Abstract
Background
Colon cancer is still the second leading cause of cancer deaths in the United States. Epigenetic gene silencing involving DNA methyltransferases (DNMTs) and histone deacetylases (HDACs) play an important role in the progression of colon cancer.
Materials and Methods
Here we found that the sensitivity of colon cancer cells to methylation plays a role in its response to alternative therapy involving the green tea polyphenol, epigallocatechin 3-gallate. HDAC and DNMT protein expression was reduced when methylation-sensitive HCT 116 human colon cancer cells was treated with EGCG, but was relatively stable in the HT-29 cell line. This decrease in expression may be partially explained by our finding that DNMT3A and HDAC3 are degraded in the methylation-sensitive colon cancer cells in part by inhibiting their association with the E3 ubiquitin ligase, UHRF1.
Conclusion
These findings provide a rationale for the development of a more targeted therapy for methylation-sensitive colon cancer that can include EGCG in combination with other DNMT and HDAC inhibitors.
Green tea is the world’s most popular beverage and substantial evidence supports its success in the prevention of carcinogenesis in animal models (11). Green tea has been found to reactivate genes in carcinogen-induced rodent models of colon cancer, which ultimately led to the suppression of intestinal tumorigenesis (25). The most active compound in green tea, epigallocatechin gallate (EGCG) induces cell cycle arrest and apoptosis of cancer cells (1, 23). In colon cancer models, EGCG has been shown to inhibit epidermal growth factor receptor (EGFR), vascular endothelial growth factor receptor (VEGFR), and cyclooxygenase-2 (COX2), as well as human epidermal growth factor 3 (HER3) (20, 21). This includes EGCG in a host of compounds that may be useful in preventing tumor metastasis through inhibiting angiogenesis, as well as supports its inclusion as an anti-inflammatory agent. EGCG also inhibits DNA methyltransferases (DNMTs), as well as reactivating key regulatory genes silenced in colon cancer (7). EGCG targets multiple signaling pathways, making it a good subject for inclusion as a chemopreventive or therapeutic agent. The inhibition of DNMTs by EGCG provides a potential mechanism as to how tumorigenesis is halted by green tea but specific details on the role of EGCG as it pertains to DNMTs or other epigenetic players is limited. It is possible that EGCG may work not only through inhibition of enzymatic activity but also through classic pathways involving protein degradation.
More recently, with categorization of the different molecular genetic profiles of colon carcinomas, it has become clear that variations in response of human cancer to different therapies may depend on genetic and epigenetic profiles. One subset of genetically distinct colon carcinomas are considered to be microsatellite instable (MSI) in which the mismatch repair gene, human MutL homolog 1 (hMLH1), is silenced due to aberrant methylation of its promoter (17). MSI is associated with colorectal cancer that has aberrant methylation in the CpG islands of genes (24). This is referred to as the CpG island methylator phenotype, or CIMP. CIMP-positive tumors in colorectal cancer exhibit methylation of tumor-suppressor genes and silencing of other regulatory genes (24). The belief is that if these regulatory genes can be de-silenced, cancer progression can be halted or reversed.
DNA hypermethylation and histone deacetylation are key epigenetic mechanisms for the silencing of many genes including tumor-suppressor genes (18), genes responsible for cell cycle regulation and control, and apoptosis and DNA repair (13). Targeting DNA hypermethylation and histone deacetylation with pharmacological inhibitors has proved successful in altering genetic expression in models of disease (4, 13). DNA methylation can lead to transcriptional inactivation by directly inhibiting the binding of transcription factors, masking the DNA sequence it recognizes, HDACs, or recruiting methyl-binding proteins that interact directly with transcription factors (2, 9). The use of common pharmacological inhibitors of DNMTs and HDACs is limited in human patients due to their toxicity (3).
We hypothesized that EGCG contributes to the degradation of DNMT3A and HDAC3 through a classic pathway involving the E3 ubiquitin ligase, Ubiquitin-like, containing plant homeo domain (PHD) and really interesting new gene (19) finger domains, 1 (UHRF1). By examining the effects of EGCG on the association of DNMT3A and HDAC3 with UHRF1 in the MSI colon cancer cell line HCT 116 and the methylation-insensitive HT-29 cell line, we observed a clear difference in the way these cells respond to EGCG treatment.
Materials and Methods
Cell lines and cell culture
The human colon cancer cell lines HT-29 and HCT 116 were obtained from the American Type Culture Collection ATCC (Manassas, VA, USA). The cells were maintained in a 5% CO2 atmosphere at 37°C in McCoys 5a medium containing 10% fetal bovine serum and 1% penicillin/streptomycin. To determine dose dependent changes in protein and gene expression, cells were treated with 50, 100, 150 μM of EGCG (Sigma Aldrich, St. Louis, MO, USA) or an equal volume of dimethyl sulfoxide (DMSO) as a vehicle control for 48 and 72 hours.
