the 80° rotation, in addition to ATP binding.
To summarize, for each catalytic site, ATP hydrolysis, Pi release, and ADP release occur at 200°, 200°, and 240° after ATP binding, respectively (Fig. 3 A). Although chemical events in the three catalytic sites occur sequentially, the F1-ATPase employs a much more intricate coordination scheme than the canonical sequential model. All three catalytic sites participate to drive each 120° rotation of the γ-subunit, and each site is involved in three consecutive 120° steps (102). The model for F1 is the first and still one of the most comprehensively characterized intersubunit coordination schemes for a ring NTPase. However, this model is not without controversies. Some studies suggested that Pi is released at 320° instead of 200°, thus after ADP is released (103,104). Additionally, under certain conditions with a slow hydrolysis mutant, rotation was found to proceed in steps of 120° and no 80° or 40° substeps were observed (105), a phenomenon also found in the related enzyme V1-ATPase (106–108). Future studies will help to clarify these points of contention.
φ29 DNA packaging motor
Using a high-resolution dual-trap optical tweezers instrument, it has been possible to observe and characterize the discrete DNA translocation cycles of the gp16 ring ATPase from the φ29 packaging motor (71). Under low external forces (<10 pN), it was found that DNA is translocated in 10-bp cycles, and that each cycle is composed of a stationary dwell phase and a DNA-translocating burst phase (Fig. 3 B). The duration of the dwell phase depends on the concentration of ATP, whereas the duration of the burst phase does not. These results suggest that ATP binding occurs during the dwell but not during the actual translocation of the substrate.
Under high external forces (30–40 pN), the translocation burst slows down as expected, revealing that each 10-bp burst is composed of four 2.5-bp steps that occur in rapid succession (71). As mentioned above, translocation is powered by the release of Pi in the φ29 motor. Thus, Pi release most likely coincides with or immediately precedes each 2.5-bp step.
Nonhydrolyzable ATP analogs, ATP-γ-S and AMP-PNP, were used to determine the timing of ATP hydrolysis in the dwell-burst cycle (58). The analog-induced pausing events interrupt the burst phase, suggesting that hydrolysis occurs during the burst, with each subunit hydrolyzing ATP before taking a 2.5-bp step. To determine where in the cycle ADP release occurs, the packaging experiments were repeated in the presence of orthovanadate, a Pi analog that forms a complex with ADP, delaying the dissociation of ADP from the binding pocket (58). In contrast to the ATP-γ-S case, ADP-vanadate-induced pausing events specifically prolong the dwell phase, suggesting that ADP release occurs during the dwell.
To investigate the coordination between ATP binding and ADP release events, which both take place during the dwell, ADP was added to the reaction mixture. It was found that increasing [ADP] gradually lengthens all dwells in a linear fashion (58). The linear dependence indicates that at any given time only one binding site is available for ADP inhibition, consistent with a model in which ADP release and ATP binding occur in an alternating fashion.
Thus, the φ29 packaging motor operates as a highly coordinated machine in which ADP release and ATP binding occur in an interlaced fashion during the dwell phase, whereas ATP hydrolysis, Pi release, and DNA translocation occur in an ordinal manner, one subunit after another, during the burst phase (Fig. 3 B). A crucial question remained: given that the homo-pentameric motor only generates four steps per cycle, what is the function of the special nontranslocating subunit during the cycle? An interesting observation shed light on this question: it was found that while an analog nucleotide remains bound to the ring, the motor can still stochastically take a few 10-bp bursts separated by pauses much longer than regular dwells, resulting in what was termed a “pause cluster” (58). This phenomenon requires that while the analog remains bound to one subunit, the remaining four subunits bind and spontaneously hydrolyze ATP to translocate DNA by 2.5 bp each, albeit much slower than the normal operation of the motor. This observation suggests that the long pauses in a pause cluster reflect the behavior of the motor when the nontranslocating subunit cannot hydrolyze the analog nucleotide. Hence, proper ATP turnover by the special, nontranslocating subunit is also necessary for the normal activity of the ring. Therefore, the function of this special subunit is regulatory, ensuring the timed and proper firing of the other four translocating subunits. This finding reveals an unprecedented division of labor among subunits in a homomeric ring ATPase: only four of the five subunits translocate the substrate, whereas the special one plays a critical regulatory role.
Is the regulatory role always performed by the same special subunit, or is that task passed around the ring in consecutive cycles? The discrepancy between the 10-bp burst size of the motor and the 10.4-bp helical pitch of B-form DNA suggests that the motor may rotate the DNA to bring the DNA into register with the motor after every cycle. If so, various scenarios for the identity of the special subunit predict different direction and/or magnitude of DNA rotation. A rotor-bead assay was developed to monitor changes of the DNA angle around its helical axis while simultaneously observing DNA translocation (74). This assay clearly showed that the DNA is indeed rotated by the motor in a left-handed direction. In the absence of DNA organization inside the capsid, the motor rotates DNA by 1.5 ± 0.2° per bp packaged. This measurement supports a model in which the same special subunit makes regulatory contacts with the DNA in successive cycles: after a 10-bp burst, the DNA backbone winds by 346°; thus a 14° rotation realigns the DNA with the subunit that contacted the DNA in the previous cycle, yielding a rotation density of 1.4°/bp.
T7 replicative helicase
Unlike the other ring NTPases discussed here, the T7 gp4 hexameric DNA helicase utilizes dTTP instead of ATP to power its operation (109). Single-molecule optical tweezers experiments showed that ATP also supports unwinding, but does so much less efficiently than dTTP, because ATP induces frequent slipping that ultimately prevents unwinding over any significant distance (45). This unique property enabled the investigation of the T7 gp4 subunit coordination mechanism by measuring the unwinding rate and processivity with different amounts of ATP and dTTP. It was found that ATP and dTTP compete for the same nucleotide binding sites and the unwinding rate is simply determined by ATP and dTTP concentrations and their respective Vmax and Km. The helicase processivity, defined here as the length unwound between slips, does not follow a simple competitive behavior; instead, it shows a cooperative dependence on the ratio of [dTTP] and [ATP]. In other words, a small fraction of dTTP mixed with ATP significantly increases the processivity. These results are inconsistent with a random model in which the subunits function independently, and support a model in which most of the six subunits act together to coordinate nucleotide binding/hydrolysis and DNA binding to ensure processivity. Similar to the φ29 packaging motor, only one subunit in the T7 helicase ring is poised to bind a nucleotide at a time while the other subunits are nucleotide-occupied.
