Summary
The transcriptional co-activator peroxisome proliferator-activated receptor-gamma co-activator-1 α (PGC-1α) regulates metabolic genes in skeletal muscle, and contributes substantially to the response of muscle to exercise. Muscle specific PGC-1α transgenic expression and exercise both increase the expression of thermogenic genes within white adipose. How the PGC-1α mediated response to exercise in muscle conveys signals to other tissues remains incompletely defined. We employed a metabolic profiling approach to examine metabolites secreted from myocytes with forced expression of PGC-1α, and identified β-aminoisobutyric acid (BAIBA) as a novel small molecule myokine. BAIBA increases the expression of brown adipocyte-specific genes in white adipose tissue and fatty acid β-oxidation in hepatocytes both in vitro and in vivo through a PPARα mediated mechanism, induces a brown adipose-like phenotype in human pluripotent stem cells, and improves glucose homeostasis in mice. In humans, plasma BAIBA concentrations are increased with exercise and inversely associated with metabolic risk factors. BAIBA may thus contribute to exercise-induced protection from metabolic diseases.
Introduction
Exercise is an effective intervention for both the prevention and treatment of obesity and type 2 diabetes (Knowler et al., 2002). Recent studies suggest that skeletal muscle integrates many of the signals contributing to the salutary effects of exercise (Bassel-Duby and Olson, 2006). The transcriptional co-activator peroxisome proliferator-activated receptor-gamma co-activator-1 α (PGC-1α) controls an extensive set of metabolic programs within skeletal muscle and in part regulates the adaptive response of muscle to exercise (Handschin and Spiegelman, 2006; Olesen et al., 2010). PGC-1α regulates these metabolic programs by binding to nuclear receptors and other transcription factors to form active transcriptional complexes (Puigserver et al., 1999; Puigserver et al., 1998). Exercise enhances the expression of PGC-1α, which results in increased mitochondrial biogenesis and fatty acid β-oxidation, greater glucose transport, and an induction of muscular fiber type switching towards a more oxidative phenotype (Lin et al., 2002b; Michael et al., 2001; Wu et al., 1999).
Transgenic mice with muscle specific PGC-1α expression show an enhanced ability to perform endurance exercise and have an increased peak oxygen uptake (Calvo et al., 2008). These transgenic mice also demonstrate increased expression of brown adipocyte-specific genes within white adipose tissue (WAT) and an increased adipose respiratory phenotype (Bostrom et al., 2012), suggesting that skeletal muscle signals to other tissues to alter their function. In addition, exercise increases mitochondrial number and brown adipocyte-specific gene expression in white adipose depots and ameliorates glucose intolerance induced by a high fat diet (Sutherland et al., 2009; Xu et al., 2011). Exercise also enhances the brown proliferative adipocyte progenitor cell population and brown fat adipogenesis (Xu et al., 2011). Cells expressing brown-adipocyte specific genes have been reported as interspersed within the WAT of rodents and humans (so-called beige or brite cells(Ishibashi and Seale, 2010; Petrovic et al., 2010; Seale et al., 2008)) and demonstrate anti-diabetic and anti-obesity effects in rodent models(Cousin et al., 1992; Kopecky et al., 1995; Lowell et al., 1993; Melnyk et al., 1997; Oberkofler et al., 1997; Seale et al., 2007). Uncoupling protein-1 (UCP-1) and Cell death-inducing DFFA-like effector a (CIDEA) are among the brown adipocyte-specific genes increased in expression in WAT by exercise and by muscle specific PGC-1α expression (Cao et al., 2011). UCP-1 uncouples the mitochondrial electron transport chain from ATP synthesis, an activity that is key to the thermogenic role of brown adipose tissue (BAT) (Enerback et al., 1997). Likewise, CIDEA is a mitochondrial brown adipocyte-specific gene with a role in the regulation of the thermogenic process.
Gene expression arrays and a bioinformatics approach recently highlighted irisin as a novel secreted protein by which PGC-1α dependent signals from muscle drive functional changes in other tissues(Bostrom et al., 2012). There is strong motivation to investigate whether additional mechanisms triggered by PGC-1α expression in muscle might confer hormone-like signals to modulate fat metabolism or contribute to the benefits of exercise, especially with regard to small organic molecules. Here we applied a liquid chromatography-mass spectrometry (LC-MS) metabolic profiling technique to identify small molecules secreted from myocytes with forced expression of PGC-1α. We then tested the effects of candidate small molecules on WAT in vitro and in vivo, and examined metabolites in the context of cardiometabolic risk factors and exercise in humans.
Results
β-aminoisobutyric acid is regulated by PGC-1α and increases expression of brown adipocyte-specific genes
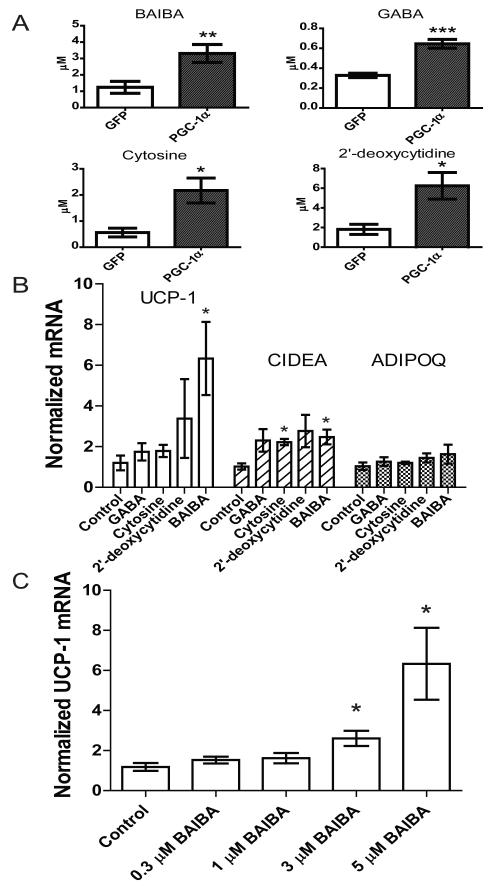
Serum free media taken from muscle cells with forced expression of PGC1-α increases mRNA levels of several brown adipocyte-specific genes when transferred to primary adipocytes(Bostrom et al., 2012). To identify candidate small molecules that might be contributing to this phenomenon, we applied LCMS metabolic profiling to this media and compared the findings to media from GFP expressing control cells. As expected, glucose levels were significantly decreased in the supernatants of the PGC-1α overexpressing cells (−15.3% P = 0.025) (Michael et al., 2001). Four metabolites, β-aminoisobutyric acid (BAIBA), γ-aminobutyric acid (GABA), cytosine, and 2′-deoxycytidine were significantly enriched in the media of the PGC-1α overexpressing myocytes (BAIBA, 2.7-fold increase, P = 0.01; GABA, 1.9-fold increase, P = 0.004; cytosine, 3.9-fold increase, P = 0.02; 2′-deoxycytidine, 3.4-fold increase, P = 0.02) (Figure 1A).
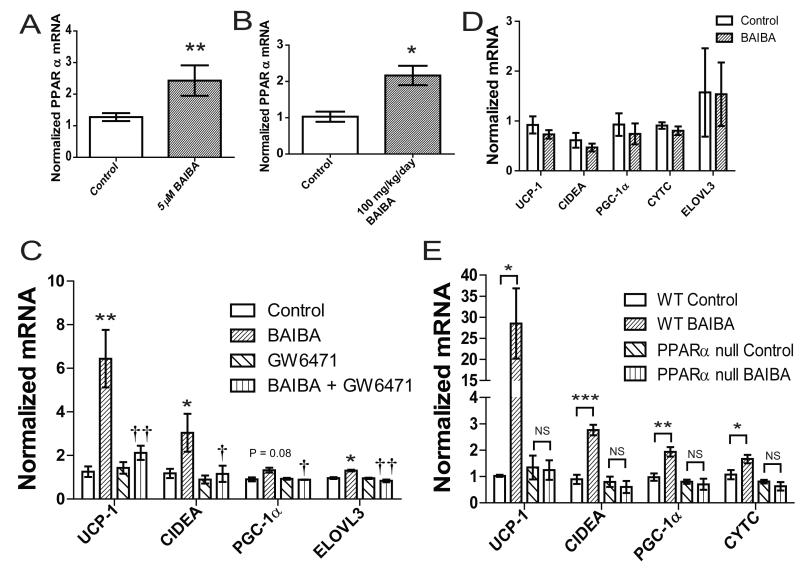
Figure 1. Metabolites accumulate in the media of myocytes as a result of forced PGC-1α expression and stimulate expression of brown adipocyte-specific genes in adipocytes.
A) Myocytes were transduced with an adenoviral vector expressing either PGC-1α (n=6) or GFP (n=6). After 24 hours of exposure to these cells, media was analyzed using an LC-MS based metabolite profiling method measuring 100 small molecules (see Methods). B) BAIBA (5 μM) induces expression of brown adipocyte-specific genes in primary adipocytes differentiated from the stromal vascular fraction isolated from inguinal WAT over 6 days. Additional metabolites tested at physiologically relevant doses included GABA (3 μM), cytosine (1 μM), and 2-deoxycytidine (15 μM). While BAIBA significantly increased the expression of the brown adipocyte-specific genes UCP-1 and CIDEA, it did not alter the expression of the white adipocyte gene adiponectin (ADIPOQ). Cumulative data from a total of 5 independent observations are shown. C) BAIBA concentrations in the low micromolar range significantly and dose-dependently increased the expression of the brown adipocyte-specific gene UCP-1. *, P ≤ 0.05, **, P ≤ 0.01, ***, P ≤ 0.001. Data are represented as Mean ± SEM.
We assessed the ability of these candidate molecules to increase the expression of brown adipocyte-specific genes using the primary stromal vascular fraction isolated from subcutaneous (inguinal) WAT of mice during 6 days of the differentiation process to mature adipocytes. BAIBA treatment enhanced UCP-1 and CIDEA mRNA by 5.3-fold and 2.25-fold, respectively, as assessed by quantitative PCR (Figure 1B). GABA, cytosine and 2′-deoxycytidine treatment did not induce concordant upregulation of brown adipocyte-specific genes, so we focused subsequent analyses on BAIBA. BAIBA concentrations in the low micromolar range significantly increased UCP-1 in the primary adipocytes in a dose-dependent manner (Figure 1C). By contrast, BAIBA did not significantly alter the expression of the canonical white adipocyte gene adiponectin (ADIPOQ), which is also expressed to a similar extent in the brite cell population (Wu et al., 2012). To strengthen the link between BAIBA secretion and PGC1-α expression we analyzed the concentration of BAIBA in the media of muscle cells in response to increased expression of PGC1-α using an adenoviral vector. Increased levels of PGC-1α lead to an increase in the concentration of BAIBA in the media (Figure S1).