Protein extraction
Nuclear protein extractions were performed as follow: Treated HT-29 and HCT 116 cells were kept on ice for 10 minutes with a low salt lysis buffer (10 mM HEPES, 10 mM KCl, 1 mM EDTA), then scraped and spun down. To harvest the nuclear fraction, 50–100 μl high-salt lysis buffer (20 mM HEPES, 0.4 M NaCl, 1 mM EDTA) was added to the cell pellet and left on ice for 30 min. The tubes were spun down and nuclear protein was collected.
Western Blot
The nuclear protein samples were loaded onto 12% acrylamide gel (50 μg per lane). After electrophoresis, the proteins were blotted onto a nitrocellulose membrane. Membranes were then blocked in a solution of 10% nonfat milk in phosphor-buffered saline with Tween 20 (PBST). Following incubation with blocking buffer, membranes were incubated with the primary antibodies: DNMT3A, herpesvirus-associated ubiquitin specific protease (HAUSP), HDAC3 and topoisomerase type II B (TOPOIIβ) (Santa Cruz Biotechnology, Santa Cruz CA, USA) and UHRF1 (Millipore, Billerica, MA, USA). After washing with PBST, the membranes were then incubated with the respective horseradish peroxidase-labeled secondary antibody and visualized using the SuperSignal West Pico Chemiluminescent Substrate Kit (Thermo, Waltham, MA, USA). Densitometry was carried out using the image processing software, ImageJ (NIH, Bethesda, MD, USA).
Immunoprecipitation
HCT 116 and HT-29 cells were treated with either DMSO or 100 μM EGCG. Cells were harvested after 24 and 48 hours and nuclear extracts were prepared as described above. Protein concentration was determined by the NanoVue Plus spectrophotometer (GE Healthcare, Piscataway, NJ, USA). Primary antibody (2 μg: anti-UHRF1) was added to nuclear protein (250 μg) and tubes were rotated at 4°C overnight. Protein G Sepharose Beads (50 μl) was added and tubes were rotated for another two hours at 4°C. Beads were then collected and washed in immunoprecipitation buffer. To separate the protein complex from beads, the tubes were heated to 95°C following addition of 20 μl Laemmli Buffer (Bio-Rad, Hercules, CA, USA).
RNA extraction and real-time polymerase chain reaction (PCR)
RNA was isolated from the cells using the High Pure RNA Isolation Kit (Roche, San Francisco, CA, USA) according to the manufacturer’s protocol. RNA purity and concentration was quantified using NanoVue Plus spectrophotometer (GE Healthcare, Piscataway, NJ, USA). Reactions were set up in triplicate for each sample. PCR was performed in a 25 μl reaction containing 12.5 μl of the 2X SYBR PCR reaction mix, 300 nM of each primer, 0.5 μL of iScript reverse transcriptase and 25 ng of RNA. The reaction protocol was as follows: initial incubation at 50°C for 10 minutes to allow for cDNA synthesis, reverse transcriptase inactivation at 95°C for 5 minutes; and then 40 cycles of PCR cycling and detection with 95°C for 10 seconds, 55°C for 30 seconds and 95°C for one minute. The melting curve was determined using 55°C for one minute and 80 cycles of 0.5°C increments from 55°C to 95°C. Primer sequences were: DNMT3A: 5′-CTGAGAGTCAGGGACTTGGC-3′ (sense) and 5′-AGTCTA GCAATCGTTGGCGT-3′ (anti-sense); DNMT3B: 5′-CAGGGAAAACTGCAAAGCTC-3′ (sense) and 5′-ATTTGTTACGTCGTGGCTCC-3′ (anti-sense); DNMT1; 5′-TGTGAGGACATGCAGCTTTC-3′ (sense) and 5′-ACCAACTCGGTACAGGATGC-3′ (anti-sense); HDAC1: 5′-CTTCCTGCTGAGTCCCTCAC-3′ (sense) and 5′-CCAGTGGGAAGGTACGAAAA-3′ (anti-sense); HDAC2: 5′-AAAACCCAGTTTTCTGCCCT-3′ (sense) and 5′-TCTGCCTCCCACTCTGTCTT-3′ (anti-sense); HDAC3: 5′-ACGTGGGCAACTTCCACTAC-3′ (sense) and 5′-GACTCTTGGTGAAGCCTTGC-3′ (anti-sense).
Protein degradation with cycloheximide
Cells were pretreated for 24 hours with either DMSO or 100 μM EGCG. Cycloheximide was added to cells for a final concentration of 10 μg/ml. Nuclear protein was extracted after 6, 12, 24, and 48 hours using low and high-salt lysis buffers as mentioned previously. DNMT3a and HDAC3 protein is then quantified and subjected to western blot analysis.
Statistics
Data were analyzed for Real-time PCR using Student’s t-test. Significance was acceptable at p<0.05.