ClpX unfoldase/translocase
Structural and single-molecule data suggest a step size of ∼1 nm per ClpX subunit (51,52,110). Recent single-molecule work found that ClpX translocates polypeptides in increments of 2, 3, and 4 nm, suggesting that multiple ClpX subunits (between two and four) fire in a coordinated fashion (53). Although ClpX translocates its substrate through a dwell-burst mechanism similar to the one employed by φ29 gp16, the reduction in velocity at low ATP concentration is achieved by ClpX and φ29 gp16 through very different means. At low ATP concentrations the φ29 motor slows down by lengthening the mean dwell duration while maintaining a constant mean burst size (71). In contrast, at low [ATP], ClpX slows down by decreasing its average burst size while maintaining a constant mean dwell duration (53). ClpX’s mechanism of slowing down at low [ATP] suggests the existence of a type of internal clock that triggers the motor to fire a burst regardless of how many ClpX subunits are bound to ATP (Fig. 3 C). At [ATP] ≤ Km, the ClpX translocation bursts are predominantly 2–3 nm in size, whereas at [ATP] ≫ Km, the burst sizes are mostly 3–4 nm and do not exceed 4 nm. These results support a model in which the ClpX hexamer contains two high-affinity and two low-affinity nucleotide binding sites, with the other two sites remaining empty, presumably due to large deformations in the ring (110,111). At low [ATP] only the two high-affinity sites bind ATP before the internal-clock mechanism triggers the ring to fire, resulting in a 2-nm burst size (Fig. 3 C). At saturating [ATP], up to four sites can load nucleotides before the burst is triggered, resulting in a maximum burst size of 4 nm. It is not known what serves as ClpX’s internal clock and triggers the burst phase.
A recent study suggests that ClpX periodically undergoes ring-resetting independent of ATP hydrolysis (112). In the resetting events, the hexameric ring isomerizes and reassigns individual subunits with apparently-new nucleotide-binding properties. Stinson et al. (112) proposed that this resetting would make it possible for ClpX to avoid stalling during the unfolding of very stable proteins, or to circumvent temporary subunit inhibition by ADP in an ADP-rich cellular environment. Such spontaneous resetting represents a plausible candidate for the internal-clock mechanism that gives rise to the [ATP]-independent dwell duration. The proposed operation model for ClpX bears some similarity to the model proposed for the φ29 packaging motor, in that one (in the case of φ29 gp16) or two (in the case of ClpX) subunit interfaces in the ATPase ring are different from the rest, implying a functional asymmetry in these homomeric rings (Fig. 3, B and C).
Structural Basis of Motor Operation
The complete elucidation of a motor’s mechanism requires a detailed understanding of the structural motifs in the motor-substrate complex that are responsible for its functional dynamics. High-resolution structures have undoubtedly shed much light on the operating mechanism of ring NTPases (6–9,91,93–95,110,113). Here we will discuss the complementary and emerging method of combining single-molecule experiments with targeted mutagenesis of the motor or specific modifications of the substrate to dissect the physical basis of motor operation.
Motor mutants
The ASCE motors share a variety of conserved sequence elements, including the phosphate-binding Walker A motif (also known as P-loop), the metal-binding Walker B motif, the catalytic glutamate residue that activates a water molecule for nucleophilic attack on the bound NTP, and the arginine finger that protrudes into the NTP binding pocket from an adjacent subunit and participates in hydrolysis and intersubunit communication (5) (Fig. 1 B). The NTP binding site is typically located at the interface between two adjacent subunits. The roles of these elements in motor operation have started to be characterized at the single-molecule level. Tsay et al. (114,115) generated a series of point mutations in the bacteriophage λ packaging motor. For example, a mutation (Y46F) in the Q-motif—a putative adenine-binding motif upstream of Walker A—not only decreased the velocity but also showed increased force sensitivity and impaired processivity. These results suggest that the Q-motif is an important part of the mechanical pathway of force generation, coupling changes in the ATP-binding pocket to DNA substrate propulsion.
With high-resolution single-molecule measurements, it is possible to pinpoint where the mutation takes effect within the mechanochemical cycle of the motor. For example, the catalytic glutamate mutant in F1-ATPase, β-E190D, specifically prolonged the 80° dwells and was used to determine the timing of hydrolysis (99). Moreover, to investigate the intersubunit coordination and functional asymmetry within a ring motor, it is advantageous to mutate only one or a few of the ring subunits rather than create a homomeric mutant ring. This task can be achieved by mixing wild-type and mutant monomers or engineering covalently linked dimers or trimers (57,102). Ideally, one would covalently connect all subunits to form a single-chain ring motor, allowing precise control of the mutation sites within the ring and ensuring sample homogeneity (97).
Substrate modifications
To determine the nature of motor-DNA interaction for the φ29 packaging motor, the motor was challenged with a variety of substrate modifications, including DNA with a neutral backbone, abasic DNA, single-stranded gaps and bulges, and even nonbiological polymers (116). The data indicate that the motor-DNA interaction is remarkably promiscuous, involving a wide variety of contacts with the nucleic acid moieties as well as contacts not specific to DNA. Moreover, it was found that the type of interaction changes during the course of the mechanochemical cycle:
-
1.
During the ATP-loading dwell phase, the motor makes specific electrostatic contacts with a pair of adjacent backbone phosphates every 10 bp. These ionic contacts have both a load-bearing role and a sensory or regulatory role, coupling mechanical and chemical events. They are also likely responsible for rectifying the symmetry mismatch between the motor and the DNA at the end of each cycle via DNA rotation. It is plausible that these specific contacts are themselves responsible for breaking the symmetry of the ring and for conferring the regulatory role on the subunit that makes these interactions.
-
2.
During the DNA-translocating burst phase, the motor makes nonspecific DNA contacts that are most likely steric. These nonspecific interactions may be the reason why the step size of φ29 gp16 is a noninteger number of basepairs.