Exposure of human induced pluripotent stem cells to BAIBA during differentiation to mature white adipocytes induces a brown adipocyte-like phenotype
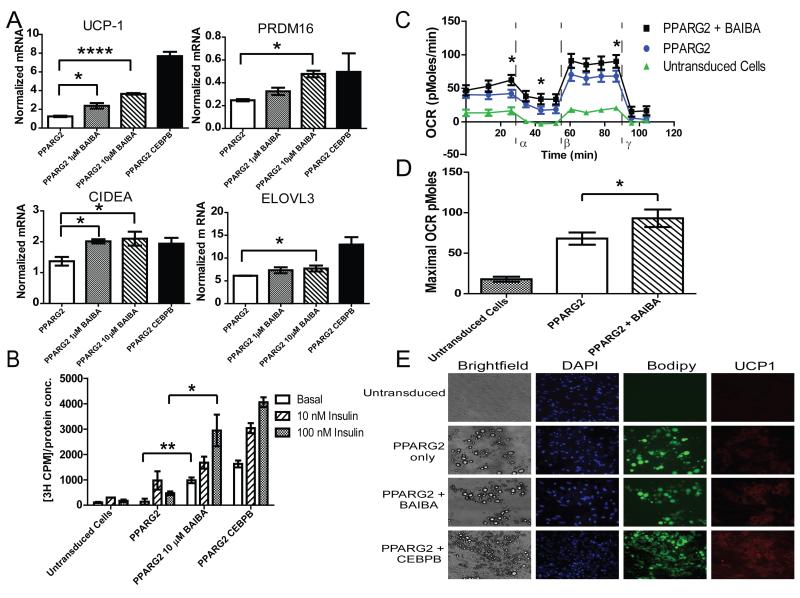
Since BAIBA induced expression of brown adipocyte specific genes in primary adipocytes differentiated from the stromal vascular fraction, we investigated whether BAIBA would induce a browning response in human pluripotent stem cells during their differentiation to mature white adipocytes (Figure S2). BJ fibroblasts reprogrammed with modified RNA (BJ RiPS) human induced pluripotent stem cells (IPSCs) were differentiated into mesenchymal progenitor cells as previously reported (Ahfeldt et al., 2012; Warren et al., 2010). Lentiviral mediated expression of PPARG2 or both PPARG2 and CCAAT/enhancer-binding protein β (CEBPB) in these human pluripotent stem cell-derived mesenchymal progenitor cells was used to program their differentiation into either white or brown mature adipocytes, respectively (Enerback et al., 1997; Kajimura et al., 2009; Tontonoz et al., 1994; Wright et al., 2000). Administration of BAIBA to IPSCs differentiated into mature white adipocytes conferred a dose-dependent increase in the expression of brown-adipocyte specific genes, including UCP-1, CIDEA, and PRDM16. Expression of ELOVL3, a critical enzyme for lipid accumulation and metabolic activity in brown adipocytes, was also slightly increased (Figure 2A). BAIBA did not significantly alter the expression of the canonical white adipocyte gene adiponectin (ADIPOQ). (Figure S3A). The effect of BAIBA on the expression of brown adipocyte-specific genes was reproduced in white adipocytes derived from other human pluripotent cell lines (Figure S3B). By contrast, mesenchymal progenitor cells differentiated into mature brown adipocytes in the presence of BAIBA did not exhibit an increase in the classical browning response genes, suggesting BAIBA does not initiate the thermogenic response in BAT in vitro (Figure S3C). One prior publication found that BAIBA does not affect UCP-1 expression in intrascapular BAT in vivo (Begriche et al., 2008), which might relate to known differences between the development of beige/brite cells and activation of classical brown fat (Frontini and Cinti, 2010; Wu et al., 2012).
Figure 2. BAIBA treatment of BJ RiPS human iPSCs induces brown adipocyte-specific gene expression and function.
A) BAIBA significantly and dose dependently increased the expression of brown-adipocyte specific genes in human induced pluripotent stem cell (IPSC) derived mature adipocytes. B) Glucose uptake in human IPSC derived adipocytes was assessed by the transport of [3H]-2-deoxy-D-glucose at basal level and during insulin stimulation (10 nM and 100 nM). BAIBA increased both basal and insulin stimulated glucose uptake (data from 3 independent observations are shown). C) Comparison of the oxygen consumption rate (OCR) of human IPSC-derived white adipocytes with and without BAIBA treatment; Untransduced cells differentiated with adipogenic media (green line), PPARG2 programmed cells (blue line), and PPARG2 programmed cells treated with BAIBA (black line). The OCR was measured over time with the addition of oligomycin (α), an ATPase inhibitor, carbonyl cyanide m-chlorophenyl hydrazone (CCCP), a proton gradient uncoupler (β) allowing determination of the maximum OCR, and antimycin (γ). D) BAIBA significantly increased the maximal OCR of PPARG2 transduced adipocytes. (Data from n = 10 independent observations are shown). E) Images of untransduced cells, PPARG2 programmed white adipocytes, PPARG2 programmed white adipocytes treated with BAIBA, and PPARG2-CEBPB programmed brown adipocytes. Shown from left to right: brightfield images illustrating the morphology of the cells; 4′,6-diamidino-2-phenylindole (DAPI) fluorescent nuclear staining (blue); fluorescent staining with the neutral lipid dye BODIPY (green); fluorescent images of immunostaining with antibodies against the marker protein UCP-1 (red) (100× magnification). *, P ≤ 0.05, **, P ≤ 0.01, ***, P ≤ 0.001,****, P < 0.0001. Data are represented as Mean ± SEM.
To establish whether the observed transcriptional changes conferred functional effects, we assessed the uptake of [3H]-2-deoxy-D-glucose in the PPARG2-programmed cells. We observed a striking increase in the basal and insulin-stimulated glucose uptake in the presence of BAIBA (Figure 2B). Furthermore, the basal oxygen consumption rate (OCR) was found to be higher in the programmed white adipocytes treated with BAIBA, as assessed by an extracellular flux analyzer. Oligomycin was then used to inhibit the ATP synthase. The OCR of white adipocytes treated with BAIBA remained higher than the untreated white adipocytes, consistent with increased uncoupling. The addition of the electron transport chain decoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP) allowed the measurement of the maximal respiratory capacity. Programmed white adipocytes treated with BAIBA showed significantly higher respiratory capacity compared to the untreated programmed white adipocytes (Figures 2C and 2D). In addition, brightfield images and BODIPY fluorescent staining demonstrate PPARG2-programmed cells contain the single large, well defined lipid droplet characteristic of mature white adipocytes; this morphology was maintained when the cells were treated with BAIBA (Figure 2E). Fluorescent immunostaining revealed a higher degree of UCP-1 staining in the PPARG2 programmed white adipocytes treated with BAIBA when compared to untreated cells. Together, these data indicate that BAIBA activates a browning gene program and increases the mitochondrial activity of human IPSCs differentiated into white adipocytes.
BAIBA induces increased expression of brown/beige adipocyte-specific genes in vivo
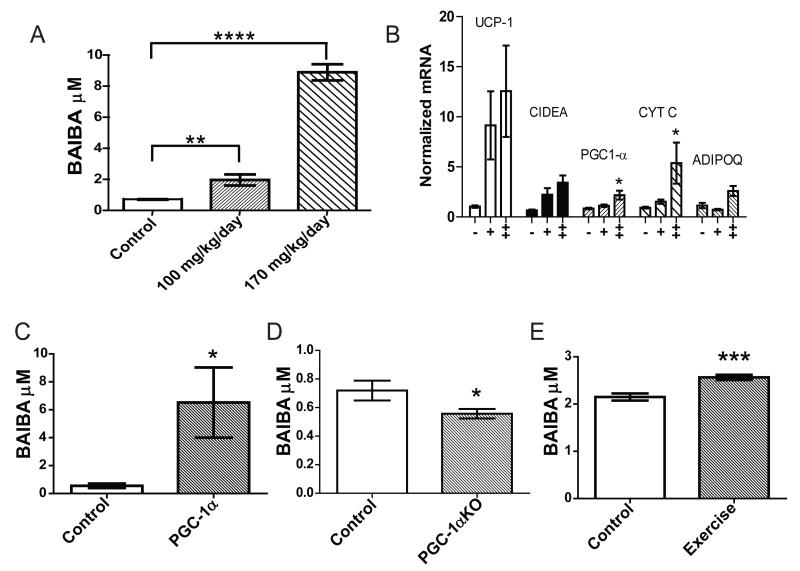
To examine whether BAIBA could dose-dependently induce the expression of brown adipocyte-specific genes in WAT in vivo, mice were treated with 100 mg/kg/day or 170 mg/kg/day of BAIBA in drinking water for 14 days based on preliminary dose escalation studies. BAIBA treatment led to a 2.7-fold (100 mg/kg/day) and 12.2-fold (170 mg/kg/day) increased plasma concentration of the metabolite by 14 days (100 mg/kg/day BAIBA; 2 ± 0.03 μM, P = 0.009, 170 mg/kg/day BAIBA; 8.9 ± 0.5 μM, P < 0.0001) (Figure 3A). Expression analysis of inguinal WAT using qPCR revealed significant increases in brown adipocyte-specific genes UCP-1 (100 mg/kg/day BAIBA; 8.8-fold increase, P = 0.03, 170mg/kg/day BAIBA; 12.1-fold increase, P = 0.02) and CIDEA (100 mg/kg/day BAIBA; 3.4-fold increase, P = 0.03, 170mg/kg/day BAIBA; 5.24-fold increase, P = 0.005), recapitulating the in vitro findings (Figure 3B). Expression of PGC-1α and Cytochrome C were also increased following BAIBA treatment (PGC-1α, 100 mg/kg/day BAIBA; 1.3-fold increase, P = 0.09, 170mg/kg/day BAIBA; 2.6-fold increase, P = 0.02; Cytochrome C, 100 mg/kg/day BAIBA; 1.64-fold increase, P = 0.03, 170mg/kg/day BAIBA; 5.8-fold increase, P = 0.04).
Figure 3. BAIBA induces expression of brown adipocyte-specific genes in WAT in vivo and muscle specific PGC-1α expression and exercise significantly increase plasma BAIBA levels.