Results
Effects of EGCG on DNMT mRNA in HCT 116 and HT-29 cells
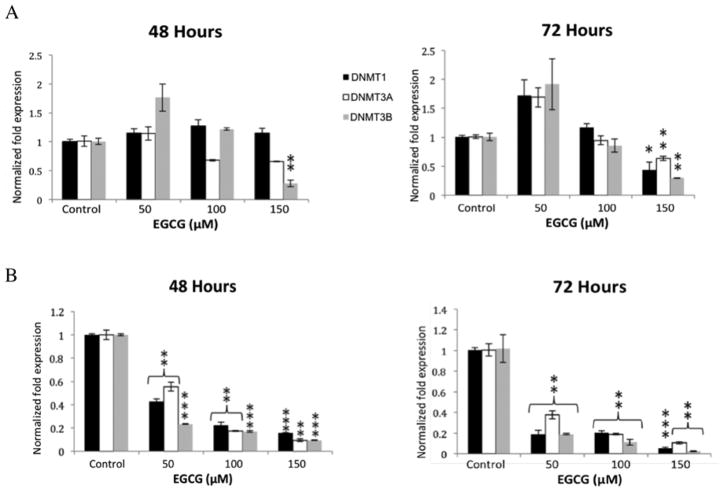
It is known that EGCG can reduce methylation of promoter regions of genes or reactivate silenced genes in various models of disease (7, 8, 18). To determine the effects of EGCG on mRNA expression of DNMTs in human colon cancer cell lines, HT-29 and HCT 116 cells were treated with EGCG (from 50 μM to 150 μM) for 48 and 72 hours. Transcript expression was evaluated by quantitative real-time PCR. The resulting graphs show a statistically significant decrease in DNMT3B transcript in HCT 116 cells at an EGCG concentration of 150 μM while all studied DNMT transcripts were found to be significantly downregulated in HT-29 cells at 48 hours at all concentrations of EGCG (Figure 1A). At 72 hours, HCT 116 cells exhibited significant down-regulation of all studied transcripts at 150 μM ECGG and HT-29 cells exhibitied a similar, but more drastic decrease in expression of all DNMT transcripts (Figure 1B).
Figure 1.
Epigallocatechin 3-gallate reduces expression of some DNA methyltransferase (DNMT) transcripts. DNMT1, -3A and -3B transcript levels were reduced in HCT 116 cells at 72 hours at 150 μM EGCG (A). All studied transcripts were significantly reduced at 72 hours in HT-29 cells at the lower concentration of 50 μM (B). Error bars represent +/− one standard deviation (n=3). *p<0.05, **p<0.02, ***p<0.01 compared with the control.
Effect EGCG on DNMT3A protein expression in HCT 116 and HT-29 cells
Given the results of RT-PCR for the DNMT transcripts, we chose DNMT3A to determine if the results found at the mRNA level would be corroborated at the protein level. HT-29 and HCT 116 cells were treated with EGCG (from 50 μM to 150 μM) for 48 and 72 hours. The nuclear protein was extracted and probed for DNMT3A and TopoIIB (loading control). The western blot shows a time and dose-dependent decrease in DNMT3A protein in the methylation-sensitive HCT 116 cell line (Figure 2A). Densitometry for the depicted blot is also shown. In the HT-29 cells, expression of DNMT3a remained relatively constant at 48 and there is only a noticeable decrease in expression at the 150 μM concentration at 72 hours (Figure 2B).
Figure 2.
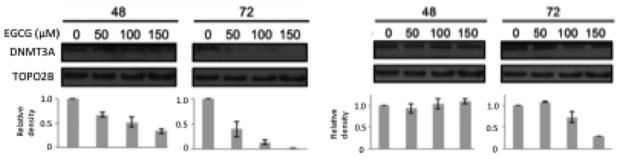
DNMT3A protein levels decrease in response to EGCG in HCT 116 cell line while expression in HT-29 cells remained constant at 48 hours. Response to EGCG treatment is both time and dose- dependent in HCT 116 cells. There is no change in DNMT3a protein in response to EGCG in HT-29 cells at 48 hours but a significant decrease at 100 and 150 μM at 72 hours. Error bars represent +/− one standard deviation.