Substrate modifications have also been used to investigate the structural basis of γ rotation in F1-ATPase. Wang and Oster (117) proposed a push-pull mechanism based on structural data, in which a nucleotide-bound β-subunit is bent toward, and pushes, the axle section of the γ-subunit, whereas an empty β-subunit retracts and pulls γ. This model requires a rigid γ-axle pivoted at the bottom. However, it was later found that an axleless γ-mutant still supports rotation in the correct direction (118). Other truncation studies further suggested that no part of the γ-subunit is required for rotation, even though some parts are needed for the generation of full speed and full torque (22,119–121). Thus, it appears that rotation does not depend on specific interactions between γ and the (αβ)3 hexamer. In this regard, as Kinosita (122) has pointed out, it would be fascinating to investigate whether F1-ATPase is able to rotate a completely foreign object such as DNA.
Biological Functions
Motor proteins are often part of larger multicomponent assemblies in vivo. When carrying out their biological tasks, these motors need to adjust their operation upon interaction with accessory factors as well as other environmental changes such as fluctuating chemical concentrations and varying mechanical loads. Below we will discuss the biological functions of several ring motors and some experimental efforts to examine how their activities are regulated during these tasks.
ATP synthesis and hydrolysis
The F0F1-ATP synthase is composed of two rotary motors—the ATP-hydrolysis-driven F1 and the proton-flow-driven F0. Depending on the metabolic state of the cell, the coupled enzyme can either synthesize ATP using the free energy from a trans-membrane proton gradient, or pump protons across the membrane using the free energy from ATP hydrolysis (Fig. 2 D). Through a series of elegant single-molecule experiments, it has been shown that the γ-subunit rotates in opposite directions during ATP hydrolysis and synthesis (123), and that the isolated F1 can produce ATP when γ is forced to rotate in reverse via mechanical manipulation (68,69). Nonetheless, the rotary mechanism for ATP synthesis is still poorly understood compared to that for ATP hydrolysis. Although reasonable as a first-order approximation, it remains to be tested whether the mechanochemical pathway for synthesis is the exact reversal of the pathway for hydrolysis (Fig. 3 A). Moreover, the largely constant output torque as a function of rotation angle implies an elastic power transmission between F1 and F0, which is considered essential for the high kinetic efficiency and robust operation of the holoenzyme consisting of two counteracting and symmetry-mismatched (C3 versus C10–14) stepping motors (124,125). The detailed coupling mechanism between these two motors is eagerly anticipated.
Viral genome packaging
In dsDNA viruses, the packaging motor needs to overcome major energetic barriers to compact the stiff and highly charged genome to near-crystalline densities inside a small capsid (Fig. 2 A). It has long been known that packaging slows down as DNA fills the capsid and generates an internal pressure working against the motor (41,43). However, the details of how capsid filling affects the operation of the packaging motor remained unclear. Recently the φ29 packaging dynamics was examined at different stages of capsid filling (74). It was found that φ29 gp16 employs a complex throttle control mechanism in which multiple parameters of the motor’s mechanochemical cycle are modified in response to the increasing capsid filling. Specifically, high filling causes the motor to bind ATP more slowly, enter long-lived pausing states more frequently, take smaller steps per cycle (∼10 bp at low filling versus ∼9 bp at high filling), and translocate DNA more slowly due to an effective internal force. The internal force generated by the compressed DNA can be inferred from the measurement of the burst phase duration as a function of force and as a function of capsid filling. This approach revealed that the internal force reaches ∼20 pN at 100% filling, which corresponds to an internal pressure of ∼20 atm (74).
Remarkably, despite all the changes that cause the net velocity to drop by two orders of magnitude at high filling, the motor maintains its strict subunit coordination and division of labor (74). This feat is achieved by the simultaneous adjustments of the step size and the amount of DNA rotation per cycle. Thus the φ29 packaging motor possesses a remarkable operational flexibility in the context of a fixed mechanism of mechanochemical coupling and intersubunit coordination to allow robust activity as the genome becomes more and more densely packed. It will be interesting to investigate whether or not such strategy is shared with other dsDNA viruses, a group that includes most tailed bacteriophages and some human pathogens such as herpes and adenoviruses.
Bacterial chromosome segregation
During cell division, FtsK and SpoIIIE are involved in segregating the circular bacterial genome between daughter cells (in the case of FtsK) or between a mother cell and a forespore (in the case of SpoIIIE) (Fig. 2 C). A central question in the field is how these DNA translocases ensure the efficient and directed segregation of a circular DNA through a division septum. As discussed above, FtsK and SpoIIIE can reverse their translocation direction in vitro and in vivo (38–40,63). If not properly regulated, such direction reversals during chromosome segregation are detrimental to the cell. It is possible that the interaction with other regulatory factors, the skewed distribution of the orientation of the recognition sequences (KOPS, SRS) in the genome, or the organization of the motor at the division septum itself, rectifies the motion of the translocase and maintains its directionality. It should be pointed out that the functional oligomeric form (hexameric versus dodecameric) of SpoIIIE is still under debate (126,127). Future studies should clarify this point.
Roadblock displacement
Many ring translocases and helicases possess the ability of displacing roadblocks bound to their substrates. Roadblock displacement activity is essential for replicative helicases because it ensures unimpeded fork progression along a dsDNA template bound with nucleosomes, RNA polymerases, and transcription factors. Single-molecule replication experiments with MCM2-7 and SV40 LTag revealed that the rate of replication fork progression is the same for sparsely and densely chromatinized DNA, suggesting that both helicases efficiently displace histones and other proteins bound to DNA (46,47). SV40 LTag was also shown to evict streptavidin from biotinylated DNA, thus disrupting one of the strongest noncovalent interactions (47). Considering the diversity of DNA-bound obstacles encountered by these helicases, their roadblock displacement activity is most likely achieved via steric extrusion. Interestingly, SV40 LTag is also capable of bypassing protein barriers cross-linked to DNA (47), presumably by cracking its ring to form a lock-washer-like structure. It remains to be determined whether MCM2-7 can use the same mechanism to bypass impediments cross-linked to DNA.