A) The plasma concentration of BAIBA in mice given 100 mg/kg/day (n = 5) or 170 mg/kg/day (n = 5) of the metabolite in their drinking water significantly increased over 14 days as compared to age matched control mice (n = 5). B) Expression of brown adipocyte-specific genes in inguinal WAT from control mice (-) (n = 5), mice treated with 100 mg/kg/day BAIBA for 14 days (+) (n = 5), or mice treated with 170 mg/kg/day BAIBA for 14 days (++) (n=5). C) Plasma from muscle specific PGC-1α transgenic mice (n = 5) was analyzed using an LC-MS metabolite profiling platform and compared to plasma from age matched control mice (n = 5). D) Plasma from PGC-1α knockout mice (n = 9) was analyzed using LC-MS and compared to plasma from age matched control mice (n = 8) E) Mice were subjected to a 3 week free wheel running exercise regimen (n = 6) or housed as sedentary controls (n=6), and plasma BAIBA levels were assessed by LC-MS. *, P ≤ 0.05, **, P ≤ 0.01, ***, P ≤ 0.001, ****, P < 0.0001. Data are represented as Mean ± SEM.
BAIBA levels are increased in the plasma of muscle PGC-1α expressing and exercising mice
Since BAIBA was elevated in the media of cultured myocytes by forced expression of PGC-1α, we tested whether plasma concentrations of this metabolite were increased in mice with muscle-specific transgenic expression of PGC-1α (MCK-PGC1α) and with chronic exercise in wild-type animals. The MCK-PGC1α transgenic mouse has 10-fold increased expression of PGC-1α in gastrocnemius (Lin et al., 2002b; Viscomi et al., 2011; Wu et al., 2011). Plasma concentrations of BAIBA were significantly increased 11-fold to 6.5 ± 2.5 μM, P = 0.03 as a result of PGC-1α muscle forced expression in vivo (Figure 3C). By contrast, the absence of PGC-1α decreases the plasma concentration of BAIBA as compared to wild type controls (0.77-fold decrease, P = 0.046) (Figure 3D).
In exercise trained wild-type mice subjected to 3 weeks of free wheel running, UCP-1 expression in the subcutaneous inguinal white adipose tissue was significantly increased by 25-fold compared to sedentary controls (Bostrom et al., 2012). LC-MS analysis of metabolites extracted from the gastrocnemius and quadriceps of exercise trained mice demonstrated a 5.2 ± 0.09-fold, P < 0.0001 and 2.2 ± 0.5-fold, P < 0.0001 increase in BAIBA concentrations respectively. Analysis of plasma from the exercise trained mice confirmed a highly significant increase in the plasma concentration of BAIBA (19% increase to 2.6 ± 0.05 μM, P = 0.001) as compared to sedentary controls (Figure 3E).
BAIBA decreases weight gain and improves glucose tolerance in mice
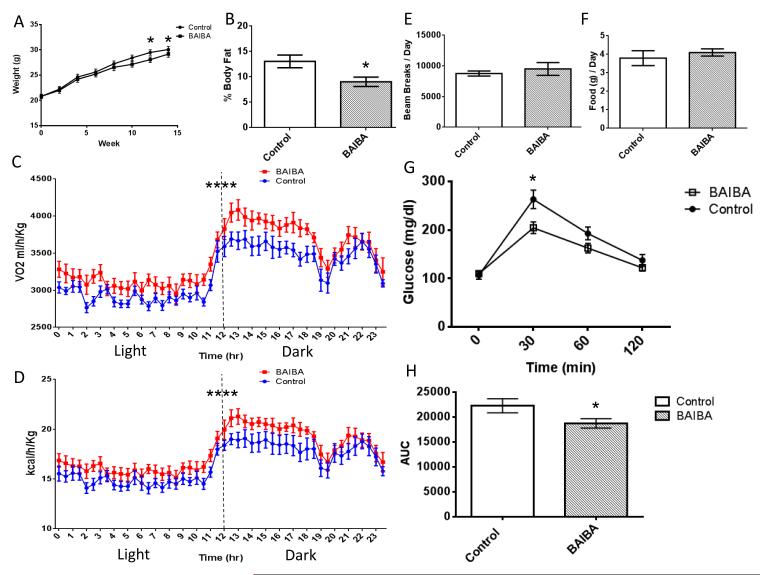
Since browning of WAT improves glucose homeostasis and reduces weight gain (Bostrom et al., 2012), we examined the functional effect of BAIBA on weight gain and glucose tolerance in vivo. Six week old mice were either treated with BAIBA (100 mg/kg/day) or remained untreated (control mice). Their weights were monitored weekly. Weight was slightly decreased in the mice by the end of BAIBA treatment (ANOVA, P = 0.01) (Figure 4A). Analysis of body composition using MRI demonstrated BAIBA treatment significantly decreased body fat in the mice (% body fat, control = 13.1 ± 1.25, BAIBA = 9 ± 0.92, P = 0.02) (Figure 4B). Consistent with the effects on thermogenic and β-oxidation gene expression and body weights, analysis with metabolic cages indicated that oxygen consumption (VO2) and whole body energy expenditure were increased in the BAIBA treated mice (VO2, Two-Way ANOVA, P ≤ 0.0001, energy expenditure, Two-Way ANOVA, P ≤ 0.0001) (Figure 4C and 4D) without any significant difference in activity (control = 8758 ± 417.5 beam breaks / day, BAIBA = 9504 ± 1043 beam breaks per day, P = 0.52) (Figure 4E) or food intake (control = 3.79 ± 0.4 g / day, BAIBA = 4.1 ± 0.2 g / day, P = 0.5) (Figure 4F). The mice were also challenged with an intraperitoneal glucose tolerance test (IPGTT) (Figure 4G). BAIBA was found to significantly improve the glucose tolerance in the mice as determined by the area under the curve of the IPGTT (−15.9%, P ≤ 0.05) (Figure 4H).
Figure 4. BAIBA decreases weight gain and improves glucose tolerance in mice.
A) The weights of mice given 100 mg/kg/day BAIBA (n = 8) in their drinking water compared to untreated controls (n = 8). B) The percentage body fat of 100 mg/kg/day BAIBA treated mice (n = 8) compared to untreated controls (n = 8). C) Diurnal oxygen consumption of control mice (n = 8) and BAIBA (100 mg/kg/day) treated mice (n = 8). D). Diurnal energy expenditure of control mice (n = 8) and BAIBA (100 mg/kg/day) treated mice (n = 8). E) Activity of control mice (n = 8) and mice treated with 100 mg/kg/day BAIBA .F) Food consumption of control mice and mice treated with 100 mg/kg/day BAIBA. G) Mice treated with 100 mg/kg/day BAIBA for 14 weeks showed significantly improved glucose tolerance as determined by an IPGTT. H) The area under the curve of an IPGTT comparing BAIBA treated mice to untreated controls (Control, n = 8, BAIBA n = 8). *, P < 0.05. Data are represented as Mean ± SEM.
PPARα mediates BAIBA-induced effects on adipose tissue in vitro and in vivo
We next examined how BAIBA may be driving the increase in thermogenic gene expression. In a focused interrogation of potential downstream mediators, we observed that BAIBA significantly increases the expression of PPARα in white adipocytes both in vitro (2.4-fold increase, Figure 5A) and in the inguinal white fat depot in vivo (2.2-fold increase, Figure 5B). PPARα is a key transcription factor known to stimulate the expression of UCP-1(Bostrom et al., 2012; Komatsu et al., 2010). We were interested to find that the selective PPARα antagonist GW6471 significantly abrogated the BAIBA-stimulated increase in thermogenic gene expression in primary adipocytes (Figure 5C). The functional interaction between the BAIBA and GW6471 treatments on thermogenic gene expression was confirmed using two-way ANOVA (P < 0.005). To further define the contribution of PPARα to the browning response of primary white adipocytes in vitro, we isolated the stromal vascular fraction from the subcutaneous WAT of PPARα null mice and differentiated the cells into mature adipocytes in the presence or absence of BAIBA. Analysis of the thermogenic gene expression in these cells using qPCR demonstrated a loss of the BAIBA-induced browning effect in the setting of PPARα deficiency (Figure 5D), consistent with the findings seen with the biochemical inhibitor.
Figure 5. PPARα functions downstream of BAIBA.
A) BAIBA (5 μM) induces expression of PPARα in primary adipocytes differentiated from the stromal vascular fraction isolated from inguinal WAT over 6 days. Cumulative data from a total of 6 independent observations are shown. B) Expression of PPARα in inguinal WAT from control mice (n = 5) and mice treated with 100 mg/kg/day BAIBA for 14 days (n = 5). C) Primary adipocytes differentiated from the stromal vascular fraction treated with BAIBA (5 μM) and/or GW6471 for 6 days. The graph shows qPCR of indicated genes. †< 0.05,†† < P0.01 compared to BAIBA treatment. D) BAIBA (5 μM) failed to induce expression of brown adipocyte-specific genes in primary adipocytes differentiated from the stromal vascular fraction isolated from inguinal WAT of PPARα null mice. Cumulative data from a total of 6 independent observations are shown. E) Expression of brown adipocyte-specific genes in inguinal WAT from wild type (WT) control mice (n = 5), WT mice treated with 100 mg/kg/day BAIBA for 14 days (n = 5), PPARα null control mice (n=5) and PPARα null mice treated with 100 mg/kg/day BAIBA for 14 days (n=5). *, P < 0.05, **, P ≤ 0.01 compared to control. Data are represented as Mean ± SEM.
The role of PPARα in the BAIBA-induced increase in thermogenic gene expression in white adipose tissue in vivo was also examined using PPARα null mice. PPARα null mice were treated with 100 mg/kg/day BAIBA in drinking water for 14 days. qPCR analysis of subcutaneous (inguinal) WAT demonstrated that BAIBA failed to increase expression of thermogenic genes, including UCP-1, CIDEA, PGC-1α and Cytochrome C, in the PPARα null mice (Figure 5E). Therefore, these results indicate that BAIBA increases expression of the browning gene program through a specific PPARα-dependent mechanism.
