Abstract
Aim
The purpose of this study was to determine the relationship of rs4444903 (EGF+61A/G) SNP genotype with colorectal cancer and tumor stage in an Iranian population.
Background
Epidermal growth factor (EGF) is one of the important proteins that determine survival of cells. EGF binds to its receptor on the cell surface and then activates some of the cell signaling pathway networks within cells that lead to activation or deactivation of factors which are responsible for growth and apoptosis of cells. In this study we assessed the association in EGF polymorphism rs4444903 with colorectal cancer (CRC) in Iranian population.
Patients and methods
We conducted case-control study to investigate the association of polymorphism rs4444903 in EGF, with colorectal cancer risk in Iranian population. Analyzed Polymorphism of EGF rs4444903 with restriction fragment length polymorphisms (RFLP) among two groups of subjects consisting of including 220 cases with colorectal cancer and 220 healthy individuals as controls. Mutations were confirmed in 10% of the samples by direct sequencing.
Results
The frequencies of AA, AG and GG genotypes among cases with colorectal cancer were 28.2, 46.8, and 25.0 % respectively and in controls genotype frequencies were 23.2, 56.4, and 20.5 %, respectively. Frequency of A allele among case group was 51.6% and for control group was 51.4%. The frequency of G allele in case and control was, respectively 48.4% and 48.6% (OR= 1.009, 95% CI= 0.775-1.315; P= 0.946). The percentage of Stage 0, I, II, III, IV were 5%, 9.35%, 38.84%, 30.21% and 16.54%, respectively, among the cases. However, no significant association between this polymorphism and CRC stage was observed (p=0.626).
Conclusion
Our data suggest a SNP rs4444903 may not represent a risk factor in the development and progression of CRC among Iranian population.
Keywords: Colorectal cancer, Epidermal growth factor, rs4444903, EGF+61A/G, Single nucleotide polymorphism
Introduction
Colorectal cancer is the second cancer leading to death after lung cancer in the developed countries (1). In 1980, 5.8 percent of all types of cancers were colorectal cancer. In 2000, this cancer has been the fourth cancer after gastric, lung, and liver cancer in Iran. Many studies have indicated that colorectal cancer is increasing in Asians (2–6). The incidence of CRC has increased during the last 25 years in the Iranian population (7). CRC is the fifth most common cancer among Iranian men and third among women (8). According to the statistics from the Iranian Ministry of Health, 1130 patients, 450 women and 680 men, died in 2006 due to colorectal cancer (9).
Cancer is caused by genetic and environmental factors. The primary method of therapy is surgery. Finding new diagnostic markers may lead to better diagnosis and treatment (10). One of the most important cancer-related genes is epidermal growth factor (EGF). Epidermal growth factor is encoded by the EGF gene, mapped to chromosome 4q25. The longest transcript of EGF gene shows that it has 24 exons and 23 introns (11). When the EGF protein binds to its receptor (EGFR) on cell surface it activates a series of intracellular signaling networks including PI3K/AKT, Ras/Erk, JAK/STAT. These networks activate or deactivate some transcription factors regulating some proteins responsible for the death or survival of cell (12–14).
Single nucleotide polymorphisms(SNP) are the most prevalent sources of human genetic variation that may be associated with increasing risk of cancer (10). The rs4444903 (EGF+61A/G) polymorphism is one of the most important polymorphisms in EGF gene, located in the EGF 5'untranslated region (UTR). rs4444903 polymorphism contains a change of Guanine base (G) whit an Adenine base (A). In a recent study it was suggested that this change causes an increase in EGF expression (15). Epidemiologic studies, based on different tests, show a close correlation between rs4444903 polymorphism and different types of cancer (16–18). Recently a study in Iranian population reported rs4444903 polymorphism has no significant association with colorectal cancer, The stage of the colorectal cancer was not considered (19).
The aim of this study was to determine the association of rs4444903 SNP in developing colorectal cancer and its relationship to the tumor stage.
Patients and Methods
Study population
Peripheral blood samples of 220 patients suffering from colorectal cancer and 220 healthy individuals were taken. All patients with colorectal cancer were recruited from Taleghani Hospital, during the period 2006-2011. Colonoscopy was performed by a gastroenterologists and diagnosis was confirmed by a pathologist. Controls were recruited from healthy individuals volunteer. Written informed consent was obtained from all the subjects.
