Abstract
Aim
The aim of the present study was to determine the species and genotypes of Cryptosporidium spp. among children with diarrhea by PCR-RFLP using the TRAP-C2 gene.
Background
Cryptosporidium is a globally distributed protozoan parasite and one of the most common causes of infection and diarrhea in humans.
Patients and methods
Four hundred and sixty nine stool samples were collected from children less than 12 years with diarrhea who had been referred to Pediatrics Medical Centers in Gazvin provinces. The presence of Cryptosporidium oocysts was determined by Ziehl-Neelsen acid fast staining, then, genomic DNA was extracted from positive samples and nested PCR-RFLP was performed to amplify the TRAP-C2 gene.
Results
The overall prevalence of Cryptosporidium infection in children was 2.5 %. Results of nested PCR amplification showed that of 12 positive children samples, 10 (83.3%) were belonged to C. parvum, followed by C. hominis in 1 (8.3%) and mixed infection in 1 isolate (8.3%).
Conclusion
This study showed that Cryptosporidium parvum (the zoonotic genotypes) is more prevalent than other Cryptosporidium species in children from this area. This suggests that zoonotic transmission is the main mode of transmission of Cryptosporidium infection in Iran.
Keywords: Cryptosporidium, Genotypes, TRAP-C2 gene
Introduction
Cryptosporidium is a protozoa infection which is a cause of diarrhea in children and immuno-compromised patients throughout the world. Currently 18 Cryptosporidium species have been recognized. Cryptosporidium parvum and C. hominis are the species predominantly found in humans. Other species found in a variety of hosts includes C. meleagridis, C. felis, C. canis, C. suis, C. muris, C. andersoni, C. baileyi (1).
In recent years, researchers have developed PCR-based techniques for detection and identification of Cryptosporidium spp. These techniques are based on the polymorphic nature of the Cryptosporidium oocyst wall protein (COWP), the dihydrofolate reductase (DHFR), thrombospondin related adhesive protein 1 (TRAP-C1), thrombospondin-related adhesive protein 2 (TRAP-C2), internally transcribed spacer 1 (ITS1), polythreonine repeat (Poly-T), small-subunit (SSU) rRNA genes and undefined genomic sequences among strains that infect humans and most animals at (2–6). Previous studies of cryptosporidiosis in Iran have been performed using microscopy (7–12) and recently studies of Cryptosporidium molecular characterization have been conducted (13–14).
In this study we identified the genotypes of the Cryptosporidium isolates from clinical samples using polymerase chain reaction (PCR) amplification and restriction fragment length polymorphism (RFLP) analyses of the TRAP-C2 gene (Thrombospondin-Related Adhesive Protein of Cryptosporidium-2).
Materials and Methods
Sampling
A total of 469 fecal samples were collected from children with diarrhea who had been referred to Pediatric Medical Centers in Ghazvin, Iran. Cryptosporidium oocysts were identified in samples after concentration by formalin–ethyl– acetate sedimentation and staining with a modified Zeihl-Neelsen technique. The positive Cryptosporidium spp. isolates were preserved in 2.5% potassium dichromate and kept at 4°C until used for DNA extraction.
DNA extraction
Approximately 300 μl of fecal suspension was washed three times with distilled water to remove trace of dichoromate and then genomic DNA was extracted using the DNAzol kit according to the manufacturer’s instructions (Invitrogen, life technologies, Cat. No 10503-027, USA) with the addition of three freeze-thaw cycles (10 minutes) after resuspending the DNA samples in lysis (to rupture the Cryptosporidium oocysts). The oocysts were frozen in liquid nitrogen. Thawing was carried out at 90° C in water bath.