BAIBA increases hepatic β-oxidation through PPARα
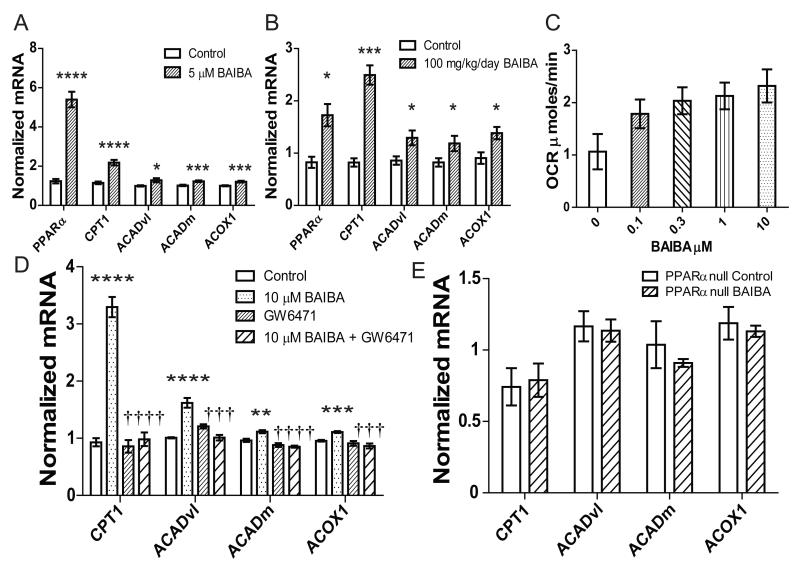
BAIBA may also function to induce additional tissue-specific salutary effects. Exercise has been shown to increase liver β-oxidation (Aoi et al., 2011; Oh et al., 2006; Rabol et al., 2011; Rector et al., 2011). Therefore we investigated whether BAIBA would directly induce β-oxidation gene expression in hepatocytes in vitro. Hepatocytes were incubated with 5μM BAIBA for 6 days. As in vivo BAIBA significantly increased the expression of PPARα (5.4-fold, P < 0.0001), carnitine palmitoyltransferase 1 (CPT1) (2.2-fold, P < 0.0001), the very-long-chain acyl-CoA dehydrogenase (ACADvl) (1.3-fold, P = 0.03), the medium-chain acyl-CoA dehydrogenase (ACADm) (1.2-fold, P = 0.005) and acyl-CoA oxidase 1 (ACOX1) (1.2-fold P = 0.004) (Figure 6A). The interaction between BAIBA and β-oxidation gene expression was also determined to be significant by two-way ANOVA (P < 0.0001).
Figure 6. BAIBA increases hepatic β-oxidation through PPARα.
A) BAIBA (5 μM) induces expression of fatty acid β-oxidation genes in hepatocytes treated for 6 days (Control = 6, BAIBA 5 μM = 6). B) BAIBA dose-dependently induces expression of hepatic fatty acid β-oxidation genes in vivo. Expression of fatty acid β-oxidation genes in the liver of control mice (n = 5) and mice treated with 100 mg/kg/day BAIBA for 14 days (n = 5). C) BAIBA dose dependently increases the respiration rate of hepatocytes. Hepatocytes were treated with a range of BAIBA concentrations (0, 0.1, 0.3, 1, 3, 10 μM) for 6 days. Maximal oxygen consumption rate (OCR) was induced using FCCP. D) qPCR of key β- oxidation genes in hepatocytes treated with BAIBA and/or PPARα antagonist GW6471 for 6 days. Cumulative data from a total of 5 independent observations are shown. †††, P ≤ 0.001, ††††, P < 0.0001 compared to BAIBA treatment. E) Expression of β-oxidation genes in liver from PPARα null control mice (n = 5) and PPARα null mice treated with 100 mg/kg/day BAIBA for 14 days (n = 5). *, P < 0.05, **, P ≤ 0.01, ***, P ≤ 0.001, ****, P < 0.0001 compared to control. All data are represented as Mean ± SEM.
As BAIBA increased the expression of β-oxidation genes in vitro, we investigated whether BAIBA would induce hepatic β-oxidation gene expression in vivo. The expression of key genes involved in fatty acid β-oxidation was measured in the liver of mice treated with 100 mg/kg/day BAIBA for 14 days using qPCR. As in vitro, BAIBA significantly increased the expression of PPARα (1.73-fold P = 0.03), CPT1 (2.5-fold P = 0.0005), ACADvl (1.3-fold P = 0.04), ACADm (1.2-fold P < 0.05), and ACOX1 (1.4-fold, P = 0.03) (Figure 6B). The functional interaction between BAIBA and β-oxidation gene expression was confirmed using two-way ANOVA (P < 0.0001).
To establish whether the observed transcriptional changes conferred functional effects we measured the respiratory rate of hepatocytes treated with BAIBA for 6 days at a range of concentrations. The addition of the electron transport chain uncoupler carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) allowed the measurement of the maximal respiratory capacity. BAIBA treatment significantly and dose dependently increased the maximal oxygen consumption rate (OCR) of the hepatocytes (ANOVA, P = 0.03) (Figure 6C). Together these data demonstrate that BAIBA induces a transcriptional change in hepatocytes leading to a more oxidative phenotype.
We then examined whether BAIBA is driving the increase in hepatic fatty acid β-oxidation through a conserved PPARα mechanism, as was observed with brown-adipocyte gene expression in white adipocytes. We show that BAIBA significantly increases the expression of PPARα both in vitro and in vivo. PPARα is known to regulate hepatic free fatty acid transport, uptake, and catabolism via β-oxidation (Berger and Moller, 2002; Brandt et al., 1998; Gulick et al., 1994). The BAIBA-induced increase in expression of the fatty acid β-oxidation genes, CPT1, ACADvl, ACADm and ACOX1 was abolished by the selective PPARα antagonist, GW6471 (Figure 6D). The functional interaction between the BAIBA and GW6471 treatments on β-oxidation gene expression was confirmed using two-way ANOVA (P < 0.0001).
The role of PPARα in the BAIBA-induced increase in hepatic fatty acid β-oxidation gene expression in in vivo was also examined using PPARα null mice. PPARα null mice were treated with 100 mg/kg/day BAIBA in drinking water for 14 days. Expression analysis of liver using qPCR demonstrated that BAIBA failed to increase expression of β-oxidation genes, including CPT1, ACADvl, ACADm and ACOX1, in the PPARα null mice (Figure 6E). Together, these results indicate that BAIBA increases hepatic fatty acid oxidation gene expression through a PPARα dependent mechanism.
BAIBA plasma concentrations are inversely correlated with cardiometabolic risk factors in humans and are increased during exercise training
We examined the association of plasma BAIBA levels with metabolic traits in a large human cohort study. In 2067 random subjects enrolled in the longitudinal, community-based Framingham Heart Study (FHS), BAIBA levels were inversely correlated with fasting glucose (P = 0.0003), insulin (P < 0.0001), the Homeostatic Model Assessment-Insulin Resistance (HOMA-IR) (P < 0.0001), triglycerides (P < 0.0001), and total cholesterol (P < 0.0001) in age and sex adjusted analyses. In addition, there was a trend towards an inverse association with BMI (P = 0.08).
We also assessed BAIBA concentrations in humans before and after an exercise training intervention. As part of the HERITAGE Family Study, sedentary subjects were recruited for a 20 week program of supervised exercise training (Table 1). Metabolomic profiling was performed on plasma from 80 subjects before and after the exercise training intervention. Following the 20 week exercise program, the average VO2 max of the subjects had increased by more than 20%. The plasma BAIBA concentration increased by 17% (+/−5% SEM, p = 0.03), a very consistent percentage increase compared with the murine exercise data.
Table 1.
Heritage exercise study demographics
| SEX (% Male) |
AGE (years) |
BMI | Baseline VO2 MAX |
Post Exercise VO2 MAX |
Δ VO2 MAX |
|---|---|---|---|---|---|
| 50 | 34.1 ± 14 | 26 ± 5.4 | 2577.3 ± 717.2 |
3007.4 ± 849.6 |
430 ± 390.3 |
BMI, body mass index. Data presented as mean ± SD. V02 max is reported in mL 02/min.
Integration of human genetic and transcriptional data highlights a role for PGC-1α in BAIBA generation
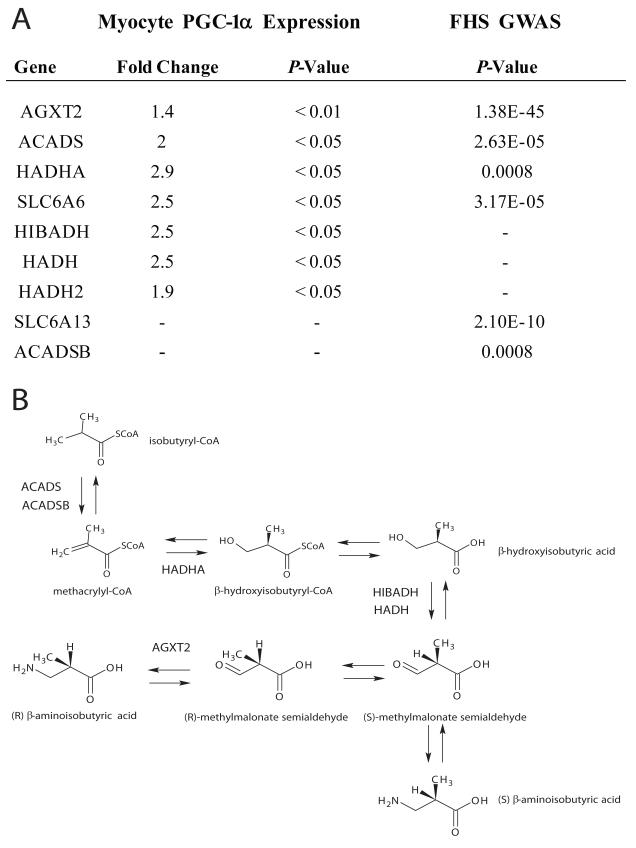
The availability of BAIBA levels and genome-wide genotyping in 1000 FHS participants allowed us to identify genes responsible for modulating metabolite levels in humans in an unbiased manner. These analyses highlighted putative enzymes involved in BAIBA generation (Figure 7A). AGXT2 encodes the enzyme alanine-glyoxylate aminotransferase 2, which catalyses the transamination between BAIBA and pyruvate. Strong association was noted (P = 1.38E-45) between the top SNP at the locus, rs37370, and BAIBA concentrations in FHS. AGXT2 has previously been associated with the urine concentration of BAIBA in humans (Suhre et al., 2011). Other significant associations involved ACADS (rs476676, P = 2.63E-05) and ACADSB (rs11248396, P = 0.0008), which encode enzymes catalyzing the reaction forming methacryl-CoA from isobutryl-CoA upstream of BAIBA. The analyses also highlighted HADHA (rs10165599, P = 0.0008), which encodes the enzyme hydroxyacyl-CoA dehydrogenase responsible for the catalysis of the reaction forming β-hydroxyisobutyryl-CoA from methacryl-CoA in the biogenesis of BAIBA. In addition to variants in the biosynthetic pathway, SNPs were also identified in genes for two solute carriers, SLC6A13 (rs2289957, P = 2.1E-10) and SLC6A6 (rs11128708, P = 3.17E-05), which encode the GABA transporter GAT2 and the taurine transporter TauT, respectively, which may also function as BAIBA transporters (Broer, 2008; Liu et al., 1999).