DNA extraction and genotyping
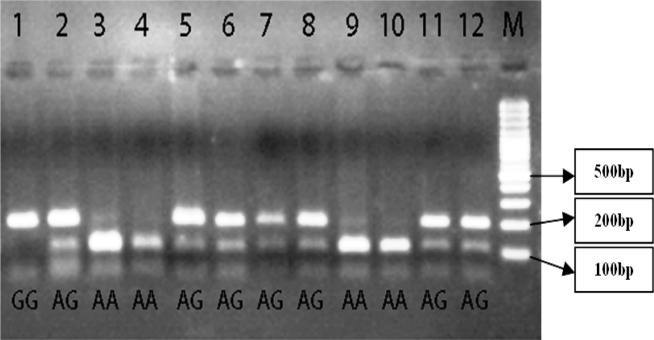
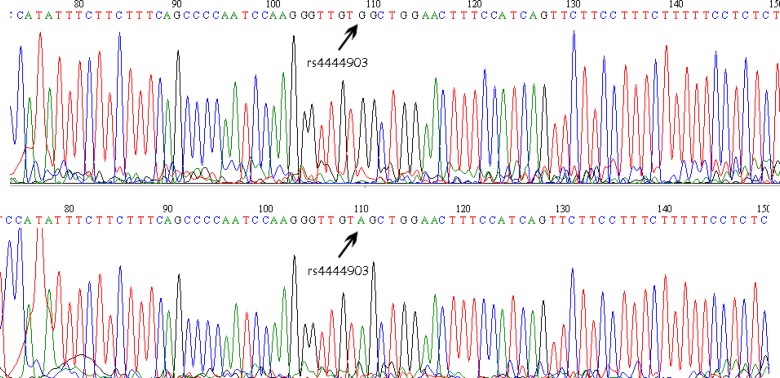
The samples were extracted using standard salting out method and the quality and quantity of DNA was evaluated by Nanodrop Spectrophotometer(20). Genotyping of rs4444903 polymorphism was performed by restriction fragment length polymorphisms analysis. To amplify DNA segment, the specific primers were used (Table 1) (18). The PCR cycle conditions consisted of an initial denaturation step at 95° C for 5 min followed by 30 cycles of 45 s at 95° C; 40 s at 60° C; 45 s at 72° C; and a final elongation at 72° C for 10 min. After PCR, product was run on in to 1% agarose gel in order to ensure a successful reproduction. The products were digested by AluI enzyme (New England Biolabs). The recognition and cutting point of this enzyme is AG/CT, in which SNP region is show by capital letters. Cutting point of enzyme can be observed by a diagonal line. After enzyme digestion, the allele A in 102+91+34+15 bp length, and allele G in 193+34+15 bp lengths were observed. The length of the genotype bands can be seen in the Table 2 (Figure 1). The digested PCR products were determined on a 3% agarose gel and stained with ethidium bromide for visualization under UV light. To confirm genotyping results, a 10% random sample representing all 3 genotypes was sequenced by automated DNA sequencing, using the ABI genetic analyzer 3130xl (Figure 2).
Table 1.
Primer sequence and resulting fragment length for growth factor gene polymerase chain reaction (PCR)
| Primer direction | Primer sequence | %GC | Resulting fragment bp | |
|---|---|---|---|---|
| 1 | Forward | 5'-TGTCACTAAAGGAAAGGAGGT-3' | 42.86 | 245 |
| 2 | Reverse | 5'-TTCACAGAGTTTAACAGCCC-3' | 45 |
Table 2.
The result of RFLP genotyping
| Row | Genotype | Restriction pattern length (bp) |
|---|---|---|
| 1 | AA | 102+91+34+15 |
| 2 | AG | 193+102+34+15 |
| 3 | GG | 193+34+15 |
Figure 1.
Electrophoresis digested products with AluI restricted enzyme on agarose gel showed different bonds. The marker (M) that used was 100 base pairs
Figure 2.
Direct DNA sequencing results for the EGF rs4444903, genotype GG and genotype AA were observed from the electropherogram.
Statistical analysis
The cases and controls were compared using a Student's t-test for the continuous variables and a χ2 test for the categorical variables. Hardy–Weinberg equilibrium (HWE) was tested using a goodness-of-fit χ2 test. Unconditional logistic regression analysis was used to calculate the odds ratios (ORs) and 95% confidence intervals (CIs), with an adjustment for possible confounders (gender, age and smoking). The statistical significance level consider lower than 0.05. The data were analyzed using of SPSS software (version 13).
