Abstract
Aim
In this study, genetic polymorphism of two tRNA-liked short tandem repeat (STR)-containing loci, R-R and S-Q, was analyzed in order to clarify further the genotypic differences among E. dispar isolates.
Background
Entamoeba dispar is closely related to the human pathogen E. histolytica, the agent of amebic dysentery and amebic liver abscesses. E. dispar is, to some extent, capable of producing variable focal intestinal lesions in animals and of destroying epithelial cell monolayers in vitro, and some have reported it to be capable of producing amoebic liver abscess in hamsters. However no evidence exists at present to link E. dispar with human disease.
Patients and methods
A total of 28 E. dispar samples from gastrointestinal disorder patients were characterized using PCR and sequencing. The sequences obtained were edited manually and aligned.
Results
Sequence analysis showed 9 and 6 different patterns of units in the repeat-containing region of R-R and S-Q, respectively. The repeat-containing regions of R-R and S-Q loci were found to be extensively polymorphic, varying in size, number and order of repeat units.
Conclusion
The results demonstrate extensive genetic variability among Iranian E. dispar clinical isolates. The genetic diversity of tRNA gene-linked STR loci shows them to be suitable for epidemiological studies such as the characterization of the routes of transmission of these parasites in Iran.
Keywords: E. dispar, STRs, Genetic diversity, Iran
Introduction
In 2005, the genome sequence of the Entamoeba histolytica strain most widely used in research laboratories, HM-1:IMSS, was completed (1). One of the striking findings was the abundance and unique organization of the tRNA genes (2). Over 10% of all the sequence reads contained tRNA genes and almost all were organized in tandem arrays. The intergenic regions are rich in A + T (about 80%) and contain non-coding short tandem repeats (STRs) (3). Diversity in these loci among E. histolytica strains are mainly due to varying numbers of STRs (4).
When a comparison is made between E. histolytica and E. dispar the loci vary not only in the number but also in the sequence and arrangement of STRs (5, 6). Ali et al. (2005) have described a system for typing E. histolytica strains based on 6 of the tRNA gene-linked STRs (7). Using this method, a recent study of E. histolytica samples collected from asymptomatic, diarrhea/dysenteric and amebic liver abscess patients in Bangladesh revealed that the parasite genotype prevalences identified in the 3 groups were significantly different from each other, suggesting that the parasite genotype plays a role in the outcome of infection in humans (8).
E. dispar is closely related to E. histolytica but has never been documented to cause disease in humans, although a recent report suggests that a few strains may be able to produce liver abscesses in hamsters (9). Genetic variation in E. dispar has not been widely studied to date. To investigate this, we have been comparing the structure of STR loci in E. dispar using strains from Iran. In this study, genetic polymorphism at two tRNA-liked STR-containing loci, R-R and SQ, was analyzed in Iranian E. dispar isolated from patients with GI symptoms using PCR and sequencing methods.
Patients and Methods
A total of 28 E. dispar strains were analyzed. Clinical information on the samples is given in Table 1. All the samples used in this study were diagnosed as positive for Entamoeba spp. by microscopic examination of fresh stools using direct smears, formalin-ether concentrated, and trichrome stained specimens (10).
Table 1.