DsDNA translocases are also capable of displacing roadblocks. Both SpoIIIE and FtsK were shown to efficiently displace ssDNA from DNA triplexes (61,63). FtsK can disrupt the biotin-streptavidin interaction (57). SpoIIIE can strip histone-like proteins and RNA polymerases from the DNA, which facilitates DNA compaction in the forespore, and helps to maintain the compartment-specific gene expression program in the mother cell and the forespore (128). To ensure maximal genome compaction, viral packaging motors also must displace DNA-bound proteins. Preliminary experiments suggest that the φ29 packaging motor can disrupt the biotin-streptavidin linkage (Fig. 4, G. Chistol, C. Bustamante, unpublished data).
Figure 4.
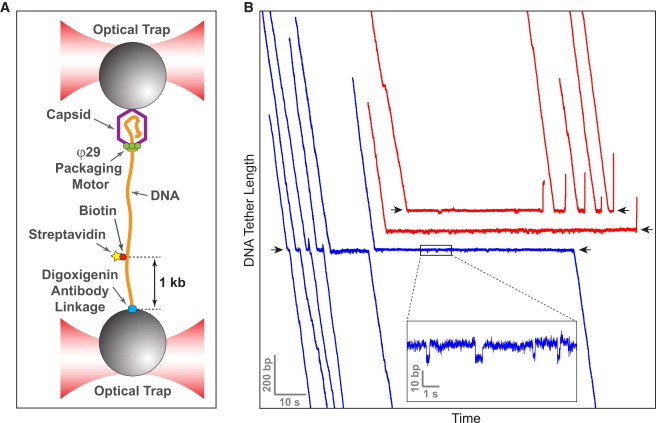
The φ29 packaging motor is capable of disrupting the biotin-streptavidin linkage. (A) The packaging complex is assembled in a dual-trap optical tweezers instrument. Streptavidin is bound to a biotin on DNA. (B) Trajectories of individual motors exhibit clear pauses at the location of the roadblock (black arrows). For clarity, some traces are offset vertically. In ∼50% of the cases, the motor successfully overcomes the roadblock (blue). In the remaining cases, the motor slips (red). Because the inner diameter of packaging motor cannot accommodate both dsDNA and streptavidin, these data suggest that the motor can disrupt the biotin-streptavidin linkage.
Protein unfolding and translocation
Protein unfoldases/translocases such as ClpX have evolved distinct features to cope with unique challenges (Fig. 2 E). Compared to nucleic acid translocases, protein translocases act on a substrate that is much more heterogeneous—polypeptides that can contain as many as 20 amino acids with different physicochemical properties. Thus, it is plausible that enzymes like ClpX interact with their polypeptide substrate via nonspecific steric interactions (110). These nonspecific contacts may be essential to ClpX’s ability to processively and unidirectionally translocate irregular and diverse substrates from either the N-terminus or the C-terminus (64).
To efficiently unravel stably folded protein substrates, unfoldases must perform not only the thermodynamic task of destabilizing the native state, but also the kinetic task of quickly capturing the unraveled polypeptide before it can refold. For example, to successfully unfold GFP, ClpX must first quickly take a 4-nm burst that dislodges and traps the β-strand 11 before this segment can refold onto the GFP barrel (53). At saturating [ATP], the time required for unfolding GFP is determined by the time that passes before the motor makes a 4-nm burst. At low [ATP], hydrolysis is often triggered before four ATP molecules can bind to the motor and thus the motor rarely succeeds in unfolding GFP. These findings are consistent with the observed nonlinear decrease of substrate unfolding probability with ATPase rate. The presence of the ClpP peptidase drastically reduces the slip frequency of ClpX—probably by providing additional contacts with unfolded polypeptides—thereby enhancing the motor’s processivity and its ability to unfold and translocate substrates (51,52).
Concluding Remarks
Organized as homo- or hetero-oligomers, ring NTPases coordinate the catalytic and mechanical activity of their monomeric constituents during operation. Through the application of single-molecule methods, scientists have begun to address and delineate how the unique properties of each motor—processivity, mechanical strength, directionality, speed, and detailed dynamics—are controlled and determined by the coordination and communication among their subunits. To date, only a handful of motors have been interrogated in this manner and their characterization is by no means close to complete. Yet, it is already clear that each of these machines has become specialized and optimized to fulfill a specific task in the cell. Whereas some of them operate as extremely coordinated devices in which all subunits perform their function in a predetermined and invariant order, others are seen to have relaxed their degree of coordination gaining instead in flexibility and efficiency. The ultimate goal of these studies is to obtain as complete a description as possible of these motors’ operation, including what parts move, when and how they move, and to map the sequence of interactions responsible for the communication among the individual ring subunits. Although some of the key interactions and structural motifs involved in this communication have been recognized, we are still far from understanding how information and energy flow among the subunits and how they trigger a response at the right location and time somewhere else in the ring. Further studies along these lines, combining structural detail at the atomic scale with high spatial and temporal resolution trajectories obtained from individual molecules, should make it possible to arrive at a more complete description of the biological designs and physical principles that underlie the operation of these important and fascinating nano machines.
Acknowledgments
The authors thank Ninning Liu, Maya Sen, Jae Yen Shin, and Sara Tafoya for critical readings of the manuscript, and members of the Bustamante laboratory for stimulating discussions.
This work was supported by the National Institutes of Health under grant No. R01-GM071552, the Department of Energy under contract No. DE-AC02-05CH11231, and the Howard Hughes Medical Institute (to C.B.). S.L. acknowledges support from the NIH Pathway to Independence Award No. K99-GM107365.
Footnotes
Shixin Liu and Gheorghe Chistol contributed equally to this work.
Gheorghe Chistol’s present address is Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA.