Figure 7. Integration of human genetic and transcriptional data highlights a role for PGC-1α in BAIBA generation.
A) A table of the transcriptional changes in genes associated with the BAIBA biosynthesis pathway in primary myocytes expressing PGC-1α as assessed by expression arrays (left panel). Right panel includes genes in the BAIBA biosynthesis pathway and the significance of their relationship to BAIBA plasma concentrations in the Framingham Heart Study (FHS). B) The BAIBA biosynthesis pathway annotated with the genes increased by forced PGC-1α expression in primary myocytes or identified by the GWAS study.
To then examine potential mechanisms by which PGC-1α expression might increase levels of BAIBA, we performed transcriptional analysis on PGC-1α overexpressing myocytes. There was striking overlap between BAIBA pathway participants highlighted by GWAS and those increased by forced PGC-1α expression. AGXT2 was found to be increased by PGC-1α overexpression in myocytes (Fold Change = 1.4, P < 0.01), as was the expression of ACADS (Fold Change = 2.0, P < 0.05) and HADHA (Fold Change = 2.9, P < 0.05). The expression of the TauT transporter, SLC6A6 was also increased by PGC-1α expression (Fold Change = 2.5, P < 0.05).
In addition, forced PGC-1α expression in muscle in turn increased expression of DLD, HIBADH, HADH and HADH2, genes encoding the enzymes dihydrolipoamide dehydrogense, 3-hydroxyisobutyrate dehydrogenase, L-3-hydroxyacyl-Coenzyme A dehydrogenase and hydroxyacyl-Coenzyme A dehydrogenase, type II, respectively (DLD Fold change 2.78, P < 0.05, HIBADH Fold change = 2.5, P < 0.05, HADH Fold Change = 2.5, P < 0.05, HADH2 Fold Change = 1.9, P < 0.05). These enzymes catalyze the formation of isobutryl-CoA from valine as part of the branched-chain alpha-keto acid dehydrogenase complex, and methylmalonate semialdehyde from β-hydroxyisobutyric acid in the pathway producing BAIBA. Thus, PGC-1α increases the expression of genes encoding the metabolic enzymes required for production and transport of BAIBA in myocytes, a number of which are genetic determinants of BAIBA plasma concentrations in humans (Figure 7).
Discussion
Transgenic mice expressing PGC-1α in their skeletal muscle display an improved capability for exercise (Calvo et al., 2008). Muscle specific PGC-1α expression in mice also increases the expression of brown adipocyte-specific genes and changes the characteristics of WAT to a more brown-like phenotype (Bostrom et al., 2012). These cells have been termed beige or brite cells (Ishibashi and Seale, 2010; Petrovic et al., 2010). A similar effect has been identified in the WAT of mice undergoing exercise programs (Bostrom et al., 2012; Sutherland et al., 2009; Xu et al., 2011). The identification of the PGC-1α dependent polypeptide hormone irisin, which is secreted into circulation from muscle and triggers the browning response of WAT, establishes one mechanism by which signals from muscle during exercise can mediate energy metabolism in other tissues (Bostrom et al., 2012). However, this recent discovery does not exclude a potential role for other mediators, especially small molecules.
BAIBA was identified in a screen of small molecules generated by myocytes expressing PGC-1α in vitro, and was subsequently found to be increased in the plasma of both chronically exercised and muscle specific PGC-1α transgenic mice. Intramuscular levels of BAIBA were also strikingly increased by exercise, though elevations of BAIBA in other tissues is also possible. BAIBA increases the expression of brown adipocyte-specific genes both in vitro and in vivo through a PPARα mediated mechanism. BAIBA also functions to increase hepatic fatty acid β-oxidation through PPARα. Treating mice with BAIBA improves glucose tolerance. We were interested to find that BAIBA treatment during differentiation of white adipocytes from BJ RiPS IPSCs also induces a brown adipocyte-like phenotype with concordant functional effects on basal and insulin stimulated glucose uptake and oxygen consumption. In humans, BAIBA plasma concentrations are increased by regular exercise and are significantly inversely correlated with multiple cardiometabolic risk factors. Finally, by integrating human genetic data and in vitro transcriptional findings we highlight a cassette of BAIBA biosynthetic enzymes that are under PGC-1α transcriptional control in muscle. We note that while the browning effect appears to be operative in our studies, our findings do not exclude the possibility that macroscopic brown adipose depots in mice increase energy expenditure and contribute to the observed effects.
BAIBA is a non-protein β-amino acid that can be generated by catabolism of the branched-chain amino acid valine. Our expression studies would seem to highlight a role for PGC-1α expression in muscle with the production of BAIBA from valine. Fasting plasma concentrations of valine are correlated with obesity and serum insulin (Felig et al., 1969; Newgard et al., 2009), and we recently identified valine plasma concentration as a predictor of future development of diabetes (Wang et al., 2011). Skeletal muscle is a major site of branched-chain amino acid utilization and, during exercise, catabolism of the branched-chain amino acids is elevated (Harper et al., 1984; Shimomura et al., 2006; Shimomura et al., 2004). Furthermore, the expression of genes in the valine degradation pathway was found to be increased in the skeletal muscle of physically active members of twin pairs compared to their inactive co-twins (Leskinen et al., 2010). Our findings suggest a possible connection between valine utilization in skeletal muscle during exercise and beneficial effects on peripheral WAT.
Independently, BAIBA treatment has been found to reduce weight gain in partially leptin deficient (ob/+) mice (Begriche et al., 2008). Glucose tolerance was improved in the ob/+ mice treated with BAIBA, consistent with the diminished weight gain. Prior work suggests that BAIBA may enhance fatty acid oxidation and reduces de novo lipogenesis in the liver (Begriche et al., 2008; Maisonneuve et al., 2004; Note et al., 2003). Interestingly, the effect of BAIBA on hepatic lipid metabolism mirrors the action of exercise, which has also been shown to increase liver fatty acid oxidation and decrease hepatic lipogenesis through PPARα (Aoi et al., 2011; Oh et al., 2006; Rabol et al., 2011; Rector et al., 2011). The effects of BAIBA on browning of white adipose depots were not evaluated in any prior studies. Our studies demonstrate the PPARα-dependent mechanism of BAIBA’s salutary effects on liver β-oxidation, and extend the prior literature by identifying its link to PGC-1α, browning of WAT, and its relationship to chronic exercise. Future work may also uncover beneficial effects of BAIBA on other tissues. Our work also highlights BAIBA as a potential disease marker in human populations.
It is notable that several enzymes of the valine degradation pathway and thus BAIBA biosynthetic pathway are shared with that of β-oxidation of fatty acids (ACADSB, ACADS, ACADM, HADHA, and HADH), a number of which we found to be transcriptionally controlled by PGC-1α in myocytes. It would seem evolutionarily advantageous to integrate the production of a metabolite myokine signal with the fatty acid oxidation pathway, the primary source of energy for muscle during endurance exercise. The HADH knockout mouse is cold intolerant and exhibits impaired adaptive thermogenesis with a decreased fat tolerance at 4 °C (Schulz et al., 2011). Similarly, the ACADS knockout mouse also shows cold intolerance and defects in thermogenesis (Guerra et al., 1998). Although the disruption to β-oxidation in both the ACADS and HADH knockout mice is likely to contribute to impaired thermogenesis, a perturbation in BAIBA production may also play a role in this process.
While we demonstrate that BAIBA increases expression of PPARα, and that BAIBA-induced browning of WAT requires this nuclear receptor, as yet the direct mechanism of action upstream of PPARα is unknown. PPARα null mice remain resistant to cold, can activate cold-induced thermogenesis and express equivalent levels of UCP-1 in intrascapular brown adipose tissue compared to wild-type controls (Kersten et al., 1999). Moreover, as PPARα null mice do not have reduced UCP1 expression in white adipose tissue compared to controls following cold exposure (Xue et al., 2005), absence of PPARα does not blunt the general browning effect. Therefore, our results indicate that BAIBA increases browning gene expression through a specific PPARα-dependent mechanism. BAIBA may function to activate a cell surface receptor since structurally similar metabolites, including butyrate and isobutyrate, activate short chain carboxylic acid receptors in white adipocytes (Brown et al., 2003). Ascertaining the cell signaling pathways by which BAIBA leads to increased PPARα expression will be a focus of future studies.
In summary, we identify BAIBA as a novel small molecule myokine representing the first in its class of non-adrenergic activators of the thermogenic program in WAT (Whittle and Vidal-Puig, 2012). The identification of BAIBA as a PGC-1α mediated and exercise triggered signal has significant implications not only for our understanding of exercise and its protective role against the development of metabolic diseases, but also for potential therapeutics for type 2 diabetes and the metabolic syndrome.
Experimental Procedures
Myocyte culture
Primary satellite cells (myoblasts) were isolated as previously described.(Bostrom et al., 2012; Megeney et al., 1996). Briefly, myoblasts were cultured in F-10 medium supplemented with 20% FBS and basic FGF. For differentiation into myotubes, cells were changed to DMEM supplemented with 5% horse serum. At day 2 of differentiation, myocytes were transduced with an adenovirus expressing either PGC-1α or GFP as previously described (St-Pierre et al., 2003). At 24 hr post transduction cells were washed with PBS and freestyle media (GIBCO/Invitrogen, Grand Island, NY). Freestyle media was added to GFP and PGC-1α expressing myocytes, and cells were incubated for 24 hours. Media was collected and cleared with centrifugation (1000g, 4 °C for 5 min × 3). The supernatant was then snap frozen in aliquots.
Culture and differentiation of the mouse primary adipocytes isolated from inguinal white adipose stromal vascular fraction
Primary white adipose stromal vascular cells were fractionated as previously described (Soukas et al., 2001). Stromal vascular cells were then cultured and induced to differentiate into adipocytes also according to published methods (Bostrom et al., 2012; Seale et al., 2011). Stromal vascular cells were cultured in DMEM/F12 containing 10% FBS. Adipocyte differentiation was induced in preadipocytes cultured by treating confluent cells for 48 hrs in medium containing 10% FBS, isobutylmethylxanthine 0.5 mM, indomethacin 125 nM, dexamethasone 1 μM, insulin 850 nM, T3 1 nM, and rosiglitazone 1 μM (Cayman Chemical). 2 days after the induction of differentiation, media was removed and replaced with maintenance media containing 10% FBS, insulin 850 nM, T3 1 nM, and rosiglitazone 1 μM. During the 6 days, cells were cultured with either saline (control), GABA 3 μM, cytosine 1 μM, 2′-deoxycytidine 15 μM and BAIBA 0.3, 1, 3 and 5 μM. All chemicals for cell culture were obtained from Sigma-Aldrich unless noted.