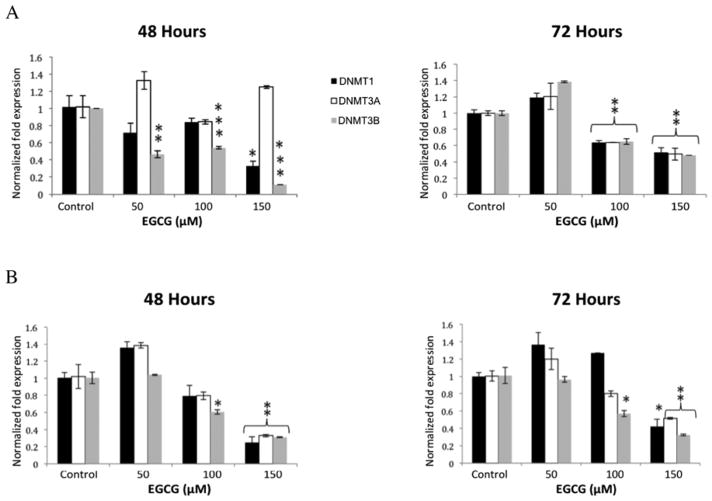
Effect of EGCG on HDAC mRNA. HDAC inhibitors can reduce expression of key regulatory genes in colon tumors (13)
To determine the effect EGCG has on HDACs, we treated HCT 116 and HT-29 cells with various concentrations of EGCG and looked at class I HDAC mRNA expression. In the HCT 116 cells, HDAC3 expression at 48 hours decreased at all concentrations of EGCG, while there was no decrease in HDAC1 expression, and only a significant decrease in HDAC2 expression at 150 μM EGCG (Figure 3A). At 72 hours, in HCT 116 cells, HDAC 1, -2, and -3 were significantly reduced at both 100 and 150 μM EGCG. Only at 150 μM EGCG were all three HDACs reduced at both 48 and 72 hours in the HT-29 cells, while HDAC3 was also reduced at 100 μM (Figure 3B).
Figure 3.
Histone deacetylase 1, -2, and -3 transcript levels decreased in HCT 116 and HT-29 cell lines when treated with EGCG. Significant results are seen at the higher concentration of 150 μM. Error bars represent +/− one standard deviation (n=3). *p<0.05, **p<0.02, ***p<0.01 compared with the control.
Effects of EGCG on HDAC protein expression
To determine if EGCG elicits the same effect on protein expression as seen on transcript levels, nuclear protein from EGCG treated cells was extracted at 48 and 72 hours. Immunoblots were probed with antibodies to HDAC1, HDAC2, and HDAC3. HCT 116 cells exhibited significant decreases in expression of HDAC2 and HDAC3, both time- and dose-dependently (Figure 4). Expression of HDAC1 remained relatively constant. There was no pattern of loss of expression of HDACs in the HT-29 cell line, but HDAC3 did show a decrease at 72 hours at 150 μM EGCG (Figure 4). Densitometry for each western blot is provided.
Figure 4.
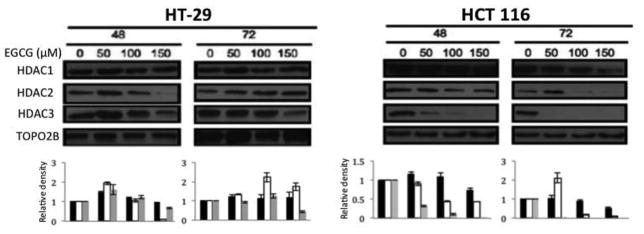
Protein expression of histone deacetylase 2 and -3 significantly decreased in a time- and dose- dependent manner in HCT 116 cells in response to EGCG treatment, while HDAC1 expression remained relatively constant except at the highest EGCG dosage and after 72 hours. In HT-29 cells, protein expression for all targets remained relatively constant.
Effects of EGCG on DNMT3A and HDAC3 degradation
Using the targets DNMT3A and HDAC3 and drug concentration (100 μM) that elicit the most consistent and dramatic result, we determined if the difference between message and protein levels was due to the effects of EGCG on protein degradation. Using the translation inhibitor cycloheximide to inhibit protein synthesis, we tested the effects of 100 μM EGCG on these targets over time. HCT 116 and HT-29 cells were pre treated for 24 hours with 100 μM EGCG of DMSO. Following pretreatment, cycloheximide was then added to cells to a final concentration of 10 μg/ml. Nuclear protein was extracted after 6, 12, 24 and 48 hours. Western blots were then probed with antibodies to DNMT3A and HDAC3. HCT 116 cells treated with EGCG showed a marked difference in expression of DNMT3A and HDAC3 when compared to cells treated with DMSO only (Figure 5aA. Expression of cyclin D1 was used to confirm inhibition of protein synthesis (data not shown). To correct for variance in the loading control and normalize each sample against expression of its control, ImageJ was used to standardize the data and the resulting graphic representation of the western blot data is shown. Expression of DNMT3A in HCT 116 cells treated with DMSO was increased while cells treated with 100 μM EGCG lended towards a decrease in expression. The same pattern was shown in the HCT 116 cells when probed for expression of HDAC3. In contrast, the expression trends of DNMT3A and HDAC3 in the HT-29 cells were similar when treated with DMSO and EGCG (Figure 5B).
Figure 5.