References
- 1.Bustamante C., Chemla Y.R., Izhaky D. Mechanical processes in biochemistry. Annu. Rev. Biochem. 2004;73:705–748. doi: 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 2.Kolomeisky A.B., Fisher M.E. Molecular motors: a theorist’s perspective. Annu. Rev. Phys. Chem. 2007;58:675–695. doi: 10.1146/annurev.physchem.58.032806.104532. [DOI] [PubMed] [Google Scholar]
- 3.Chowdhury D. Modeling stochastic kinetics of molecular machines at multiple levels: from molecules to modules. Biophys. J. 2013;104:2331–2341. doi: 10.1016/j.bpj.2013.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iyer L.M., Leipe D.D., Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 5.Erzberger J.P., Berger J.M. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 6.Abrahams J.P., Leslie A.G., Walker J.E. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 7.Enemark E.J., Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 8.Thomsen N.D., Berger J.M. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itsathitphaisarn O., Wing R.A., Steitz T.A. The hexameric helicase DnaB adopts a nonplanar conformation during translocation. Cell. 2012;151:267–277. doi: 10.1016/j.cell.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapanidis A.N., Strick T. Biology, one molecule at a time. Trends Biochem. Sci. 2009;34:234–243. doi: 10.1016/j.tibs.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 11.van Oijen A.M. Honey, I shrunk the DNA: DNA length as a probe for nucleic-acid enzyme activity. Biopolymers. 2007;85:144–153. doi: 10.1002/bip.20624. [DOI] [PubMed] [Google Scholar]
- 12.Greenleaf W.J., Woodside M.T., Block S.M. High-resolution, single-molecule measurements of biomolecular motion. Annu. Rev. Biophys. Biomol. Struct. 2007;36:171–190. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joo C., Balci H., Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu. Rev. Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 14.Moffitt J.R., Chemla Y.R., Bustamante C. Recent advances in optical tweezers. Annu. Rev. Biochem. 2008;77:205–228. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 15.De Vlaminck I., Dekker C. Recent advances in magnetic tweezers. Annu. Rev. Biophys. 2012;41:453–472. doi: 10.1146/annurev-biophys-122311-100544. [DOI] [PubMed] [Google Scholar]
- 16.Finkelstein I.J., Greene E.C. Supported lipid bilayers and DNA curtains for high-throughput single-molecule studies. Methods Mol. Biol. 2011;745:447–461. doi: 10.1007/978-1-61779-129-1_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puchner E.M., Gaub H.E. Force and function: probing proteins with AFM-based force spectroscopy. Curr. Opin. Struct. Biol. 2009;19:605–614. doi: 10.1016/j.sbi.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Walter N.G., Huang C.-Y., Sobhy M.A. Do-it-yourself guide: how to use the modern single-molecule toolkit. Nat. Methods. 2008;5:475–489. doi: 10.1038/nmeth.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Veigel C., Schmidt C.F. Moving into the cell: single-molecule studies of molecular motors in complex environments. Nat. Rev. Mol. Cell Biol. 2011;12:163–176. doi: 10.1038/nrm3062. [DOI] [PubMed] [Google Scholar]
- 20.Ando T., Uchihashi T., Kodera N. High-speed AFM and applications to biomolecular systems. Annu. Rev. Biophys. 2013;42:393–414. doi: 10.1146/annurev-biophys-083012-130324. [DOI] [PubMed] [Google Scholar]
- 21.Kodera N., Yamamoto D., Ando T. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature. 2010;468:72–76. doi: 10.1038/nature09450. [DOI] [PubMed] [Google Scholar]
- 22.Uchihashi T., Iino R., Noji H. High-speed atomic force microscopy reveals rotary catalysis of rotorless F1-ATPase. Science. 2011;333:755–758. doi: 10.1126/science.1205510. [DOI] [PubMed] [Google Scholar]
- 23.Noi K., Yamamoto D., Ogura T. High-speed atomic force microscopic observation of ATP-dependent rotation of the AAA+ chaperone p97. Structure. 2013;21:1992–2002. doi: 10.1016/j.str.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 24.Heller I., Sitters G., Wuite G.J. STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA. Nat. Methods. 2013;10:910–916. doi: 10.1038/nmeth.2599. [DOI] [PubMed] [Google Scholar]
- 25.Comstock M.J., Ha T., Chemla Y.R. Ultrahigh-resolution optical trap with single-fluorophore sensitivity. Nat. Methods. 2011;8:335–340. doi: 10.1038/nmeth.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long X., Parks J.W., Stone M.D. Mechanical unfolding of human telomere G-quadruplex DNA probed by integrated fluorescence and magnetic tweezers spectroscopy. Nucleic Acids Res. 2013;41:2746–2755. doi: 10.1093/nar/gks1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singleton M.R., Dillingham M.S., Wigley D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 28.Lyubimov A.Y., Strycharska M., Berger J.M. The nuts and bolts of ring-translocase structure and mechanism. Curr. Opin. Struct. Biol. 2011;21:240–248. doi: 10.1016/j.sbi.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enemark E.J., Joshua-Tor L. On helicases and other motor proteins. Curr. Opin. Struct. Biol. 2008;18:243–257. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iino R., Noji H. Intersubunit coordination and cooperativity in ring-shaped NTPases. Curr. Opin. Struct. Biol. 2013;23:229–234. doi: 10.1016/j.sbi.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 31.Hetherington C.L., Moffitt J.R., Bustamante C. Viral DNA packaging motors. In: Goldman Y.E., Ostap E.M., editors. Molecular Motors and Motility. Elsevier; Oxford, UK: 2012. pp. 420–446. [Google Scholar]
- 32.Rao V.B., Feiss M. The bacteriophage DNA packaging motor. Annu. Rev. Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 33.Patel S.S., Pandey M., Nandakumar D. Dynamic coupling between the motors of DNA replication: hexameric helicase, DNA polymerase, and primase. Curr. Opin. Chem. Biol. 2011;15:595–605. doi: 10.1016/j.cbpa.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaimer C., Graumann P.L. Players between the worlds: multifunctional DNA translocases. Curr. Opin. Microbiol. 2011;14:719–725. doi: 10.1016/j.mib.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 35.Kinosita K., Jr., Adachi K., Itoh H. Rotation of F1-ATPase: how an ATP-driven molecular machine may work. Annu. Rev. Biophys. Biomol. Struct. 2004;33:245–268. doi: 10.1146/annurev.biophys.33.110502.132716. [DOI] [PubMed] [Google Scholar]