Background of E. dispar isolates
| No. | Isolates | Isolation | Clinical symptoms | Sex | Age (yrs) | |
|---|---|---|---|---|---|---|
| date | location | |||||
| 1 | NH1IR | 2006 | Tehran | Abdominal pain, diarrhea | F | 20 |
| 2 | NH2IR | 2006 | Tehran | Abdominal pain | M | 6 |
| 3 | NH3IR | 2006 | Tehran | Abdominal pain, bloating | M | 22 |
| 4 | NH4IR | 2006 | Tehran | Abdominal pain | M | 32 |
| 5 | NH5IR | 2006 | Tehran | Abdominal pain, vomiting | F | 27 |
| 6 | NH6IR | 2006 | Tehran | Abdominal pain | M | 63 |
| 7 | NH7IR | 2007 | Tehran | Diarrhea, bloating | M | 33 |
| 8 | NH8IR | 2007 | Tehran | Abdominal pain, diarrhea | F | 24 |
| 9 | NH9IR | 2007 | Tehran | diarrhea | F | 36 |
| 10 | NH10IR | 2007 | Tehran | Abdominal pain | F | 38 |
| 11 | NH11IR | 2007 | Tehran | Abdominal pain, bloating | F | 63 |
| 12 | NH12IR | 2007 | Tehran | Abdominal pain | M | 64 |
| 13 | NH13IR | 2007 | Tehran | Abdominal pain | M | 42 |
| 14 | NH14IR | 2007 | Tehran | Abdominal pain, vomiting | M | 54 |
| 15 | NH15IR | 2007 | Tehran | Abdominal pain | M | 53 |
| 16 | NH16IR | 2007 | Tehran | Abdominal pain, bloating | F | 8 |
| 17 | NH17IR | 2007 | Tehran | Diarrhea, vomiting | M | 14 |
| 18 | NH18IR | 2007 | Tehran | Abdominal pain | F | 12 |
| 19 | NH19IR | 2007 | Tehran | Abdominal pain, vomiting | F | 20 |
| 20 | NH20IR | 2007 | Tehran | Abdominal pain, diarrhea | F | 31 |
| 21 | NH21IR | 2007 | Tehran | Abdominal pain, diarrhea | F | 8 |
| 22 | SHN3IR | 2004 | Zahedan | Abdominal pain | F | 25 |
| 23 | SHN4IR | 2004 | Zahedan | Abdominal pain, vomiting | M | 42 |
| 24 | SHN7IR | 2004 | Zahedan | Abdominal pain, vomiting | M | 32 |
| 25 | NHM1IR | 2005 | Gonbad | Asymptomatic | F | 28 |
| 26 | NHM2IR | 2005 | Gonbad | Asymptomatic | M | 31 |
| 27 | NHM3IR | 2005 | Gonbad | Asymptomatic | M | 31 |
| 28 | NHM4IR | 2005 | Gonbad | Asymptomatic | M | 31 |
The genomic DNA was extracted directly from stool and samples were identified as E. dispar by loci D-A and A-L based PCR analysis, as previously described (11, 12). For genotype analysis, loci R-R and S-Q were amplified by PCR using two E. dispar specific pairs of oligonucleotides previously described (7). PCR products were analyzed by electrophoresis using 1.8% agarose gels (Fermentas, #R0491) in Tris-boric acid-EDTA buffer containing ethidium bromide after which the gels were photographed under ultraviolet light (UVIdoc, UVItec Limited, Cambridge, United Kingdom). The PCR products were sequenced using the amplification primers and an Applied Biosystems (ABI) BigDye® Terminator V3.1 Cycle Sequencing Kit, and analysed on an ABI 3130xl Genetic Analyzer.
The sequences obtained were edited manually and aligned using Gene Runner software (version 3.05). Nucleotide sequences, except for the forward and reverse primer regions, were aligned with the only previously available locus R-R and S-Q sequences from E. dispar in GenBank (EF421343 and AY842971). All new sequences were submitted to the GenBank/EMBL/DDBJ database under accession numbers HQ439911-30 for S-Q Loci and HQ439959-83 for R-R loci.
Results
Polymorphism in nucleotide sequences of the noncoding STR locus R-R
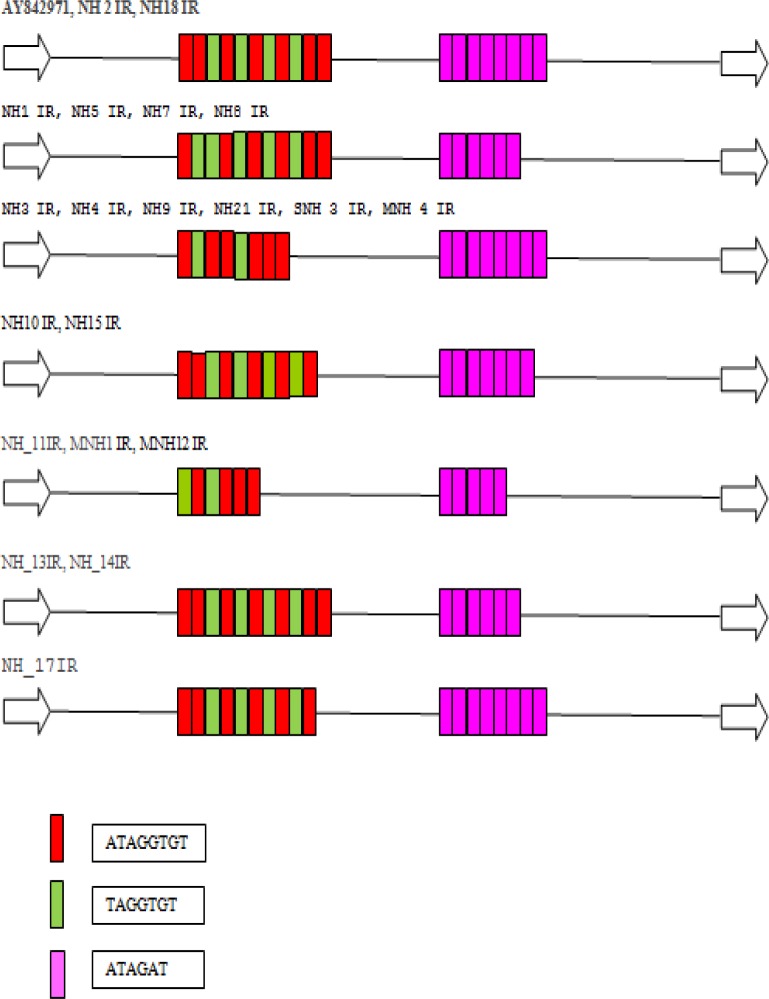