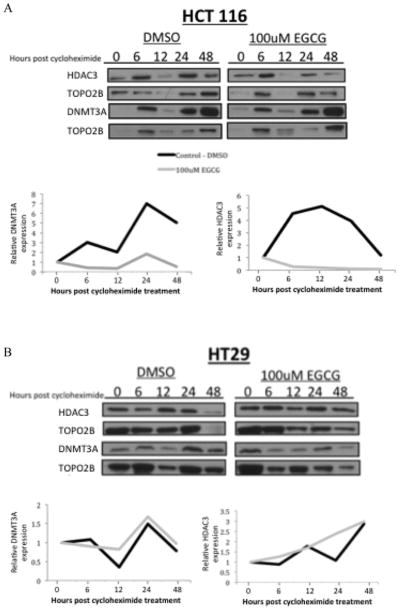
Treatment with cycloheximide to inhibit protein synthesis lends decreased expression of histone deacetylase 3 and DNA methyltransferase 3A in extracts from HCT 116 cells co-treated with 100 μM EGCG in (5A). HT-29 cells co-treated with EGCG and cycloheximide showed no difference in protein expression (5B).
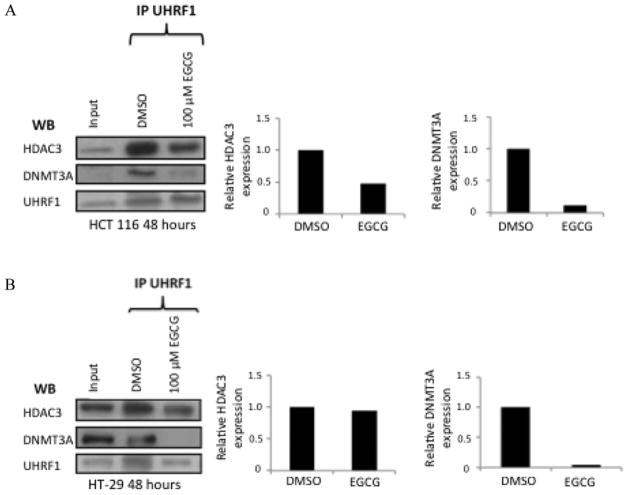
Effects of EGCG on association between UHRF1, DNMT3A and HDAC3
Still focusing on 100 μM EGCG, we decided to look for changes in association between the ubiquitin ligase, UHRF1, and DNMT3A as well as its association with HDAC3. We incubated the nuclear lysates of HCT 116 and HT-29 cells with primary antibody against UHRF1 following treatment of cells with DMSO or 100uM EGCG for 48 hours, and used protein G sepharose beads to capture the antibody complex. The beads were heated to 95 degrees C in loading dye to separate the antibody complex from the beads. Input (cell lysate containing no UHRF1 antibody) was run alongside the pulldown samples. The western blot was probed for DNMT3A, HDAC3A and UHRF1 (control). The results show a decreased association between UHRF1 and DNMT3 in cells treated with EGCG for both HCT 116 and HT-29 cell lines (Figure 6). The results also show a decreased association between UHRF1 and HDAC3 in only the HCT 116 cell line (Figure 6a).
Figure 6.
Immunoprecipitation of UHRF1 reveals an association of both histone deacetylase 3 and DNA methyltransferase 3A. HCT 116 cells treated with 100 μM EGCG exhibitied a decrease in association between UHRF1 and both HDAC3 and DNMT3a (6A). In HT-29 cells there is only a decerased association between UHRF1 and DMT3a in response to EGCG (6B).
Discussion
There are various factors that contribute to cancer development. Epigenetic events, including DNA methylation and histone modifications, that help regulate gene expression and cellular function are now conjectured to play a significant role in cancer development and present possible targets for cancer prevention (5, 14, 15). Aberrant DNA methylation is a hallmark of cancer and leads to silencing of tumor suppressor genes in many different types of cancer and in animal models thereof. Green tea intake is linked to a lower relative risk for a wide range of human cancer types, including colon cancer (29). Chemopreventive studies performed in rodent models of colorectal cancer indicate that green tea inhibited, to some degree, the cancer initiation process (10, 12). The most active green tea polyphenol, EGCG, has been shown to be involved in epigenetic modifications to actually slow the progression of various types of cancers.
One of the most intriguing roles for EGCG is its ability to reverse hypermethylation and reactivate silenced genes. It is thought that EGCG has some role in inhibiting class 1 HDACs and DNMTs (6, 22). It still is not definitively known how the gene silencing actions of HDACs and DNMTs are inhibited by EGCG. Computer modeling shows that there could be direct binding between EGCG and DNMTs (16). There is evidence that supports the idea that EGCG contributes to the degradation of key regulatory proteins such as cyclin D1, leading to increased expression of p21 in colon cancer (32). Here we specifically assayed the effects EGCG has on the message and protein levels of a specific DNA methyltransferases as well has class I HDACs. Class I HDACs are of a particular interest because they are commonly overexpressed in colon cancer (28).