TRAP-C2 gene amplification
Nested PCR was used to amplify a fragment (266-366 bp) of the TRAP-C2 gene using two sets of oligonucleotideprimers: CATATTCCCTGTCCCTTGAGTTGT and TGGACAACCCAAA TGCAGAC for primary PCR and GGTAATTGGTCACGA and CCAAGTTCAGGCTTA for secondary PCR, as described previously (2, 5). PCR products were visualized on 1% agarose gel after ethidium bromide staining.
RFLP analysis
For restriction fragment length polymorphism (RFLP) analysis of secondary PCR products, we used restriction enzymes BstEII, HaeIII for TRAP-C2 gene. The restriction digestion products were visualized by electrophoresis on 2% agarose gel after ethidium bromide staining.
Results
In this study Cryptosporidium isolated from children with diarrhea in Gazvin province of Iran were characterized by PCR-RFLP. From all isolates thought to possibly contain Cryptosporidium, oocysts were detected and then stained by modified Ziehl-Neelsen method.
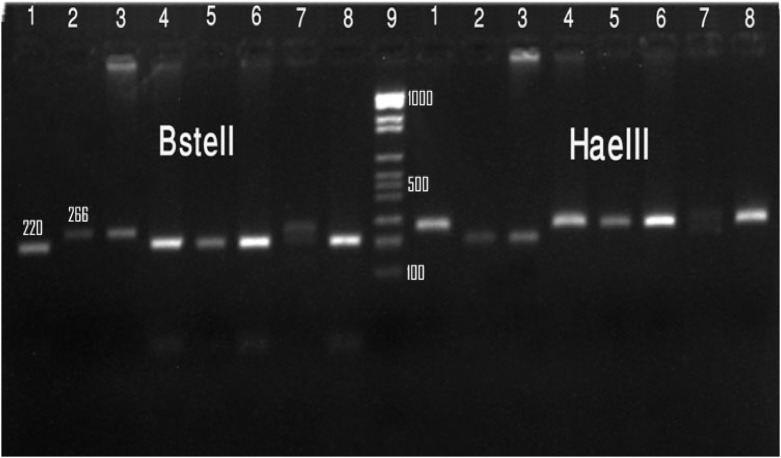
From the 469 samples, a total of 12 Cryptosporidium isolates were characterized. The TRAP-C2 gene fragment of all isolates was successfully amplified from 12 positive samples. RFLP analysis of the nested PCR products by using two enzymes (BstEII , HaeIII) showed that 10 out of 12 isolates (83.3%) were C. parvum, 1 (8.3%) C. hominis and one isolate (8.3%) showed mix infection pattern of C. parvum and C. hominis (Fig. 1 and Fig. 2).
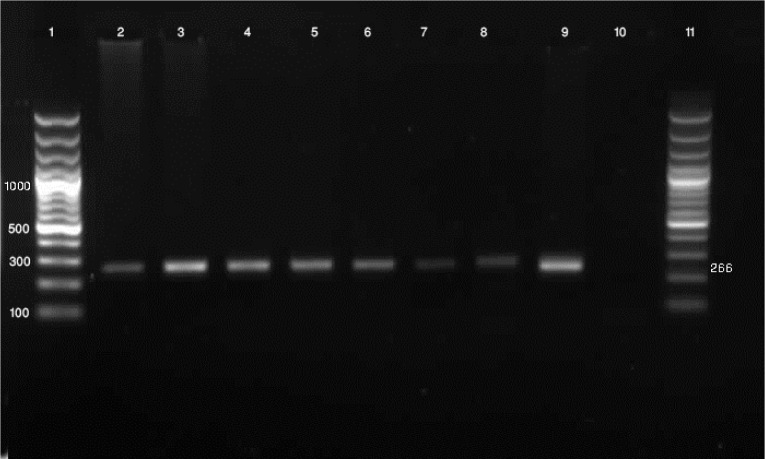
Figure 1.
Molecular diagnosis of Cryptosporidium spp. by a nested PCR based on TRAP-C2 gene. Lane 1: 100 bp
Figure 2.