Maintenance of pluripotent cells, generation of mesenchymal progenitor cells and adipocyte differentiation
Human induced pluripotent stem cells were maintained and differentiated into mature brown and white adipocytes as previously described(Ahfeldt et al., 2012; Schinzel et al., 2011). Briefly, human induced pluripotent stem cells were cultured feeder free on Geltrex (Invitrogen) in the chemically defined medium mTESR1 (Stem Cell Technologies). Human induced pluripotent stem cells were disaggregated with dispase into small clumps containing 5-10 cells and transferred to low-adhesion plastic 6-well plates (Costar Ultra Low Attachment; Corning Life Sciences) in growth medium containing DMEM, 15% FBS, and 1% Glutamax. After 7 days in suspension culture, embryoid bodies were collected and re-plated on gelatin-coated 6-well plates in medium containing DMEM, 10% FBS, and 1% Glutamax. After cells reached confluency (approximately 5 days), they were trypsinized (0.25% trypsin) and re-plated on cell culture dishes containing mesenchymal progenitor cell growth medium containing DMEM, 15% FBS, 1% Glutamax, and bFGF 2.5 ng/ml (Aldevron). Cells were passaged with a 1:3 split ratio and differentiated after at least 3 passages and prior to passage 10. Adipogenic differentiation was carried out for 21 days using an adipogenic differentiation medium containing DMEM, 7.5% knockout serum replacement (KOSR; Invitrogen), 7.5% human plasmanate, 0.5% nonessential amino acids, 1% penicillin/streptomycin, dexamethasone 0.1 μM, insulin 10 μg/ml, and rosiglitazone 0.5 μM. Differentiation was carried out either in the absence of BAIBA, in the case of controls, and with two concentrations of BAIBA at 1 μM or 10 μM in the differentiation media. Adipogenic differentiation medium was supplemented for 16 days with doxycyline 700 ng/ml, and afterwards cells were maintained in culture in the absence of doxycycline until day 21.
Production of lentivirus and transduction
Lentivirus production and transduction of the cells was performed as previously described (Ahfeldt et al., 2012). A third-generation, Tat-free packaging system (Tiscornia et al., 2006) was used to produce recombinant lentivirus. The vectors—either Lenti-rtTA plasmid (Stadtfeld et al., 2008) or Lenti-PPARG2 plasmid—together with the two packaging plasmids—pMDL, pREV—and the plasmid coding for VSV-G envelope were transfected into HEK 293T cells using calcium chlorate as previously described (al Yacoub et al., 2007). Cells were transduced with lentiviral supernatant 24 hr after passaging at about 40% confluency.
Glucose uptake assay
Adipocytes were serum-starved in 2% BSA DMEM overnight. Cells were then incubated in KRH buffer (NaCl 121 mM, KCl 4.9 mM, MgSO4 1.2 mM, CaCl2 0.33 mM, HEPES 12 mM, pH7.4) for 4 hrs at 37 oC, followed by washing three times in KRH buffer. Glucose uptake was measured by incubating cells with 2-deoxy-D-[3H] glucose 0.5μCi/ml (Perkin-Elmer) for 5 min at 37 °C. After cold PBS washing three times, cells were lysed with 1% Triton X-100 solution and subjected to scintillation counting. Non-specific uptake was measured in the presence of cytochalasin B 10 μM and subtracted from total uptake.
Measurement of cellular OCR
Cells were plated in gelatin-coated XF24-well cell culture microplates (Seahorse Bioscience) and differentiated into adipocytes. Cells were incubated in pre-warmed unbuffered DMEM medium (DMEM containing GlutaMax 2 mM, sodium pyruvate 1 mM, NaCl 1.85 g/L, and glucose 25 mM) for 1h. The oxygen consumption was measured by the XF24 Extracellular Flux Analyzer (Seahorse Biosciences). Mitochondrial function was profiled by injecting, oligomycin 2 μM, CCCP 0.5 μM (carbonyl cyanide trifluoromethoxyphenylhydrazone), and antimycin A 5 μM in succession. OCR was determined by plotting the oxygen tension of the medium in the chamber as a function of time (pMoles/min).
Hepatocyte culture
H4IIE hepatocytes (ATCC) were seeded at 200,000 cells per well in a 24 well collagen I coated plate. The cells were incubated in MEM media (ATCC supplemented with 10% FBS (Sigma) and Penicillin/Streptomycin) with 5 μM BAIBA (n = 6), 1 μM GW6471 PPARα antagonist (n = 6), and 1 μM GW6471 and 5 μM BAIBA (n = 6) for 6 days.
Hepatocyte respirometry
H4IIE hepatocytes were grown for 6 days in Oxoplates (PreSens) with MEM + 10% FBS media (Sigma Aldrich). Cells were plated at 40,000 cells per well and BAIBA or BAIBA and 1 μM GW6471 PPARα antagonist were added on the same day. The OxoPlate OP96F (PreSens, Regens- burg, Germany) contains oxygen-sensitive particles PSLi-Pt-1 (Opto-Sense, Worth, Germany), which consist of small polystyrene particles. Sulforhodamin is covalently attached to these particles as reference dye, and a platinum porphin is incorporated as indicator dye. The sensor has a thickness of about 10 μm and is fixed at the bottom of each well of a 96-flat bottom-well plate (Greiner, Frickenhausen, Germany). Basal serum-free MEM media (100 μl) containing BAIBA in a range of concentrations (0-10 μM) (n = 6 per dose) and BAIBA in the presence of 1 μM GW6471 PPARα antagonist (n = 6 per dose) was added into the wells of an OxoPlate. The media was exchanged every two days with the addition of fresh compounds. After 6 days each well was then overlaid with 250 μl of mineral oil after the addition of 5 μM of final concentration of Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP). Wells containing oxygen-free water (cal 0) and air-saturated water (cal 100) served as standards. Oxygen-free water was prepared by dissolving 1 g of sodium sulfite in 100 mL of water. Water is oxygen free after approximately 1 min. Air-saturated water was prepared by shaking 100 mL of water vigorously for 2 min. The oxygen concentration in each well was measured right after mineral oil addition, 15 min and 30 min after mineral oil addition. Fluorescence of each well was measured in dual kinetic mode (Multiskan Ascent CF, Thermo Lab- systems, Vantaa, Finland). Filter pair 1 (544/650 nm) detects fluorescence of the indicator dye. The second filter pair (544/590 nm) measures fluorescence of the reference dye. Oxygen tension was calculated according to the Stern-Volmer equation and transformed into nmol of oxygen . The difference between baseline, 15 min and 30 min oxygen tension was used to calculate the oxygen consumption per min per well.
Animal experimentation
Muscle specific PGC-1α transgenic mice were generated and maintained as previously described (Bostrom et al., 2012; Lin et al., 2002a). For the exercise experiments, 12-week-old B6 mice were used (Jackson Laboratory, Bar Harbor, ME). Endurance exercise was carried out using free wheel running for 3 weeks (n = 6) (Rasbach et al., 2010). Controls were age matched sedentary littermates (n = 6). For the short term BAIBA treatment cohort, 6 week old C57BL6/J mice (Jackson Laboratory, Bar Harbor, ME) were weight-matched and assigned to groups for treatment. Mice were treated with either 100 mg/kg/day or 170 mg/kg/day BAIBA in their drinking water for 2 weeks and fed a standard chow diet ad libitum (Prolab RMH 3000-5P75, Labdiet, Brentwood, MO). 129S4/SvJae-Pparatm1Gonz/J mice (Jackson Laboratory, Bar Harbor, ME) were weight-matched and assigned to groups for treatment. Mice were treated with 100 mg/kg/day BAIBA in their drinking water for 2 weeks and fed a standard chow diet ad libitum. For the long term BAIBA treatment cohort, 6 week old C57BL6/J mice (Jackson Laboratory, Bar Harbor, ME) were weight-matched and assigned to groups for treatment (n=11 per group). Mice were treated with 100 mg/kg/day BAIBA in their drinking water for 14 weeks and fed a high fat diet ad libitum (60% Fat, DIO-VHFD Research Diets Inc. New Brunswick, NJ).
Study mice were fasted, sacked and plasma was collected via left ventricular puncture at completion of the study (Week 16). Inguinal WAT and liver was rapidly dissected, snap frozen in liquid nitrogen and stored at snap frozen in liquid nitrogen and stored at80 °C until mRNA extraction. All mice were housed in a controlled temperature, lighting and humidity environment.
Indirect calorimetry
All experiments were performed with 6-week old mice treated with either BAIBA (100 mg/kg/day) or water for 14 weeks (n=8 mice per group). A PhenoMaster system (TSE Systems, Calo-(D)/Feed/BWXZ, 16 mice) was used to monitor oxygen consumption, carbon dioxide production, food intake, daily body mass, and locomotory activity. The PhenoMaster system was calibrated before each experiment. Animals were subjected to a 7-day acclimation period in a training cage without monitoring to habituate to the environment of the metabolic cages. Animals were maintained in normal cedar bedding at 22°C throughout the monitoring period. Twice hourly measurements for each animal were obtained for oxygen and carbon dioxide with ad libitum access to food and water (or water plus BAIBA) on a controlled 12-hour light/dark cycle. Cages contained one mass sensor to monitor food intake and a second sensor attached to a glass housing to measure body mass. Oxygen consumption is expressed normalized to body mass.
Intraperitoneal glucose tolerance test
Mice were fasted for 6 hours with free access to water prior to baseline glucose measurements (n=11 mice per group). Administration of glucose (Sigma, St. Louis, MO) was performed by intraperitoneal injection (glucose 1.5mg /gram of body weight; glucose solution 150mg/ml). Blood was obtained from the tail vein immediately prior to glucose injection and then at 30, 60 and 120 minutes post injection. Glucose levels were measured using a Bayer Contour Glucose Meter (Bayer Healthcare, Mishawaka, IL).
Gene expression analysis
Total RNA from human cell lines, mouse inguinal WAT stromal vascular fraction derived primary adipocytes, hepatocytes, and mouse WAT and liver was extracted with Trizol (Invitrogen) and purified via the RNeasy mini kit (Qiagen) according to the manufacturer’s instructions. The RNA yield was determined using the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). RNA was normalized and converted to cDNA using the Superscript First-Strand Kit (Invitrogen). Quantitative RT-PCR was performed using a Realplex Mastercycler (Eppendorf) with the Quantifast-SYBR Green PCR mix (Qiagen). All data were normalized to 18SrRNA or HPRT and quantitative measures obtained using the Δ-Δ-CT method.