We used the human colon cancer cell lines HCT 116 and HT-29 as an in vitro model for the methylation-sensitive and methylation-insensitive colon cancers respectively. First we showed that the methylation insensitive cell line showed a more robust response to EGCG treatment when looking at DNMT transcript levels using RT-PCR. These results were consistent across the 48 and 72 hour timepoints. In the HCT 116 cells, expression of the mRNA transcripts were only decreased at the highest concentration at 72 hours. This suggests that EGCG non discriminately inhibits transcription of DNMTs. We then examined the effect of EGCG on the expression of DNMT3A. Increased DNMT3A activity promoted tumorigenesis in a mouse model of colon cancer (19). We wanted to determine if the decrease found in transcript of DNMT3A translated to a decrease in protein levels. The data reveal that although EGCG reduced mRNA levels of DNMT3A in both HCT 116 and HT-29 cell lines, this change only translated to a decrease in protein levels for the methylation-sensitive, HCT 116 cell line. We then questioned whether EGCG plays a role in degrading DNMT3a in the HCT 116 cells
We also wanted to determine if a similar pattern of expression of HDACs occurred in response to EGCG treatment. DNMTs recruit HDACs to assist with gene silencing (2, 9). We found a decrease of all HDAC transcripts in the HT-29 cell line at the highest concentration of EGCG by 48 hours, however, there was a significant decrease in HDAC3 in the HCT 116 cell line at 50 μM at 48 hours. Thus, we also see that EGCG indiscriminately works to reduce HDAC transcription. We again looked to see if the results would be the same for the protein as it was for the mRNA. The western blots show relatively stable expression of HDACs in the HT-29 cells but a decrease in HDAC2 and -3 in the HCT 116 cells. This result also suggests that EGCG plays some role in protein degradation or stability. It is our thought that if the message remains constant but the protein decreases, degradation occurs. The opposite effect, where mRNA decreases and the remaining protein remains, it is somehow stabilized.
To determine if, in fact, EGCG contributed to the degradation of the proteins in the cells sensitive to methylation, we used the classic protein synthesis inhibitor, cycloheximide in a time course to monitor expression of our proteins of interest. At 100 μM the data demonstrate a difference between the HCT 116 and HT-29 cell lines in stability of the HDAC3 and DNMT3A proteins. There was no difference in expression of either target in HT-29 cells between the EGCG treatment and DMSO control group. On the other hand, the HCT 116 cell line showed a different response. Expression of these proteins of interest generally decreased over time when cells were treated with 100 μM EGCG. This suggests that EGCG plays a role, particularly in methylation-sensitive cancer, in degrading DNMT3A and HDAC3 in order to modulate the epigenetic profile. We looked, briefly, at a mechanism involving the E3 ubiquitin ligase, UHFR1, to see if EGCG had any effects on the association of this protein with the epigenetic regulators. UHRF1 facilitates colorectal cancer progression by recruiting DNMTs and aiding in gene silencing (27). DNMT3A directly interacts with UHRF1 (31). We found that EGCG reduces the association between the proteins possibly aiding in HDAC3 and DNMT3A degradation.
In summary, we report, as far as we know, for the first time that EGCG reduces expression of DNMT3A and HDAC3 in methylation-sensitive HCT 116 cells by targeting these proteins for degradation. We also report a difference in the way these two separate types of colon cancer respond to the same treatment. This finding has some implications for diseases such as human non polyposis colorectal cancer (HNPCC), where the hallmark of this subset of colon cancers is aberrant methylation of the human mismatch repair gene hMLH1 (17). Considerable controversy exists over whether the consistent findings for effects of EGCG in concentrations tested in vitro, where considerable salutary effects on cancer are observed, will translate to the same effects in vivo. This is a topic of continuing research in the field, and applies to other natural products. There are, however, numerous studies in other models that suggest the optimum concentration for use of EGCG in therapeutic application is greater than 50 μM (26, 30, 33). Avenues of enhancing EGCG biodistribution may allow future applicability in humans. Our findings are also of importance in the search for alternative cancer therapies that can reduce or eliminate toxicity usually associated with common pharmacological inhibitors of DNMTs and HDACs in treatment of colon cancer. Currently in our laboratory, we are assessing the efficacy of combining EGCG with major pharmacological inhibitors to determine the combined effects of inhibiting DNMT and HDAC expression on colon cancer cell growth.
Acknowledgments
Funding
This work is supported by National Institute of Health grants R01CA96694 and P30CA138313.
This work benefitted from the input of Dr. Jessica Bohonowych and Dr. Robin Muise-Helmericks.
Footnotes
Conflict of Interest Statement
None Declared.