In order to better understand the nature of the polymorphisms among the Iranian strains, we amplified and sequenced the individual products from locus R-R from all 28 isolates. PCR of the Iranian isolates amplified a product of between 586 and 726 bp in all 28 samples. Sequences revealed a complex inter-isolate polymorphism in length, location, and number of the repeat units (Figure 1). Based on the sequences of locus R-R, the 28 Iranian isolates were divided into 9 distinct types (a to h), with genotype ‘h’ being the dominant type (35.7%).
Figure 1.
Schematic representation of the STR polymorphisms in locus S-Q of E. dispar. The 6 distinct sequence types are shown as well as the identification tag for the isolates that matched each type; also shown is the structure of locus S-Q sequence in the standard isolate, E. dispar SAW760 (AY842971). The sequences of each of the 3 repeat types are shown beside their corresponding colored block. The conserved non-repeated regions are shown as a single line.
Polymorphism in nucleotide sequences of the noncoding DNA locus S-Q
PCR of locus S-Q in Iranian isolates amplified a fragment of between 381 and 437 bp in 20 samples. No amplification was observed at this locus in 8 isolates. Polymorphisms in the type, location, and number of repeat units were observed in the repeat-containing region of locus S-Q (Figure 2).
Figure 2.
Schematic representation of the STR polymorphisms in locus R-R of E. dispar. The 9 distinct sequence types are shown as well as the identification tag for the isolates that matched each type; also shown is the structures of locus R-R sequence in the standard isolate, E. dispar SAW760 (EF421343). The sequences of each of the nine repeat types are shown beside their corresponding colored block. The conserved non-repeated regions are shown as a single line.
However, locus S-Q appeared to be less polymorphic than locus R-R. Based on the nucleotide sequences obtained, the 20 Iranian isolates were divided into six distinct S-Q types (1 to 6), with genotype 5 being the dominant type (30%).
Discussion
The ability to identify strains of Entamoeba dispar may lead to insights into the population structure and epidemiology of the organism. Zaki et al. (5, 13) showed that using two STR-loci in combination allowed differentiation of a majority of the E. histolytica and E. dispar isolates studied based on product size, and they proposed that these loci had the potential to be used as polymorphic molecular markers for investigating the epidemiology of these organisms. Independently and using a repeat region of a gene coding for a serine-rich antigen (SREHP), Ayeh-Kumi et al. (2001) showed the majority of E. histolytica could be differentiated using the repeat region of this gene and that samples from liver abscess patients had polymorphisms which were not present in the intestinal isolates from the same geographic area (14).
From 79 E.histolytica strains isolated from patients with and without symptom in at least 8 different countries, Haghighi et al. (2002, 2003) sequenced PCR products from 4 loci. Limited PCR product size variation has seen in the chitinase gene but fairly superior sequence variety, with a total of 9 sequence types and an identical number of predicted peptide sequences. Thirty seven sequence types and 31 different peptide sequences seen in the majority of polymorphism with SREHP gene. The two tRNA-linked STRs showed intermediate polymorphism with 13 to 15 sequence types.