Framingham Heart Study
The Framingham Offspring Study was initiated in 1971, when 5,124 individuals enrolled into a longitudinal cohort study to examine risk factors for cardiovascular disease (Wang et al., 2011). We studied individuals attending a routine examination of this cohort that took place between 1991 and 1995. Of 3,799 attendees to the examination, 2,067 were eligible for the present investigation because they were free of diabetes and cardiovascular disease, and had measurement of BAIBA concentrations in fasting plasma samples. Fasting insulin and glucose were also measured as previously described (Wang et al., 2011).
Genotyping was performed on the Affymetrix GeneChip Human Mapping 500K Array SetR and 50K Human Gene Focused PanelR. After filtering out 15586 SNPs with Hardy-Weinberg p<1e-6, 64511 SNPs with missing rate >3%, 45361 SNPs with mishap test p<1e-9 (mishap test in PLINK, http://pngu.mgh.harvard.edu/purcell/plink/), 4857 SNPs with >100 Mendel errors, 67269 SNPs with minor allele frequency < 0.01, 2 SNPs due to strandedness issues upon merging data with HapMap, and a further 13394 SNPs not present on HapMap, a total of 378,163 SNPs were used in the imputation for 2,543,887 autosomal SNPs. MACH software (http://www.sph.umich.edu/csg/abecasis/MACH) version 1.0.15 with HapMap release 22, build 26, as a reference panel was used.
Imputation on X chromosome: After filtering out 159 SNPs with Hardy-Weinberg p<1e-6, 450 SNPs with missing rate > 3%, 1,851 SNPs with minor allele frequency < 0.01, 12 SNPs with a male heterozygote count > 45, and 619 SNPs not included in the HapMap legend files, 7,795 SNPs were used in the imputation for 62,876 X chromosome SNPs. We used IMPUTE software (https://mathgen.stats.ox.ac.uk/impute/impute.html) version 0.5.0 with Hapmap Build 35 release 21 as a reference panel.
The human study protocols were approved by the Institutional Review Boards of Boston University Medical Center and Massachusetts General Hospital, and all participants provided written informed consent.
HERITAGE clinical exercise study
The HERITAGE Family Study is a clinical trial that enrolled 557 individuals of various ages (16–65 y) to determine the effects of 20-weeks of highly-controlled endurance training on physiologic measures and risk factors for cardiometabolic disease. Only individuals who were previously sedentary, free of pre-existing disease, and not taking any medications that would affect any of the outcome variables were allowed to enter the study. Details of the aims, experimental design, and measurement protocols of the HERITAGE Family Study were presented in detail in a previous publication (Bouchard et al., 1995). Endurance training was conducted (3 d/wk for a total of 60 exercise sessions) on cycle ergometers that were computer controlled to maintain the participants’ heart rates at fixed percentages of their aerobic capacity (VO2max). The training program started at 55% of VO2max for 30 min/session and gradually increased to 75% of VO2max for 50 min/session, where it was maintained during the last 6 wk of training. Peripheral plasma samples collected from 80 HERITAGE participants before and after the 20-week endurance training program were subjected to metabolomic profiling.
Metabolic profiling
Metabolic profiling of amino acids, biogenic amines and other polar plasma metabolites were analyzed by LC-MS as previously described (Roberts et al., 2012; Wang et al., 2011). In brief, formic acid, ammonium acetate, LC-MS–grade solvents and valine-d8 were purchased from Sigma-Aldrich. Phenylalanine-d8 was purchased from Cambridge Isotope Laboratories. Plasma and media samples were prepared for LC-MS analyses via protein precipitation with the addition of nine volumes of 74.9:24.9:0.2 vol/vol/vol acetonitrile/methanol/formic acid containing two additional stable isotope-labeled internal standards for valine-d8 and phenylalanine-d8. The samples were centrifuged (10 min, 15,000g, 4 °C), and the supernatants were injected directly. Metabolite concentrations were determined using the standard addition method.
Statistical analyses
For metabolite analyses in FHS, log transformation of BAIBA concentrations was applied to approximate a normal distribution. Partial correlation coefficients were estimated between BAIBA and the following metabolic variables, after adjustment for age and sex: body mass index (BMI), fasting glucose, fasting insulin, total cholesterol, triglycerides, homeostasis model assessment of insulin resistance (HOMA-IR) and the homeostasis model assessment of β-cell function (HOMA-B), calculated as previously described (Wang et al., 2011).
For FHS GWAS analyses a linear mixed effects model that accounts for familial relatedness with an additive genetic model with one degree of freedom was used (Chen and Yang, 2010).
For animal studies, all results, unless otherwise stated, are expressed as means, and error bars depict SEMs. A two tailed Student’s t test or ANOVA was used to determine P values.
Supplementary Material
Highlights.
β-Aminoisobutyric acid (BAIBA) is secreted from PGC-1α expressing myocytes.
BAIBA activates the thermogenic program in white adipocytes via PPARα.
Circulating BAIBA levels in mice and humans are increased with exercise.
BAIBA is inversely correlated with cardiometabolic risk factors in humans.
Acknowledgments
This work was supported by NIH R01 DK081572, NIH R01 HL098280, the Leducq Foundation, and the American Heart Association (to REG) and NIH DK 31405 to BMS. The Framingham Heart Study is supported by NIH/NHLBI contract N01-HC-25195. Dr. Roberts is supported by a Leducq Foundation Career Development Award. Dr. Boström is supported by the Wenner-Gren Foundation and the Swedish Heart and Lung Foundation. The HERITAGE Family Study is currently supported by NIHRO1-HL045670. Dr. Bouchard is supported by the John W. Barton Sr. Chair in Genetics and Nutrition.
References
- Ahfeldt T, Schinzel RT, Lee YK, Hendrickson D, Kaplan A, Lum DH, Camahort R, Xia F, Shay J, Rhee EP, et al. Programming human pluripotent stem cells into white and brown adipocytes. Nat Cell Biol. 2012;14:209–219. doi: 10.1038/ncb2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- al Yacoub N, Romanowska M, Haritonova N, Foerster J. Optimized production and concentration of lentiviral vectors containing large inserts. J Gene Med. 2007;9:579–584. doi: 10.1002/jgm.1052. [DOI] [PubMed] [Google Scholar]
- Aoi W, Naito Y, Hang LP, Uchiyama K, Akagiri S, Mizushima K, Yoshikawa T. Regular exercise prevents high-sucrose diet-induced fatty liver via improvement of hepatic lipid metabolism. Biochem Biophys Res Commun. 2011;413:330–335. doi: 10.1016/j.bbrc.2011.08.097. [DOI] [PubMed] [Google Scholar]
- Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- Begriche K, Massart J, Abbey-Toby A, Igoudjil A, Letteron P, Fromenty B. Beta-aminoisobutyric acid prevents diet-induced obesity in mice with partial leptin deficiency. Obesity (Silver Spring) 2008;16:2053–2067. doi: 10.1038/oby.2008.337. [DOI] [PubMed] [Google Scholar]
- Berger J, Moller DE. The mechanisms of action of PPARs. Annu Rev Med. 2002;53:409–435. doi: 10.1146/annurev.med.53.082901.104018. [DOI] [PubMed] [Google Scholar]
- Bostrom P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, Rasbach KA, Bostrom EA, Choi JH, Long JZ, et al. A PGC1-alpha-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature. 2012;481:463–U472. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Leon AS, Rao DC, Skinner JS, Wilmore JH, Gagnon J. The HERITAGE family study. Aims, design, and measurement protocol. Med Sci Sports Exerc. 1995;27:721–729. [PubMed] [Google Scholar]
- Brandt JM, Djouadi F, Kelly DP. Fatty acids activate transcription of the muscle carnitine palmitoyltransferase I gene in cardiac myocytes via the peroxisome proliferator-activated receptor alpha. J Biol Chem. 1998;273:23786–23792. doi: 10.1074/jbc.273.37.23786. [DOI] [PubMed] [Google Scholar]
- Broer S. Amino acid transport across mammalian intestinal and renal epithelia. Physiol Rev. 2008;88:249–286. doi: 10.1152/physrev.00018.2006. [DOI] [PubMed] [Google Scholar]
- Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- Calvo JA, Daniels TG, Wang X, Paul A, Lin J, Spiegelman BM, Stevenson SC, Rangwala SM. Muscle-specific expression of PPAR gamma coactivator-1 alpha improves exercise performance and increases peak oxygen uptake. J Appl Physiol. 2008;104:1304–1312. doi: 10.1152/japplphysiol.01231.2007. [DOI] [PubMed] [Google Scholar]
- Cao L, Choi EY, Liu X, Martin A, Wang C, Xu X, During MJ. White to brown fat phenotypic switch induced by genetic and environmental activation of a hypothalamic-adipocyte axis. Cell Metab. 2011;14:324–338. doi: 10.1016/j.cmet.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;26:580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin B, Cinti S, Morroni M, Raimbault S, Ricquier D, Penicaud L, Casteilla L. Occurrence of Brown Adipocytes in Rat White Adipose-Tissue - Molecular and Morphological Characterization. J Cell Sci. 1992;103:931–942. doi: 10.1242/jcs.103.4.931. [DOI] [PubMed] [Google Scholar]
- Enerback S, Jacobsson A, Simpson EM, Guerra C, Yamashita H, Harper ME, Kozak LP. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature. 1997;387:90–94. doi: 10.1038/387090a0. [DOI] [PubMed] [Google Scholar]
- Felig P, Marliss E, Cahill GF., Jr. Plasma amino acid levels and insulin secretion in obesity. N Engl J Med. 1969;281:811–816. doi: 10.1056/NEJM196910092811503. [DOI] [PubMed] [Google Scholar]
- Frontini A, Cinti S. Distribution and development of brown adipocytes in the murine and human adipose organ. Cell Metab. 2010;11:253–256. doi: 10.1016/j.cmet.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Guerra C, Koza RA, Walsh K, Kurtz DM, Wood PA, Kozak LP. Abnormal nonshivering thermogenesis in mice with inherited defects of fatty acid oxidation. J Clin Invest. 1998;102:1724–1731. doi: 10.1172/JCI4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulick T, Cresci S, Caira T, Moore DD, Kelly DP. The peroxisome proliferator-activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc Natl Acad Sci U S A. 1994;91:11012–11016. doi: 10.1073/pnas.91.23.11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- Harper AE, Miller RH, Block KP. Branched-chain amino acid metabolism. Annu Rev Nutr. 1984;4:409–454. doi: 10.1146/annurev.nu.04.070184.002205. [DOI] [PubMed] [Google Scholar]