References
- 1.Ahmad N, Cheng P, Mukhtar H. Cell cycle dysregulation by green tea polyphenol epigallocatechin-3-gallate. Biochem Biophys Res Commun. 2000;275:328–334. doi: 10.1006/bbrc.2000.3297. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 3.Blum W, Klisovic RB, Hackanson B, Liu Z, Liu S, Devine H, Vukosavljevic T, Huynh L, Lozanski G, Kefauver C, Plass C, Devine SM, Heerema NA, Murgo A, Chan KK, Grever MR, Byrd JC, Marcucci G. Phase I study of decitabine alone or in combination with valproic acid in acute myeloid leukemia. J Clin Oncol. 2007;25:3884–3891. doi: 10.1200/JCO.2006.09.4169. [DOI] [PubMed] [Google Scholar]
- 4.Borutinskaite VV, Magnusson KE, Navakauskiene R. Histone deacetylase inhibitor BML-210 induces growth inhibition and apoptosis and regulates HDAC and DAPC complex expression levels in cervical cancer cells. Molecular biology reports. 2012 doi: 10.1007/s11033-012-1892-5. [DOI] [PubMed] [Google Scholar]
- 5.Counts JL, Goodman JI. Alterations in DNA methylation may play a variety of roles in carcinogenesis. Cell. 1995;83:13–15. doi: 10.1016/0092-8674(95)90228-7. [DOI] [PubMed] [Google Scholar]
- 6.Fang M, Chen D, Yang CS. Dietary polyphenols may affect DNA methylation. The Journal of nutrition. 2007;137:223S–228S. doi: 10.1093/jn/137.1.223S. [DOI] [PubMed] [Google Scholar]
- 7.Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, Welsh W, Yang CS. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer research. 2003;63:7563–7570. [PubMed] [Google Scholar]
- 8.Gao Z, Xu Z, Hung MS, Lin YC, Wang T, Gong M, Zhi X, Jablon DM, You L. Promoter demethylation of WIF-1 by epigallocatechin-3-gallate in lung cancer cells. Anticancer research. 2009;29:2025–2030. [PubMed] [Google Scholar]
- 9.Gilbert ER, Liu D. Flavonoids Influence Epigenetic-Modifying Enzyme Activity: Structure – Function Relationships and the Therapeutic Potential for Cancer. Curr Med Chem. 2010 doi: 10.2174/092986710791111161. [DOI] [PubMed] [Google Scholar]
- 10.Issa AY, Volate SR, Muga SJ, Nitcheva D, Smith T, Wargovich MJ. Green tea selectively targets initial stages of intestinal carcinogenesis in the AOM-ApcMin mouse model. Carcinogenesis. 2007;28:1978–1984. doi: 10.1093/carcin/bgm161. [DOI] [PubMed] [Google Scholar]
- 11.Issa AY, Volate SR, Muga SJ, Nitcheva D, Smith T, Wargovich MJ. Green tea selectively targets initial stages of intestinal carcinogenesis in the AOM-ApcMin mouse model. Carcinogenesis. 2007;28:1978–1984. doi: 10.1093/carcin/bgm161. [DOI] [PubMed] [Google Scholar]
- 12.Jia XD, Han C. Chemoprevention of tea on colorectal cancer induced by dimethylhydrazine in Wistar rats. World Journal of Gastroenterology. 2000;6:699–703. doi: 10.3748/wjg.v6.i5.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin JS, Tsao TY, Sun PC, Yu CP, Tzao C. SAHA inhibits the growth of colon tumors by decreasing histone deacetylase and the expression of cyclin D1 and survivin. Pathology oncology research: POR. 2012;18:713–720. doi: 10.1007/s12253-012-9499-7. [DOI] [PubMed] [Google Scholar]
- 14.Jones PA. DNA methylation and cancer. Oncogene. 2002;21:5358–5360. doi: 10.1038/sj.onc.1205597. [DOI] [PubMed] [Google Scholar]
- 15.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature reviews Genetics. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 16.Lee WJ, Shim JY, Zhu BT. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Molecular pharmacology. 2005;68:1018–1030. doi: 10.1124/mol.104.008367. [DOI] [PubMed] [Google Scholar]
- 17.Lynch HT, Smyrk T. Hereditary nonpolyposis colorectal cancer (Lynch syndrome). An updated review. Cancer. 1996;78:1149–1167. doi: 10.1002/(SICI)1097-0142(19960915)78:6<1149::AID-CNCR1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 18.Nandakumar V, Vaid M, Katiyar SK. (−)-Epigallocatechin-3-gallate reactivates silenced tumor suppressor genes, Cip1/p21 and p16INK4a, by reducing DNA methylation and increasing histones acetylation in human skin cancer cells. Carcinogenesis. 2011;32:537–544. doi: 10.1093/carcin/bgq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samuel MS, Suzuki H, Buchert M, Putoczki TL, Tebbutt NC, Lundgren-May T, Christou A, Inglese M, Toyota M, Heath JK, Ward RL, Waring PM, Ernst M. Elevated Dnmt3a activity promotes polyposis in Apc(Min) mice by relaxing extracellular restraints on Wnt signaling. Gastroenterology. 2009;137:902–913. 913 e901–911. doi: 10.1053/j.gastro.2009.05.042. [DOI] [PubMed] [Google Scholar]