Outside of the repeat regions in the 4 loci, no single nucleotide polymorphisms (SNPs) were detected, and also there was no correlation between clinical and sequence types (15, 16).
As the studied populations in the studies of Haghighi et al. collected from different geographically regions and obtained during consequences years, they did not detect a link between genotype and symptoms. In contrast Ali et al. (2007) found dissimilarity among sample groups used a geographically and temporally control group (8). Eventually any correlations would be lost as new genotypes arise in the population due to instability of criteria that can measure. Therefore, they must be mutating to generate the diversity in populations.
In the current study the links between diversity and virulence have shown using size and sequencing of tRNA-linked STRs.
Flow samples of stool and liver abscess from 18 ALA patients from USA, Italy and Bangladesh showed that the genotypes of the intestinal amebae were dissimilar to the corresponding flow samples of the same patient (3). The mechanism of this result is vague, but it suggests that only small numbers of multiple genotypes in intestinal could migrate to the liver or recombination of DNA procedures are arranged prior to or during migration of the amebae from the intestine to the liver.
The genetic polymorphisms in loci D-A and A-L in 28 isolates of E. dispar from three different geographic regions of Iran were previously studied using PCR and sequencing. The sequenced products revealed 12 and 7 novel E. dispar genotypes respectively and showed the loci to have the potential for use in epidemiological studies, such as the identifying the routes of transmission of these parasites in Iran (11, 12).
Use of an additional two targets for amplification in this study (loci S-Q and R-R) means that fingerprinting of 28 Iranian isolates has been completed at four loci (Table 2).
Table 2.
Fingerprinting of 28 Iranian isolates at four loci (S-Q , R-R, A-L and D-A)
| No. | Isolates | Locus S-Q | Type | locus A-L | Type | Locus R-R | Type | Type | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | NH1IR | S-Q | 3 | HQ439911 | A-L | F | HQ439931 | R-R | f | HQ439959 | locus D-A | IV |
| 2 | NH2IR | S-Q | R | HQ439912 | A-L | F | HQ439932 | R-R | a' | HQ439960 | D | VII |
| 3 | NH3IR | S-Q | 5 | HQ439913 | A-L | F | HQ439933 | R-R | h | HQ439961 | D | II |
| 4 | NH4IR | S-Q | 5 | HQ439914 | A-L | B | HQ439934 | R-R | h | HQ439962 | D | I |
| 5 | NH5IR | S-Q | 3 | HQ439915 | A-L | A | HQ439935 | R-R | a | HQ439963 | D | I |
| 6 | NH6IR | - | ---- | A-L | E | HQ439936 | R-R | h | HQ439964 | D | VI | |
| 7 | NH7IR | S-Q | 3 | HQ439916 | A-L | C | HQ439937 | R-R | a | HQ439965 | D | IV |
| 8 | NH8IR | S-Q | 3 | HQ439917 | A-L | C | HQ439938 | R-R | a | HQ439966 | D | III |
| 9 | NH9IR | S-Q | 5 | HQ439918 | A-L | B | HQ439939 | R-R | h | HQ439967 | D | VI |
| 10 | NH10IR | S-Q | 4 | HQ439919 | A-L | A | HQ439940 | R-R | c | HQ439968 | D | III |
| 11 | NH11IR | S-Q | 6 | HQ439920 | A-L | B | HQ439941 | R-R | c | HQ439969 | D | III |
| 12 | NH12IR | - | ---- | A-L | C | HQ439942 | R-R | d | HQ439970 | D | X | |
| 13 | NH13IR | S-Q | 2 | HQ439921 | A-L | A | HQ439943 | R-R | g | HQ439971 | D | V |
| 14 | NH14IR | S-Q | 2 | HQ439922 | A-L | D | HQ439944 | R-R | b | HQ439972 | D | I |
| 15 | NH15IR | S-Q | 4 | HQ439923 | A-L | E | HQ439945 | R-R | d | HQ439973 | D | X |