Gel electrophoresis of Cryptosporidium species with TRAP-C2 gene based on PCR-RFLP technique by digestion of the secondary PCR products in clinical isolates with HaeIII, BstEII enzyme. Lane 1, 4, 5, 6, 8: C. parvum, Lane 2, 3: C. huminis, Lane 7: mixed infection both genotype
DNA marker, Lane 2-9: PCR product, Lane10: negative control
Discussion
In developing countries, the association of Cryptosporidium with acute and persistent diarrhea in children is well recognized. To date, a variety of genotypic markers are available to differentiate some species and subspecies within the genus Cryptosporidium. Nested-PCR/RFLP for TRAP-C2 gene has been reported as a recognized technique for the differentiation of strains of Cryptosporidium The ability to genotype each of the 12 Cryptosporidium isolates in this study reflects the specificity and sensitivity of the PCR-RFLP method, as described previously (2, 4–5).
High rates of infection have been reported in Egypt (17%), Uganda (5.9%), Kenya (25%), Turkey (3.5%), Pakistan (10.3%) and 8.2% in Indonesia (14–19). Previous studies performed in children within Iran have reported a prevalence of 2.4 % in Tehran, 11.6% in Kermanshah province and 2 % in Shahrekord province (1, 8–9).
A recent genotype analysis of Cryptosporidium among HIV positive and negative patients in Iran using 18s rRNA gene showed that 76% of isolates was C. parvum and 24% of the isolates was belonged to C. hominis (12).
In the present study the prevalence of Cryptosporidium in children was 2.25 %. In our study the RFLP analysis indicated that C. parvum, with a rate of 83.4 %, is the predominant species in children. Our study is therefore in agreement with a previous similar study performed in Iran (1); and the results are comparable to other studies performed in countries such as the United Kingdom, Kuwait, France, Switzerland and the Netherlands (20–24).
The predominance of C. parvum in humans may be due to high prevalence of bovine cryptosporidiosis in this area. By comparison C. hominis is the predominant species found in similar studies in other nations such as Brazil, Kenya, Malawi, the United States, Thailand, Japan and South Africa (25–28). We can conclude that the C.parvum is the predominant cause of cryptosporidium infection in Iran.
Furthermore the use of TRAP-C2 primers could be an alternative diagnostic method to identify human infection with Cryptosporidium. Information generated from this diagnostic approach would be useful, not only in diagnosis and identification of the sources of contamination, but also in controlling the disease.
Further molecular characterization on human and animals is needed, to increase our knowledge about Cryptosporidium and its transmission.
Acknowledgements
We acknowledge the very helpful assistance of Prof. Lihua Xiao from Division of Parasitic Diseases, National Center for Infectious Diseases, Centers for Disease Control and Prevention, Public Health Services, U.S.
(Please cite as: Nazemalhosseini Mojarad E, Keshavarz A, Taghipour N, Haghighi N, Kazemi B, Athari A. Genotyping of Cryptosporidium spp. in clinical samples: PCR-RFLP analysis of the TRAP-C2 gene. Gastroenterology and Hepatology From Bed to Bench 2011; 4(1): 29-33).