- Ishibashi J, Seale P. Beige Can Be Slimming. Science. 2010;328:1113–1114. doi: 10.1126/science.1190816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, Walker EA, Nathan DM. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346:393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Tong Y, Li Y, Nakajima T, Li G, Hu R, Sugiyama E, Kamijo Y, Tanaka N, Hara A, et al. Multiple roles of PPARalpha in brown adipose tissue under constitutive and cold conditions. Genes Cells. 2010;15:91–100. doi: 10.1111/j.1365-2443.2009.01368.x. [DOI] [PubMed] [Google Scholar]
- Kopecky J, Clarke G, Enerback S, Spiegelman B, Kozak LP. Expression of the mitochondrial uncoupling protein gene from the aP2 gene promoter prevents genetic obesity. J Clin Invest. 1995;96:2914–2923. doi: 10.1172/JCI118363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leskinen T, Rinnankoski-Tuikka R, Rintala M, Seppanen-Laakso T, Pollanen E, Alen M, Sipila S, Kaprio J, Kovanen V, Rahkila P, et al. Differences in muscle and adipose tissue gene expression and cardio-metabolic risk factors in the members of physical activity discordant twin pairs. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002a;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu ZD, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002b;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Liu M, Russell RL, Beigelman L, Handschumacher RE, Pizzorno G. beta-alanine and alpha-fluoro-beta-alanine concentrative transport in rat hepatocytes is mediated by GABA transporter GAT-2. Am J Physiol. 1999;276:G206–210. doi: 10.1152/ajpgi.1999.276.1.G206. [DOI] [PubMed] [Google Scholar]
- Lowell BB, V SS, Hamann A, Lawitts JA, Himms-Hagen J, Boyer BB, Kozak LP, Flier JS. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature. 1993;366:740–742. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- Maisonneuve C, Igoudjil A, Begriche K, Letteron P, Guimont MC, Bastin J, Laigneau JP, Pessayre D, Fromenty B. Effects of zidovudine, stavudine and beta-aminoisobutyric acid on lipid homeostasis in mice: possible role in human fat wasting. Antivir Ther. 2004;9:801–810. doi: 10.1177/135965350400900513. [DOI] [PubMed] [Google Scholar]
- Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- Melnyk A, Harper ME, HimmsHagen J. Raising at thermoneutrality prevents obesity and hyperphagia in BAT-ablated transgenic mice. Am J Physiol-Reg I. 1997;272:R1088–R1093. doi: 10.1152/ajpregu.1997.272.4.R1088. [DOI] [PubMed] [Google Scholar]
- Michael LF, Wu ZD, Cheatham RB, Puigserver P, Adelmant G, Lehman JJ, Kelly DP, Spiegelman BM. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. P Natl Acad Sci USA. 2001;98:3820–3825. doi: 10.1073/pnas.061035098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Note R, Maisonneuve C, Letteron P, Peytavin G, Djouadi F, Igoudjil A, Guimont MC, Biour M, Pessayre D, Fromenty B. Mitochondrial and metabolic effects of nucleoside reverse transcriptase inhibitors (NRTIs) in mice receiving one of five single- and three dual-NRTI treatments. Antimicrob Agents Chemother. 2003;47:3384–3392. doi: 10.1128/AAC.47.11.3384-3392.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberkofler H, Dallinger G, Liu YM, Hell E, Krempler F, Patsch W. Uncoupling protein gene: quantification of expression levels in adipose tissues of obese and non-obese humans. J Lipid Res. 1997;38:2125–2133. [PubMed] [Google Scholar]
- Oh KS, Kim M, Lee J, Kim MJ, Nam YS, Ham JE, Shin SS, Lee CM, Yoon M. Liver PPARalpha and UCP2 are involved in the regulation of obesity and lipid metabolism by swim training in genetically obese db/db mice. Biochem Biophys Res Commun. 2006;345:1232–1239. doi: 10.1016/j.bbrc.2006.04.182. [DOI] [PubMed] [Google Scholar]
- Olesen J, Kiilerich K, Pilegaard H. PGC-1alpha-mediated adaptations in skeletal muscle. Pflugers Arch. 2010;460:153–162. doi: 10.1007/s00424-010-0834-0. [DOI] [PubMed] [Google Scholar]
- Petrovic N, Walden TB, Shabalina IG, Timmons JA, Cannon B, Nedergaard J. Chronic Peroxisome Proliferator-activated Receptor gamma (PPAR gamma) Activation of Epididymally Derived White Adipocyte Cultures Reveals a Population of Thermogenically Competent, UCP1- containing Adipocytes Molecularly Distinct from Classic Brown Adipocytes. Journal of Biological Chemistry. 2010;285:7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Adelmant C, Wu ZD, Fan M, Xu JM, O’Malley B, Spiegelman BM. Activation of PPAR gamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Rabol R, Petersen KF, Dufour S, Flannery C, Shulman GI. Reversal of muscle insulin resistance with exercise reduces postprandial hepatic de novo lipogenesis in insulin resistant individuals. Proc Natl Acad Sci U S A. 2011;108:13705–13709. doi: 10.1073/pnas.1110105108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasbach KA, Gupta RK, Ruas JL, Wu J, Naseri E, Estall JL, Spiegelman BM. PGC-1alpha regulates a HIF2alpha-dependent switch in skeletal muscle fiber types. Proc Natl Acad Sci U S A. 2010;107:21866–21871. doi: 10.1073/pnas.1016089107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rector RS, Uptergrove GM, Morris EM, Borengasser SJ, Laughlin MH, Booth FW, Thyfault JP, Ibdah JA. Daily exercise vs. caloric restriction for prevention of nonalcoholic fatty liver disease in the OLETF rat model. Am J Physiol Gastrointest Liver Physiol. 2011;300:G874–883. doi: 10.1152/ajpgi.00510.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts LD, Souza AL, Gerszten RE, Clish CB. Targeted metabolomics. Curr Protoc Mol Biol. 2012;32:31–24. doi: 10.1002/0471142727.mb3002s98. Chapter 30, Unit 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinzel RT, Ahfeldt T, Lau FH, Lee YK, Cowley A, Shen T, Peters D, Lum DH, Cowan CA. Efficient culturing and genetic manipulation of human pluripotent stem cells. PLoS One. 2011;6:e27495. doi: 10.1371/journal.pone.0027495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz N, Himmelbauer H, Rath M, van Weeghel M, Houten S, Kulik W, Suhre K, Scherneck S, Vogel H, Kluge R, et al. Role of medium- and short-chain L-3-hydroxyacyl-CoA dehydrogenase in the regulation of body weight and thermogenesis. Endocrinology. 2011;152:4641–4651. doi: 10.1210/en.2011-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang WL, Kajimura S, Chin S, Kuang SH, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–U927. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura Y, Honda T, Shiraki M, Murakami T, Sato J, Kobayashi H, Mawatari K, Obayashi M, Harris RA. Branched-chain amino acid catabolism in exercise and liver disease. J Nutr. 2006;136:250S–253S. doi: 10.1093/jn/136.1.250S. [DOI] [PubMed] [Google Scholar]
- Shimomura Y, Murakami T, Nakai N, Nagasaki M, Harris RA. Exercise promotes BCAA catabolism: effects of BCAA supplementation on skeletal muscle during exercise. J Nutr. 2004;134:1583S–1587S. doi: 10.1093/jn/134.6.1583S. [DOI] [PubMed] [Google Scholar]
- Soukas A, Socci ND, Saatkamp BD, Novelli S, Friedman JM. Distinct transcriptional profiles of adipogenesis in vivo and in vitro. J Biol Chem. 2001;276:34167–34174. doi: 10.1074/jbc.M104421200. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Lin J, Krauss S, Tarr PT, Yang R, Newgard CB, Spiegelman BM. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1alpha and 1beta (PGC-1alpha and PGC-1beta) in muscle cells. J Biol Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M, Maherali N, Breault DT, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Wallaschofski H, Raffler J, Friedrich N, Haring R, Michael K, Wasner C, Krebs A, Kronenberg F, Chang D, et al. A genome-wide association study of metabolic traits in human urine. Nat Genet. 2011;43:565–569. doi: 10.1038/ng.837. [DOI] [PubMed] [Google Scholar]
- Sutherland LN, Bomhof MR, Capozzi LC, Basaraba SAU, Wright DC. Exercise and adrenaline increase PGC-1 alpha mRNA expression in rat adipose tissue. J Physiol-London. 2009;587:1607–1617. doi: 10.1113/jphysiol.2008.165464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- Viscomi C, Bottani E, Civiletto G, Cerutti R, Moggio M, Fagiolari G, Schon EA, Lamperti C, Zeviani M. In vivo correction of COX deficiency by activation of the AMPK/PGC-1alpha axis. Cell Metab. 2011;14:80–90. doi: 10.1016/j.cmet.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. Highly Efficient Reprogramming to Pluripotency and Directed Differentiation of Human Cells with Synthetic Modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle AJ, Vidal-Puig A. NPs -- heart hormones that regulate brown fat? J Clin Invest. 2012;122:804–807. doi: 10.1172/JCI62595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright HM, Clish CB, Mikami T, Hauser S, Yanagi K, Hiramatsu R, Serhan CN, Spiegelman BM. A synthetic antagonist for the peroxisome proliferator-activated receptor gamma inhibits adipocyte differentiation. J Biol Chem. 2000;275:1873–1877. doi: 10.1074/jbc.275.3.1873. [DOI] [PubMed] [Google Scholar]
- Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Ruas JL, Estall JL, Rasbach KA, Choi JH, Ye L, Bostrom P, Tyra HM, Crawford RW, Campbell KP, et al. The unfolded protein response mediates adaptation to exercise in skeletal muscle through a PGC-1alpha/ATF6alpha complex. Cell Metab. 2011;13:160–169. doi: 10.1016/j.cmet.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZD, Puigserver P, Andersson U, Zhang CY, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Xu XH, Ying ZK, Cai M, Xu ZB, Li YJ, Jiang SY, Tzan K, Wang AX, Parthasarathy S, He GL, et al. Exercise ameliorates high-fat diet-induced metabolic and vascular dysfunction, and increases adipocyte progenitor cell population in brown adipose tissue. Am J Physiol-Reg I. 2011;300:R1115–R1125. doi: 10.1152/ajpregu.00806.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B, Coulter A, Rim JS, Koza RA, Kozak LP. Transcriptional synergy and the regulation of Ucp1 during brown adipocyte induction in white fat depots. Mol Cell Biol. 2005;25:8311–8322. doi: 10.1128/MCB.25.18.8311-8322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.