| 16 | NH16IR | - | ---- | A-L | G | HQ439946 | R-R | d | HQ439974 | D | IV | |
| 17 | NH17IR | S-Q | 1 | HQ439924 | A-L | E | HQ439947 | R-R | h | HQ439975 | D | I |
| 18 | NH18IR | S-Q | R | HQ439925 | A-L | E | HQ439948 | R-R | a | HQ439976 | D | III |
| 19 | NH19IR | - | ---- | A-L | B | HQ439949 | R-R | b | HQ439977 | D | X | |
| 20 | NH20IR | - | ---- | A-L | B | HQ439950 | R-R | h | HQ439978 | D | X | |
| 21 | NH21IR | S-Q | 5 | HQ439926 | A-L | A | HQ439951 | R-R | a '' | HQ439979 | D | X |
| 22 | SHN3IR | S-Q | 5 | HQ439930 | A-L | E | HQ439956 | R-R | h | HQ439984 | D | IX |
| 23 | SHN4IR | - | ------ | A-L | E | HQ439957 | R-R | h | HQ439985 | D | IX | |
| 24 | SHN7IR | - | ------ | A-L | E | HQ439958 | R-R | e | HQ439986 | D | XII | |
| 25 | NHM1IR | S-Q | 6 | HQ439927 | A-L | E | HQ439952 | R-R | h | HQ439980 | D | XI |
| 26 | NHM2IR | S-Q | 6 | HQ439928 | A-L | E | HQ439953 | R-R | h | HQ439981 | D | VII |
| 27 | NHM3IR | - | ------ | A-L | E | HQ439954 | R-R | e | HQ439982 | D | VII | |
| 28 | NHM4IR | S-Q | 5 | HQ439929 | A-L | E | HQ439955 | R-R | h | HQ439983 | D | XI |
Sequence analysis showed 9,12,7 and 6 different patterns based on variation of units in this repeat containing region of R-R, D-A, A-L and S-Q loci, respectively.
The tRNA gene regions in loci S-Q and R-R similar to loci D-A and A-L are conserved and are the site of the primers used, but in the middle there are repeat units of between 6 and 8 nucleotides which vary among isolates.
Elimination, duplication and substitution of units in this repeat-containing region are the basis of polymorphisms detected in the two species.
By the simultaneous investigation of locus A-L and locus D-A (11, 12), 26 subtypes out of 28 E.dispar isolates were distinguished (the molecular patterns of NH19IR and NH20IR, also NHM2IR and NHM3IR are not different in two loci).
NH 19IR and NH 20IR isolated from Tehran, SHN 3IR and SHN 4IR isolated from Zahedan, NHM 1IR and NHM 2IR isolated from Gonbad and also NHM 1IR and NHM 4IR are similar in 3 loci and differentiation of those isolates based on fourth loci are possible.
The results demonstrate an extensive genetic variability among E. dispar clinical isolates. The repeat containing regions of R-R, D-A, A-L and S-Q loci was found extensively polymorphic in size, number and also in order of repeat units. The genetic diversity of tRNA Gene-Linked short Repeat loci shows them to be suitable for epidemiological studies.
In conclusion, we propose that molecular typing and analysis of genotypes of E. histolytica and E. dispar isolates from a variety of locations will help in determining the geographic origins of isolates and routes of transmission.
Acknowledgments
The authors would like to express special thanks to Dr. C. Graham Clark from the Department of Infectious and Tropical Diseases, London School of Hygiene and Tropical Medicine, Keppel Street, London WC1E 7HT, England for editing and helpful comments on this manuscript. This work was financially supported by Gastroenterology and Liver Diseases Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran, by grant number 505.
(Please cite as: Nazemalhosseini Mojarad E, Azimirad M, Bayat M, Hellaly A, Asadzadeh Aghdaei H, Mohaghegh Shalmani H. Polymorphism in two short tandem repeat loci (R-R and S-Q) linked to tRNA genes in Entamoeba dispar isolates. Gastroenterol Hepatol Bed Bench 2012;5(4):202-208).