References
- 1.Keshavarz A, Athari A, Haghighi A, Kazami B, Abadi A, Nazemalhosseini Mojarad E, et al. Genetic characterization of Cryptosporidium spp. among children with diarrhea in Tehran and Qazvin provinces, Iran. Iranian J Parasitol. 2008;3:30–36. [Google Scholar]
- 2.Sulaiman IM, Lal AA, Xiao L. Molecular phylogeny and evolutionary relationships of Cryptosporidium parasites at the actin locus. J Parasitol. 2002;88:388–94. doi: 10.1645/0022-3395(2002)088[0388:MPAERO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 3.Kato S, Lindergard G, Mohammed HO. Utility of the Cryptosporidium oocysts wall protein (COWP) gene in a nested PCR approach for detection infection in cattle. Vet Parasitol. 2003;111:153–59. doi: 10.1016/s0304-4017(02)00353-9. [DOI] [PubMed] [Google Scholar]
- 4.Xiao L, Ryan UM. Cryptosporidiosis: an update in molecular epidemiology. Curr Opin Infect. 2004;17:483–90. doi: 10.1097/00001432-200410000-00014. [DOI] [PubMed] [Google Scholar]
- 5.Peng MM, Xiao L, Freeman AR, Arwood MJ, Escalante AA, Weltman AC, et al. Genetic polymorphism among Cryptosporidium parvum isolates evidence of two distinct human transmission cycles. Emerg Infect Dis. 1997;3:567–73. doi: 10.3201/eid0304.970423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spano F, Putignani L, Guida S, Crisanti A. Cryptosporidium parvum: PCR-RFLP analysis of the TRAP-C1 (Thrombospondin-Related adhesive protein of Cryptosporidium-1) gene discriminates between two alleles differentially associated with parasite isolates of animal and human origin. Experment Parasitol. 1998;90:195–98. doi: 10.1006/expr.1998.4324. [DOI] [PubMed] [Google Scholar]
- 7.Moghadam AA. Symptomatic and asymptomatic cryptosporidiosis in young children in Iran. Pak J biol Sci. 2008;10:1108–12. doi: 10.3923/pjbs.2007.1108.1112. [DOI] [PubMed] [Google Scholar]
- 8.Mirzaei M. Prevalence of Cryptosporidium spp. Infection in diarrheic and non diarrheic human in Iran. Korean J Parasitol. 2007;45:133–37. doi: 10.3347/kjp.2007.45.2.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khalili B, Mardani M. Frequency of Cryptosporidium and risk factors related to cryptosporidiosis in under 5–year old hospitalized children due to diarrhea. Iranian J Clil Infec Dis. 2009;4:151–55. [Google Scholar]
- 10.Hoghooghi-Rad N. Some epidemiological aspects of cryptosporidiosis in Ahvaz, center of Khoozestan Province, Islamic Republic of Iran. Med J Islam Repub Iran. 1994;1:17–22. [Google Scholar]
- 11.Nouri M, Moghadam A, Haghighatnia H. Cryptosporidium infection in human diarrhea patients in West Azerbaijan, Iran. Med J Islam Repub Iran. 1991;2:35–38. [Google Scholar]
- 12.Meamar AR, Rezaian M, Rezaie S, Mohraz M, Mohebali M, Mohammad K, et al. SSU-rRNA gene analysis of Cryptosporidium spp. in HIV positive and negative patients. Iranian J Publ Health. 2006;35:1–7. [Google Scholar]
- 13.Pirestani M, Sadraei J, Dalimi asl A, Zavvar M, Vaeznia H. Molecular characterization of Cryptosporidium isolates from human and bovine using 18s rRNA gene in Shahriar county of Tehran, Iran. Parasitol Res. 2008;103:467–72. doi: 10.1007/s00436-008-1008-2. [DOI] [PubMed] [Google Scholar]
- 14.Abdel-Messih IA, Wierzba TF, Abu-Elyazeed R, Ibrahim AF, Ahmed SF, Kamal K, et al. Diarrhea associated with Cryptosporidium parvum among young children of the Nile River Delta in Egypt. J Trop Pediatr. 2005;51:154–59. doi: 10.1093/tropej/fmh105. [DOI] [PubMed] [Google Scholar]
- 15.Tumwine JK, Kekitiinwa A, Bakeera-Kitaka S, Ndeezi G, Downing R, FengX, et al. Cryptosporidiosis and microsporidiosis in Ugandan children with persistent diarrhea with and without concurrent infection with the human immunodeficiency virus. Am J Trop Med Hyg. 2005;73:921–25. [PubMed] [Google Scholar]