- 36.Boyer P.D. The ATP synthase—a splendid molecular machine. Annu. Rev. Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 37.Baker T.A., Sauer R.T. ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim. Biophys. Acta. 2012;1823:15–28. doi: 10.1016/j.bbamcr.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saleh O.A., Pérals C., Allemand J.-F. Fast, DNA-sequence independent translocation by FtsK in a single-molecule experiment. EMBO J. 2004;23:2430–2439. doi: 10.1038/sj.emboj.7600242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pease P.J., Levy O., Cozzarelli N.R. Sequence-directed DNA translocation by purified FtsK. Science. 2005;307:586–590. doi: 10.1126/science.1104885. [DOI] [PubMed] [Google Scholar]
- 40.Lee J.Y., Finkelstein I.J., Greene E.C. Single-molecule imaging of DNA curtains reveals mechanisms of KOPS sequence targeting by the DNA translocase FtsK. Proc. Natl. Acad. Sci. USA. 2012;109:6531–6536. doi: 10.1073/pnas.1201613109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith D.E., Tans S.J., Bustamante C. The bacteriophage straight φ29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 42.Rickgauer J.P., Fuller D.N., Smith D.E. Portal motor velocity and internal force resisting viral DNA packaging in bacteriophage φ29. Biophys. J. 2008;94:159–167. doi: 10.1529/biophysj.107.104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fuller D.N., Raymer D.M., Smith D.E. Measurements of single DNA molecule packaging dynamics in bacteriophage λ reveal high forces, high motor processivity, and capsid transformations. J. Mol. Biol. 2007;373:1113–1122. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuller D.N., Raymer D.M., Smith D.E. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. Proc. Natl. Acad. Sci. USA. 2007;104:16868–16873. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun B., Johnson D.S., Wang M.D. ATP-induced helicase slippage reveals highly coordinated subunits. Nature. 2011;478:132–135. doi: 10.1038/nature10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yardimci H., Loveland A.B., Walter J.C. Uncoupling of sister replisomes during eukaryotic DNA replication. Mol. Cell. 2010;40:834–840. doi: 10.1016/j.molcel.2010.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yardimci H., Wang X., Walter J.C. Bypass of a protein barrier by a replicative DNA helicase. Nature. 2012;492:205–209. doi: 10.1038/nature11730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delagoutte E., von Hippel P.H. Molecular mechanisms of the functional coupling of the helicase (gp41) and polymerase (gp43) of bacteriophage T4 within the DNA replication fork. Biochemistry. 2001;40:4459–4477. doi: 10.1021/bi001306l. [DOI] [PubMed] [Google Scholar]
- 49.Stano N.M., Jeong Y.-J., Patel S.S. DNA synthesis provides the driving force to accelerate DNA unwinding by a helicase. Nature. 2005;435:370–373. doi: 10.1038/nature03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pandey M., Syed S., Patel S.S. Coordinating DNA replication by means of priming loop and differential synthesis rate. Nature. 2009;462:940–943. doi: 10.1038/nature08611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aubin-Tam M.-E., Olivares A.O., Lang M.J. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 2011;145:257–267. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maillard R.A., Chistol G., Bustamante C. ClpXP generates mechanical force to unfold and translocate its protein substrates. Cell. 2011;145:459–469. doi: 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sen M., Maillard R.A., Bustamante C. The ClpXP protease unfolds substrates using a constant rate of pulling but different gears. Cell. 2013;155:636–646. doi: 10.1016/j.cell.2013.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ribeck N., Kaplan D.L., Saleh O.A. DnaB helicase activity is modulated by DNA geometry and force. Biophys. J. 2010;99:2170–2179. doi: 10.1016/j.bpj.2010.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kottadiel V.I., Rao V.B., Chemla Y.R. The dynamic pause-unpackaging state, an off-translocation recovery state of a DNA packaging motor from bacteriophage T4. Proc. Natl. Acad. Sci. USA. 2012;109:20000–20005. doi: 10.1073/pnas.1209214109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chemla Y.R., Aathavan K., Bustamante C. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 57.Crozat E., Meglio A., Sherratt D.J. Separating speed and ability to displace roadblocks during DNA translocation by FtsK. EMBO J. 2010;29:1423–1433. doi: 10.1038/emboj.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chistol G., Liu S., Bustamante C. High degree of coordination and division of labor among subunits in a homomeric ring ATPase. Cell. 2012;151:1017–1028. doi: 10.1016/j.cell.2012.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee S.J., Syed S., Joshua-Tor L. Dynamic look at DNA unwinding by a replicative helicase. Proc. Natl. Acad. Sci. USA. 2014;111:E827–E835. doi: 10.1073/pnas.1322254111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bigot S., Saleh O.A., Barre F.-X. Oriented loading of FtsK on KOPS. Nat. Struct. Mol. Biol. 2006;13:1026–1028. doi: 10.1038/nsmb1159. [DOI] [PubMed] [Google Scholar]
- 61.Levy O., Ptacin J.L., Cozzarelli N.R. Identification of oligonucleotide sequences that direct the movement of the Escherichia coli FtsK translocase. Proc. Natl. Acad. Sci. USA. 2005;102:17618–17623. doi: 10.1073/pnas.0508932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Graham J.E., Sherratt D.J., Szczelkun M.D. Sequence-specific assembly of FtsK hexamers establishes directional translocation on DNA. Proc. Natl. Acad. Sci. USA. 2010;107:20263–20268. doi: 10.1073/pnas.1007518107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ptacin J.L., Nollmann M., Bustamante C. Sequence-directed DNA export guides chromosome translocation during sporulation in Bacillus subtilis. Nat. Struct. Mol. Biol. 2008;15:485–493. doi: 10.1038/nsmb.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barkow S.R., Levchenko I., Sauer R.T. Polypeptide translocation by the AAA+ ClpXP protease machine. Chem. Biol. 2009;16:605–612. doi: 10.1016/j.chembiol.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee C., Schwartz M.P., Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol. Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- 66.Hoskins J.R., Yanagihara K., Wickner S. ClpAP and ClpXP degrade proteins with tags located in the interior of the primary sequence. Proc. Natl. Acad. Sci. USA. 2002;99:11037–11042. doi: 10.1073/pnas.172378899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bustamante C., Bryant Z., Smith S.B. Ten years of tension: single-molecule DNA mechanics. Nature. 2003;421:423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 68.Itoh H., Takahashi A., Kinosita K., Jr. Mechanically driven ATP synthesis by F1-ATPase. Nature. 2004;427:465–468. doi: 10.1038/nature02212. [DOI] [PubMed] [Google Scholar]