Results
In this study 220 patients diagnosed with sporadic CRC were recruited. The patients had an average age of 61.02±12.09. 220 healthy subjects with average age of 41.99±14.35 were recruited as controls (Table 3). The genotype frequencies of the EGF rs4444903 A > G polymorphism among the cases and controls are shown in Table 4. Genotype and allele frequencies were in Hardy–Weinberg equilibrium in two groups. There was no significant deviation in the distribution of the genotypes between the cases and the controls. When the more common rs4444903 GG genotype was used as the reference, the rs4444903 AG and AA genotypes were not significantly associated with the risk of colorectal cancer (adjusted OR=0.727, 95% CI: 0.4 - 1.322; and adjusted OR=1.000, 95% CI: 0.502 - 1.992, respectively). Although the frequency of GG, AA genotypes were lower in stage IV and stage I respectively. There was no apparent association between the EGF polymorphism and stage of colorectal cancer (Table 5).
Table 3.
Characteristics of the study population
| Variable | CRC patients (n=220) | Controls (n=220) |
|---|---|---|
| Age ( years ± SD ) | 61.02±12.09 | 41.99±14.35 |
| Sex (%) | ||
| Male | 118 (53.6%) | 93 (41.5%) |
| Female | 102 (46.4%) | 127 (58.5%) |
| Smoking (%) | ||
| Ever | 15 (6.8%) | 14 (6.4%) |
| Never | 205(93.2%) | 206 (93.6%) |
Table 4.
The genotype and allele frequencies of EGF rs4444903 among CRC patients and controls
| Genotypes | CRC patients n=220(%) | Controls n=220(%) | P-value | Unadjusted OR (95%CI) | Adjusted OR (95%CI)* |
|---|---|---|---|---|---|
| GG | 55(25.0%) | 45(20.5%) | 1.00 (Ref) | 1.00 (Ref) | |
| AG | 103(46.8%) | 124(56.4%) | 0.296 | 0.680 (0.424 - 1.090) | 0.727 (0.4 - 1.322) |
| AA | 62(28.2%) | 51(23.2%) | 1.000 | 0.995 (0.579 - 1.708) | 1.000(0.502 - 1.992) |
| Alleles | CRC patients n=220(%) | Controls n=220(%) | p | OR (95%CI) | |
| G | 213(48.4%) | 214(48.6%) | 1.00(Ref) | ||
| A | 227(51.6%) | 226(51.4%) | 0.946 | 1.009 (0.775 – 1.315) | |
OR (95%CI): odds ratio (95% confidence interval)
Table 5.
Tumor-stage specific distribution of EGF rs4444903 genotypes among colorectal cancer patients
| Genotype | Stage 0 | Stage I | Stage II | Stage III | Stage IV | P value |
|---|---|---|---|---|---|---|
| AA | 0(0%) | 2(15.4%) | 19(35.2%) | 13(31.0%) | 8(34.8%) | 0.626 |
| AG | 4(51.1%) | 6(46.2%) | 20(37.0%) | 19(45.2%) | 10(43.5%) | |
| GG | 3(42.9%) | 5(38.5%) | 15(27.8%) | 10(23.8%) | 5(21.7%) |
Discussion
This study showed no significant association between rs4444903 polymorphism of the EGF gene with risk of colorectal cancer in the studied population. Our findings support a study reported by a similar Iranian study reported by Daraei et al (19). Frequencies of the GG, GA, and AA genotypes in our study were similar to other study in Iran (19). This polymorphism has been studied in glioma, breast, lung, gastric, colon and melanoma cancers (20–26). In most studies investigating the association between this polymorphism and susceptibility of cancer, conflicting results have been reported. Studies in different population showed that no significance association between this polymorphism and risk of cancer (23, 27, 28). In a study reported by Goto et al in 2005, patients suffering from gastric cancer and control group were analysed for polymorphisms in the EGFR gene, no significant correlation was found (27). In a study reported by Gao et al, from China, a uniform distribution of genotypes in two groups of patients and controls was reported, suggesting that there is no significant difference between the patients suffering from esophagus cancer and healthy individuals (28). In 2007 Kang et al studied Korean patients afflicted with lung cancer and reported no significant association between the disease and rs4444903 (23). Studies are also showed association with this polymorphism and other cancers (26, 29, 30). A meta-analysis study was conducted by Zhang et al in 2010. This meta analysis reviewed research conducted on 23 groups, consisting of 5578 patients suffering from a variety of cancers and 7306 healthy individuals - using electronic searches. It showed that allele G is related to the probable increase of cancer (30). Xu et al propose that the EGF rs4444903 polymorphism may be associated with an increased glioma risk among Asians, but a decreased glioma risk among Caucasians(31). The reason for these contradictory findings is not clear. Ethnic heterogeneity, genotype distributions, gene environment interactions and different sample size are may be the probable of this discrepancy (25, 31, 32). Spindler et al studied EGF rs4444903 polymorphism to measure EGF gene expression among healthy subjects. The results showed that GG genotype provides more EGF gene expression (15). Lurje et al reported allele A of the rs44444903 polymorphism is related to a reduction of EGF factor in serum (33). Perhaps, the findings of Lurje and Spindler suggest a possible mechanism regarding allele G and the reason of increased the risk of colorectal cancers(34). In a meta-analysis research that was performed by Wue et al, found that allele A of rs4444903 polymorphism was associated with a decreased susceptibility to cancer among Asian and Americans subjects. Allele A may be a protective factor for gastric, esophagus cancer, and liver tumors (35).