References
- 1.Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, et al. The genome of the protest parasite Entamoeba histolytica . Nature. 2005;433:865–68. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 2.Clark CG, Ali IKM, Zaki M, Loftus BJ, Hall N. Unique organisation of tRNA genes in Entamoeba histolytica . Mol Biochem Parasitol. 2006;146:24–29. doi: 10.1016/j.molbiopara.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 3.Ali IKM, Solaymani-Mohammadi S, Akhter J, Roy S, Gorrini C, Calderaro A, et al. Tissue invasion by Entamoeba histolytica: evidence of genetic selection and/or DNA reorganization events in organ tropism. PLoS Negl Trop Dis. 2008;2:e219. doi: 10.1371/journal.pntd.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaki M, Clark CG. Isolation and characterization of polymorphic DNA from Entamoeba histolytica. J Clin Microbiol. 2001;39:897–905. doi: 10.1128/JCM.39.3.897-905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zaki M, Meelu P, Sun W, Clark CG. Simultaneous differentiation and typing of Entamoeba histolytica and Entamoeba dispar . J Clin Microbiol. 2002;40:1271–76. doi: 10.1128/JCM.40.4.1271-1276.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tawari B, Ali IKM, Scott C, Quail MA, Berriman M, Hall N, et al. Patterns of evolution in the unique tRNA gene arrays of the genus Entamoeba. Mol Biol Evol. 2008;25:187–198. doi: 10.1093/molbev/msm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ali IKM, Zaki M, Clark CG. Use of PCR amplification of tRNA gene-linked short tandem repeats for genotyping Entamoeba histolytica . J Clin Microbiol. 2005;43:5842–47. doi: 10.1128/JCM.43.12.5842-5847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ali IKM, Mondal U, Roy S, Haque R, Petri WA, Jr, Clark CG. Evidence for a link between parasite genotype and outcome of infection with Entamoeba histolytica . J Clin Microbiol. 2007;45:285–89. doi: 10.1128/JCM.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shibayama M, Dolabella SS, Silva EF, Tsutsumi V. A Brazilian species of Entamoeba dispar (ADO) produces amoebic liver abscess in hamsters. Ann Hepatol. 2007;6:117–18. [PubMed] [Google Scholar]
- 10.Nazemalhosseini-Mojarad E, Nochi Z, Sahebekhtiari N, Dabiri H, Rostami-Nejd M, Zali MR, et al. Discrimination of Entamoeba moshkovskii in patients with gastrointestinal disorders by singleround PCR. Jpn J Infect Dis. 2010;63:136–38. [PubMed] [Google Scholar]
- 11.Nazemalhosseini-Mojarad E, Haghighi A, Kazemi B, Rostami –Nejd M, Abadi A, Zali MR. High genetic diversity among Iranian Entamoeba dispar isolates based on the noncoding short tandem repeat locus D-A. Acta Trop. 2009;111:133–36. doi: 10.1016/j.actatropica.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Nazemalhosseini-Mojarad E, Azimirad M, Nochi Z, Romani S, Tajbakhsh M, Rostami-Nejad M, et al. Investigation of sequence diversity in tRNA gene Locus A-L among clinical isolates of Entamoeba dispar . Iranian J Parasitol. 2012;7:97–103. [PMC free article] [PubMed] [Google Scholar]
- 13.Zaki M, Reddy SG, Jackson TFHG, Ravdin JI, Clark CG. Genotyping of Entamoeba species in South Africa: diversity, stability, and transmission patterns within families. J Infect Dis. 2003;187:1860–69. doi: 10.1086/375349. [DOI] [PubMed] [Google Scholar]
- 14.Ayeh-Kumi PF, Ali IKM, Lockhart LA, Gilchrist CA, Petri WA, Jr, Haque R. Entamoeba histolytica: genetic diversity of clinical isolates from Bangladesh as demonstrated by polymorphisms in the serine-rich gene. Exp Parasitol. 2001;99:80–88. doi: 10.1006/expr.2001.4652. [DOI] [PubMed] [Google Scholar]
- 15.Haghighi A, Kobayashi S, Takeuchi T, Masuda G, Nozaki T. Remarkable genetic polymorphism among Entamoeba histolytica isolates from a limited geographic area. J Clin Microbiol. 2002;40:4081–90. doi: 10.1128/JCM.40.11.4081-4090.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haghighi A, Kobayashi S, Takeuchi T, Thammapalerd N, Nozaki T. Geographic diversity among genotypes of Entamoeba histolytica field isolates. J Clin Microbiol. 2003;41:3748–56. doi: 10.1128/JCM.41.8.3748-3756.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samie A, Obi CL, Bessong PO, Houpt E, Stroup S, Njayou M, et al. Entamoeba histolytica: genetic diversity of African strains based on the polymorphism of the serine-rich protein gene. Exp Parasitol. 2008;118:354–61. doi: 10.1016/j.exppara.2007.09.008. [DOI] [PubMed] [Google Scholar]