- 69.Rondelez Y., Tresset G., Noji H. Highly coupled ATP synthesis by F1-ATPase single molecules. Nature. 2005;433:773–777. doi: 10.1038/nature03277. [DOI] [PubMed] [Google Scholar]
- 70.Yasuda R., Noji H., Yoshida M. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120° steps. Cell. 1998;93:1117–1124. doi: 10.1016/s0092-8674(00)81456-7. [DOI] [PubMed] [Google Scholar]
- 71.Moffitt J.R., Chemla Y.R., Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Syed S., Pandey M., Ha T. Single-molecule fluorescence reveals the unwinding stepping mechanism of replicative helicase. Cell Rep. 2014;6:1037–1045. doi: 10.1016/j.celrep.2014.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mallik R., Carter B.C., Gross S.P. Cytoplasmic dynein functions as a gear in response to load. Nature. 2004;427:649–652. doi: 10.1038/nature02293. [DOI] [PubMed] [Google Scholar]
- 74.Liu, S., G. Chistol, …, C. Bustamante. A viral packaging motor varies its DNA rotation and step size to preserve subunit coordination as the capsid fills. Cell. http://dx.doi.org/10.1016/j.cell.2014.02.034. [DOI] [PMC free article] [PubMed]
- 75.Wang M.D., Schnitzer M.J., Block S.M. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 76.Keller D., Bustamante C. The mechanochemistry of molecular motors. Biophys. J. 2000;78:541–556. doi: 10.1016/S0006-3495(00)76615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oster G., Wang H. Reverse engineering a protein: the mechanochemistry of ATP synthase. Biochim. Biophys. Acta. 2000;1458:482–510. doi: 10.1016/s0005-2728(00)00096-7. [DOI] [PubMed] [Google Scholar]
- 78.Adachi K., Oiwa K., Kinosita K., Jr. Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation. Cell. 2007;130:309–321. doi: 10.1016/j.cell.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 79.Yasuda R., Noji H., Itoh H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature. 2001;410:898–904. doi: 10.1038/35073513. [DOI] [PubMed] [Google Scholar]
- 80.Patel S.S., Picha K.M. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 81.Yodh J.G., Schlierf M., Ha T. Insight into helicase mechanism and function revealed through single-molecule approaches. Q. Rev. Biophys. 2010;43:185–217. doi: 10.1017/S0033583510000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lohman T.M., Bjornson K.P. Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 83.Singleton M.R., Wigley D.B. Modularity and specialization in superfamily 1 and 2 helicases. J. Bacteriol. 2002;184:1819–1826. doi: 10.1128/JB.184.7.1819-1826.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.von Hippel P.H., Delagoutte E. A general model for nucleic acid helicases and their “coupling” within macromolecular machines. Cell. 2001;104:177–190. doi: 10.1016/s0092-8674(01)00203-3. [DOI] [PubMed] [Google Scholar]
- 85.Betterton M.D., Jülicher F. Opening of nucleic-acid double strands by helicases: active versus passive opening. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2005;71:011904. doi: 10.1103/PhysRevE.71.011904. [DOI] [PubMed] [Google Scholar]
- 86.Lionnet T., Spiering M.M., Croquette V. Real-time observation of bacteriophage T4 gp41 helicase reveals an unwinding mechanism. Proc. Natl. Acad. Sci. USA. 2007;104:19790–19795. doi: 10.1073/pnas.0709793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Johnson D.S., Bai L., Wang M.D. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007;129:1299–1309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cheng W., Dumont S., Bustamante C. NS3 helicase actively separates RNA strands and senses sequence barriers ahead of the opening fork. Proc. Natl. Acad. Sci. USA. 2007;104:13954–13959. doi: 10.1073/pnas.0702315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Manosas M., Xi X.G., Croquette V. Active and passive mechanisms of helicases. Nucleic Acids Res. 2010;38:5518–5526. doi: 10.1093/nar/gkq273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Adelman J.L., Jeong Y.-J., Patel S.S. Mechanochemistry of transcription termination factor Rho. Mol. Cell. 2006;22:611–621. doi: 10.1016/j.molcel.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 91.Sun S., Kondabagil K., Rao V.B. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251–1262. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 92.Crampton D.J., Mukherjee S., Richardson C.C. DNA-induced switch from independent to sequential dTTP hydrolysis in the bacteriophage T7 DNA helicase. Mol. Cell. 2006;21:165–174. doi: 10.1016/j.molcel.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 93.Mancini E.J., Kainov D.E., Stuart D.I. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell. 2004;118:743–755. doi: 10.1016/j.cell.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 94.Massey T.H., Mercogliano C.P., Löwe J. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol. Cell. 2006;23:457–469. doi: 10.1016/j.molcel.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 95.Singleton M.R., Sawaya M.R., Wigley D.B. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 96.Gai D., Zhao R., Chen X.S. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell. 2004;119:47–60. doi: 10.1016/j.cell.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 97.Martin A., Baker T.A., Sauer R.T. Rebuilt AAA+ motors reveal operating principles for ATP-fueled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- 98.Noji H., Yasuda R., Kinosita K., Jr. Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 99.Shimabukuro K., Yasuda R., Yoshida M. Catalysis and rotation of F1 motor: cleavage of ATP at the catalytic site occurs in 1 ms before 40° substep rotation. Proc. Natl. Acad. Sci. USA. 2003;100:14731–14736. doi: 10.1073/pnas.2434983100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nishizaka T., Oiwa K., Kinosita K., Jr. Chemomechanical coupling in F1-ATPase revealed by simultaneous observation of nucleotide kinetics and rotation. Nat. Struct. Mol. Biol. 2004;11:142–148. doi: 10.1038/nsmb721. [DOI] [PubMed] [Google Scholar]