- 20.Shimizu M, Deguchi A, Joe AK, McKoy JF, Moriwaki H, Weinstein IB. EGCG inhibits activation of HER3 and expression of cyclooxygenase-2 in human colon cancer cells. Journal of experimental therapeutics & oncology. 2005;5:69–78. [PubMed] [Google Scholar]
- 21.Shimizu M, Shirakami Y, Sakai H, Yasuda Y, Kubota M, Adachi S, Tsurumi H, Hara Y, Moriwaki H. (−)-Epigallocatechin gallate inhibits growth and activation of the VEGF/VEGFR axis in human colorectal cancer cells. Chemico-biological interactions. 2010;185:247–252. doi: 10.1016/j.cbi.2010.03.036. [DOI] [PubMed] [Google Scholar]
- 22.Thakur VS, Gupta K, Gupta S. Green tea polyphenols increase p53 transcriptional activity and acetylation by suppressing class I histone deacetylases. International journal of oncology. 2012;41:353–361. doi: 10.3892/ijo.2012.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thangapazham RL, Singh AK, Sharma A, Warren J, Gaddipati JP, Maheshwari RK. Green tea polyphenols and its constituent epigallocatechin gallate inhibits proliferation of human breast cancer cells in vitro and in vivo. Cancer Lett. 2007;245:232–241. doi: 10.1016/j.canlet.2006.01.027. [DOI] [PubMed] [Google Scholar]
- 24.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volate SR, Muga SJ, Issa AY, Nitcheva D, Smith T, Wargovich MJ. Epigenetic modulation of the retinoid X receptor alpha by green tea in the azoxymethane-Apc Min/+ mouse model of intestinal cancer. Molecular carcinogenesis. 2009;48:920–933. doi: 10.1002/mc.20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vu HA, Beppu Y, Chi HT, Sasaki K, Yamamoto H, Xinh PT, Tanii T, Hara Y, Watanabe T, Sato Y, Ohdomari I. Green tea epigallocatechin gallate exhibits anticancer effect in human pancreatic carcinoma cells via the inhibition of both focal adhesion kinase and insulin-like growth factor-I receptor. Journal of biomedicine & biotechnology. 2010;2010:290516. doi: 10.1155/2010/290516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang F, Yang YZ, Shi CZ, Zhang P, Moyer MP, Zhang HZ, Zou Y, Qin HL. UHRF1 promotes cell growth and metastasis through repression of p16(ink(4)a) in colorectal cancer. Ann Surg Oncol. 2012;19:2753–2762. doi: 10.1245/s10434-011-2194-1. [DOI] [PubMed] [Google Scholar]
- 28.Wilson AJ, Byun DS, Popova N, Murray LB, L’Italien K, Sowa Y, Arango D, Velcich A, Augenlicht LH, Mariadason JM. Histone deacetylase 3 (HDAC3) and other class I HDACs regulate colon cell maturation and p21 expression and are deregulated in human colon cancer. J Biol Chem. 2006;281:13548–13558. doi: 10.1074/jbc.M510023200. [DOI] [PubMed] [Google Scholar]
- 29.Yang G, Shu XO, Li H, Chow WH, Ji BT, Zhang X, Gao YT, Zheng W. Prospective cohort study of green tea consumption and colorectal cancer risk in women. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2007;16:1219–1223. doi: 10.1158/1055-9965.EPI-07-0097. [DOI] [PubMed] [Google Scholar]
- 30.Zhang D, Al-Hendy M, Richard-Davis G, Montgomery-Rice V, Rajaratnam V, Al-Hendy A. Antiproliferative and proapoptotic effects of epigallocatechin gallate on human leiomyoma cells. Fertility and sterility. 2010;94:1887–1893. doi: 10.1016/j.fertnstert.2009.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J, Gao Q, Li P, Liu X, Jia Y, Wu W, Li J, Dong S, Koseki H, Wong J. S phase-dependent interaction with DNMT1 dictates the role of UHRF1 but not UHRF2 in DNA methylation maintenance. Cell Res. 2011;21:1723–1739. doi: 10.1038/cr.2011.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Min KW, Wimalasena J, Baek SJ. Cyclin D1 degradation and p21 induction contribute to growth inhibition of colorectal cancer cells induced by epigallocatechin-3-gallate. Journal of cancer research and clinical oncology. 2012;138:2051–2060. doi: 10.1007/s00432-012-1276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou L, Elias RJ. Antioxidant and pro-oxidant activity of (−)-epigallocatechin-3-gallate in food emulsions: Influence of pH and phenolic concentration. Food chemistry. 2013;138:1503–1509. doi: 10.1016/j.foodchem.2012.09.132. [DOI] [PubMed] [Google Scholar]