Finally we could not find any evidence to support an association between the rs4444903 polymorphism and colorectal cancer. We also found no correlation between this polymorphism and tumor stage.
Acknowledgements
We would like to thanks all patients who participated in this study. The project is supported by Research Center for Gastroenterology and Liver Diseases, grant No:559. This article is resulted from the Vahid Chaleshi MSc thesis project.
(Please cite as: Chaleshi V, Montazer Haghighi M, Javadi GR, Fatemi SR, Vahedi M, Zali MR. The effect of 5′untranslated region polymorphism in EGF gene, rs4444903, on colorectal cancer. Gastroenterol Hepatol Bed Bench 2013;6(3):129-135).
References
- 1.Colas C, Coulet F, Svrcek M, Collura A, Fléjou J, Duval A, et al. Lynch or not Lynch? Is that always a question? Adv Cancer Res. 2012;113:121. doi: 10.1016/B978-0-12-394280-7.00004-X. [DOI] [PubMed] [Google Scholar]
- 2.Cheung DY, Kim TH, Kim CW, Kim JI, Cho SH, Park S-H, et al. The Anatomical Distribution of Colorectal Cancer in Korea: Evaluation of the Incidence of Proximal and Distal Lesions and Synchronous Adenomas. Intern Med. 2008;47:1649–54. doi: 10.2169/internalmedicine.47.1269. [DOI] [PubMed] [Google Scholar]
- 3.Ji B-T, Devesa SS, Chow W-H, Jin F, Gao Y-T. Colorectal cancer incidence trends by subsite in urban Shanghai, 1972-1994. Cancer Epidemiol Biomarkers Prev. 1998;7:661–66. [PubMed] [Google Scholar]
- 4.Kuriki K, Tajima K. Increasing Incidence of Colorectal Cancer and the Preventive Strategy in Japan. Asian Pac J Cancer Prev. 2006;7:495. [PubMed] [Google Scholar]
- 5.Yee YK, Tan VP, Chan P, Hung IF, Pang R, Wong BC. Epidemiology of colorectal cancer in Asia. J Gastroenterol Hepatol. 2009;24:1810–16. doi: 10.1111/j.1440-1746.2009.06138.x. [DOI] [PubMed] [Google Scholar]
- 6.Yiu HY, Whittemore AS, Shibata A. Increasing colorectal cancer incidence rates in Japan. Int J Cancer. 2004;109:777–81. doi: 10.1002/ijc.20030. [DOI] [PubMed] [Google Scholar]
- 7.Azadeh S, Moghimi-Dehkordi B, Fatem S, Pourhoseingholi M, Ghiasi S, Zali M. Colorectal cancer in Iran: an epidemiological study. Asian Pac J Cancer Prev. 2008;9:123. [PubMed] [Google Scholar]
- 8.Moghimi Dehkordi B, Safaee A, Pourhoseingholi MA, Vahedi M, Habibi M, Pourhoseingholi A, et al. Prevalence of positive family history of colorectal cancer in the Iranian general population. Iranian Journal of Cancer Prevention. 2010;3:28–31. [Google Scholar]
- 9.Islamic Republic of Iran, Ministry of Health and Medical Education, Office of Deputy Minister for Health Center for Disease Control, Cancer Office. Iranian Annual National Cancer Registration Report, 2006-2007; Tehran: Iranian Ministry of Health and Medical Education; 2007. [Google Scholar]
- 10.Wu G-y, Hasenberg T, Magdeburg R, Bönninghoff R, Sturm JW, Keese M. Association Between EGF, TGF-ß1, VEGF Gene Polymorphism and Colorectal Cancer. World J Surg. 2008;33:124–29. doi: 10.1007/s00268-008-9784-5. [DOI] [PubMed] [Google Scholar]
- 11.Morton C, Byers M, Nakai H, Bell G, Shows T. Human genes for insulin-like growth factors I and II and epidermal growth factor are located on 12q22→ q24. 1, 11p15, and 4q25→ q27, respectively. Cytogenet Cell Genet. 1986;41:245–49. doi: 10.1159/000132237. [DOI] [PubMed] [Google Scholar]