- 101.Shimo-Kon R., Muneyuki E., Kinosita K., Jr. Chemo-mechanical coupling in F1-ATPase revealed by catalytic site occupancy during catalysis. Biophys. J. 2010;98:1227–1236. doi: 10.1016/j.bpj.2009.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ariga T., Muneyuki E., Yoshida M. F1-ATPase rotates by an asymmetric, sequential mechanism using all three catalytic subunits. Nat. Struct. Mol. Biol. 2007;14:841–846. doi: 10.1038/nsmb1296. [DOI] [PubMed] [Google Scholar]
- 103.Okazaki K., Hummer G. Phosphate release coupled to rotary motion of F1-ATPase. Proc. Natl. Acad. Sci. USA. 2013;110:16468–16473. doi: 10.1073/pnas.1305497110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Watanabe R., Iino R., Noji H. Phosphate release in F1-ATPase catalytic cycle follows ADP release. Nat. Chem. Biol. 2010;6:814–820. doi: 10.1038/nchembio.443. [DOI] [PubMed] [Google Scholar]
- 105.Shimabukuro K., Muneyuki E., Yoshida M. An alternative reaction pathway of F1-ATPase suggested by rotation without 80°/40° substeps of a sluggish mutant at low ATP. Biophys. J. 2006;90:1028–1032. doi: 10.1529/biophysj.105.067298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Imamura H., Takeda M., Yokoyama K. Rotation scheme of V1-motor is different from that of F1-motor. Proc. Natl. Acad. Sci. USA. 2005;102:17929–17933. doi: 10.1073/pnas.0507764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Furuike S., Nakano M., Yokoyama K. Resolving stepping rotation in Thermus thermophilus H+-ATPase/synthase with an essentially drag-free probe. Nat. Commun. 2011;2:233. doi: 10.1038/ncomms1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Minagawa Y., Ueno H., Iino R. Basic properties of rotary dynamics of the molecular motor Enterococcus hirae V1-ATPase. J. Biol. Chem. 2013;288:32700–32707. doi: 10.1074/jbc.M113.506329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Donmez I., Patel S.S. Mechanisms of a ring shaped helicase. Nucleic Acids Res. 2006;34:4216–4224. doi: 10.1093/nar/gkl508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Glynn S.E., Martin A., Sauer R.T. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hersch G.L., Burton R.E., Sauer R.T. Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: allosteric control of a protein machine. Cell. 2005;121:1017–1027. doi: 10.1016/j.cell.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 112.Stinson B.M., Nager A.R., Sauer R.T. Nucleotide binding and conformational switching in the hexameric ring of a AAA+ machine. Cell. 2013;153:628–639. doi: 10.1016/j.cell.2013.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Strycharska M.S., Arias-Palomo E., Berger J.M. Nucleotide and partner-protein control of bacterial replicative helicase structure and function. Mol. Cell. 2013;52:844–854. doi: 10.1016/j.molcel.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tsay J.M., Sippy J., Smith D.E. The Q motif of a viral packaging motor governs its force generation and communicates ATP recognition to DNA interaction. Proc. Natl. Acad. Sci. USA. 2009;106:14355–14360. doi: 10.1073/pnas.0904364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tsay J.M., Sippy J., Smith D.E. Mutations altering a structurally conserved loop-helix-loop region of a viral packaging motor change DNA translocation velocity and processivity. J. Biol. Chem. 2010;285:24282–24289. doi: 10.1074/jbc.M110.129395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Aathavan K., Politzer A.T., Bustamante C. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature. 2009;461:669–673. doi: 10.1038/nature08443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang H., Oster G. Energy transduction in the F1 motor of ATP synthase. Nature. 1998;396:279–282. doi: 10.1038/24409. [DOI] [PubMed] [Google Scholar]
- 118.Furuike S., Hossain M.D., Kinosita K., Jr. Axle-less F1-ATPase rotates in the correct direction. Science. 2008;319:955–958. doi: 10.1126/science.1151343. [DOI] [PubMed] [Google Scholar]
- 119.Hossain M.D., Furuike S., Kinosita K., Jr. Neither helix in the coiled coil region of the axle of F1-ATPase plays a significant role in torque production. Biophys. J. 2008;95:4837–4844. doi: 10.1529/biophysj.108.140061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Müller M., Pänke O., Engelbrecht S. F1-ATPase, the C-terminal end of subunit γ is not required for ATP hydrolysis-driven rotation. J. Biol. Chem. 2002;277:23308–23313. doi: 10.1074/jbc.M201998200. [DOI] [PubMed] [Google Scholar]
- 121.Kohori A., Chiwata R., Kinosita K., Jr. Torque generation in F1-ATPase devoid of the entire amino-terminal helix of the rotor that fills half of the stator orifice. Biophys. J. 2011;101:188–195. doi: 10.1016/j.bpj.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kinosita K., Jr. F1-ATPase: a prototypical rotary molecular motor. In: Rossmann M.G., Rao V.B., editors. Viral Molecular Machines. Springer; New York: 2012. pp. 5–16. [DOI] [PubMed] [Google Scholar]
- 123.Diez M., Zimmermann B., Gräber P. Proton-powered subunit rotation in single membrane-bound F0F1-ATP synthase. Nat. Struct. Mol. Biol. 2004;11:135–141. doi: 10.1038/nsmb718. [DOI] [PubMed] [Google Scholar]
- 124.Pänke O., Cherepanov D.A., Junge W. Viscoelastic dynamics of actin filaments coupled to rotary F-ATPase: angular torque profile of the enzyme. Biophys. J. 2001;81:1220–1233. doi: 10.1016/S0006-3495(01)75780-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Junge W., Sielaff H., Engelbrecht S. Torque generation and elastic power transmission in the rotary F0F1-ATPase. Nature. 2009;459:364–370. doi: 10.1038/nature08145. [DOI] [PubMed] [Google Scholar]
- 126.Fleming T.C., Shin J.Y., Pogliano K. Dynamic SpoIIIE assembly mediates septal membrane fission during Bacillus subtilis sporulation. Genes Dev. 2010;24:1160–1172. doi: 10.1101/gad.1925210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fiche J.-B., Cattoni D.I., Nöllmann M. Recruitment, assembly, and molecular architecture of the SpoIIIE DNA pump revealed by superresolution microscopy. PLoS Biol. 2013;11:e1001557. doi: 10.1371/journal.pbio.1001557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Marquis K.A., Burton B.M., Rudner D.Z. SpoIIIE strips proteins off the DNA during chromosome translocation. Genes Dev. 2008;22:1786–1795. doi: 10.1101/gad.1684008. [DOI] [PMC free article] [PubMed] [Google Scholar]