- 16.Gatei W, Wamae CN, Mbae C, Mulinge E, Waithera T, Gatika SM, et al. Cryptosporidiosis: prevalence, genotype analysis and symptoms associated with infections and children in Kenya. Am J Trop Med Hyg. 2006;75:78–82. [PubMed] [Google Scholar]
- 17.Akyon Y, Erguven S, Arikan S, Yurdakok K, Gunalp A. Cryptosporidium parvum prevalence in a group of Turkish children. Turk J Pediatr. 1999;41:189–96. [PubMed] [Google Scholar]
- 18.Iqbal J, Munir MA. Cryptosporidium infection in young children with diarrhea in Rawalpindi, Pakistan. Am J Trop Med Hyg. 1999;60:868–70. doi: 10.4269/ajtmh.1999.60.868. [DOI] [PubMed] [Google Scholar]
- 19.Katsumata T, Hosea D, Wasito EB, Kohno S, Soeparto P, Ranuh IG. Cryptosporidiosis in Indonesia: a hospital based study and a community based survey. Am J Trop Med Hyg. 1998;59:628–32. doi: 10.4269/ajtmh.1998.59.628. [DOI] [PubMed] [Google Scholar]
- 20.McLauchlin J, Amar C, Pedraza-Diaz S, Nichols GL. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1705 fecal samples from humans and 105 fecal samples from livestock animals. J Clin Microbiol. 2000;38:3984–90. doi: 10.1128/jcm.38.11.3984-3990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sulaiman IM, Hira PR, Zhou L, Al-Ali FM, Al-Shelahi FA, Shweiki HM, et al. Unique endemicity of cryptosporidiosis in children in Kuwait. J Clin Microbiol. 2005;43:2805–809. doi: 10.1128/JCM.43.6.2805-2809.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guyot K, Follet-Dumoulin A, Lelievre E, Sarfati C, Rabodonirina M, Nevez G, et al. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J Clin Microbiol. 2001;39:3472–80. doi: 10.1128/JCM.39.10.3472-3480.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan U, Weber R, Xiao L, Sulaiman I, Thompson RCA, Ndiritu W, et al. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United states. J Clin Microbiol. 2000;38:1180–83. doi: 10.1128/jcm.38.3.1180-1183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wielinga PR, de Vries A, Van der Goot TH, Mank T, Mars MH, Kortbeek LM, et al. Molecular epidemiology of Cryptosporidium in humans and cattle in the Netherlands. Int J Parasitol. 2008;38:809–17. doi: 10.1016/j.ijpara.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 25.Gatei W, Greensill J, Ashford RW, Cuevas LE, Parry CM, Cunliffe NA, et al. Molecular analysis of the 18S rRNA gene of Cryptosporidium parasites from patients with or without human immunodeficiency virus infections living in Kenya, Malawi, Brazil, the United states, and Vietnam. J Clin Microbiol. 2003;41:1458–62. doi: 10.1128/JCM.41.4.1458-1462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samie A, Bessong PO, Obi CL, Sevilleja JE, Stroup S, Houpt E, et al. Cryptosporidium species: preliminary descriptions of the prevalence and genotype distribution among school children and hospital patients in the Venda region, Limpopo Province, South Africa. Exp Parasitol. 2006;114:314–22. doi: 10.1016/j.exppara.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 27.Tiangtip R, Jongwutiwes S. Molecular analysis of Cryptosporidium species isolated from HIV-infected patients in Thailand. Trop Med Int Health. 2002;7:357–64. doi: 10.1046/j.1365-3156.2002.00855.x. [DOI] [PubMed] [Google Scholar]
- 28.Yagita K, Izumiyama S, Tachibana H, Masuda G, Iseki M, Furuya K, et al. Molecular characterization of Cryptosporidium isolates obtained from human and bovine infections in Japan. Parasitol Res. 2001;87:950–55. doi: 10.1007/s004360100480. [DOI] [PubMed] [Google Scholar]