- 12.Henson ES, Gibson SB. Surviving cell death through epidermal growth factor (EGF) signal transduction pathways: Implications for cancer therapy. Cell Signal. 2006;18:2089–97. doi: 10.1016/j.cellsig.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 13.Normanno N, Bianco C, De Luca A, Salomon DS. The role of EGF-related peptides in tumor growth. Front Biosci. 2001;6:685–707. doi: 10.2741/normano. [DOI] [PubMed] [Google Scholar]
- 14.Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol. 1995;19:183. doi: 10.1016/1040-8428(94)00144-i. [DOI] [PubMed] [Google Scholar]
- 15.Spindler K-LG, Nielsen JN, Ornskov D, Brandslund I, Jakobsen A. Epidermal growth factor (EGF) A61G polymorphism and EGF gene expression in normal colon tissue from patients with colorectal cancer. Acta Oncol. 2007;46:1113–17. doi: 10.1080/02841860701338853. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs EJ, Hsing AW, Bain EB, Stevens VL, Wang Y, Chen J, et al. Polymorphisms in angiogenesis-related genes and prostate cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:972–77. doi: 10.1158/1055-9965.EPI-07-2787. [DOI] [PubMed] [Google Scholar]
- 17.Araujo A, Ribeiro R, Azevedo I, Coelho A, Soares M, Sousa B, et al. Genetic polymorphisms of the epidermal growth factor and related receptor in non-small cell lung cancer. a review of the literature. Oncologist. 2007;12:201–10. doi: 10.1634/theoncologist.12-2-201. [DOI] [PubMed] [Google Scholar]
- 18.Shahbazi M, Pravica V, Nasreen N, Fakhoury H, Fryer AA, Strange RC, et al. Association between functional polymorphism in EGF 61 G/A gene and malignant melanoma. Lancet. 2002;359:397–401. doi: 10.1016/S0140-6736(02)07600-6. [DOI] [PubMed] [Google Scholar]
- 19.Daraei A, Salehi R, Salehi M, Emami MH, Jonghorbani M, Mohamadhashem F, et al. Effect of rs6983267 polymorphism in the 8q24 region and rs4444903 polymorphism in EGF gene on the risk of sporadic colorectal cancer in Iranian population. Med Oncol. 2012;29:1044–49. doi: 10.1007/s12032-011-9980-2. [DOI] [PubMed] [Google Scholar]
- 20.Miller S, Dykes D, Polesky H. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S, Zhao Y, Ruan Z, Chen H, Fan W, Chen J, et al. Association between EGF +61 G/A and glioma risk in a Chinese population. BMC Cancer. 2010;10:221. doi: 10.1186/1471-2407-10-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Araújo AP, Ribeiro R, Pinto D, Pereira D, Sousa B, Mauricio J, et al. Epidermal growth factor genetic variation, breast cancer risk, and waiting time to onset of disease. DNA Cell Biol. 2009;28:265–69. doi: 10.1089/dna.2008.0823. [DOI] [PubMed] [Google Scholar]
- 23.Kang H-G, Choi JE, Lee WK, Kam S, Cha SI, Kim CH, et al. +61A > G polymorphism in the EGF gene does not increase the risk of lung cancer. Respirology. 2007;12:902–905. doi: 10.1111/j.1440-1843.2007.01152.x. [DOI] [PubMed] [Google Scholar]
- 24.Hamai Y, Matsumura S, Matsusaki K, Kitadai Y, Yoshida K, Yamaguchi Y, et al. A single nucleotide polymorphism in the 5' untranslated region of the EGF gene is associated with occurrence and malignant progression of gastric cancer. Pathobiology. 2005;72:133–38. doi: 10.1159/000084116. [DOI] [PubMed] [Google Scholar]
- 25.Kovar FM, Thallinger C, Marsik CL, Perkmann T, Puhalla H, Haslacher H, et al. The EGF 61A/G polymorphism – a predictive marker for recurrence of liver metastases from colorectal cancer. Wien Klin Wochenschr. 2009;121:638–43. doi: 10.1007/s00508-009-1250-3. [DOI] [PubMed] [Google Scholar]
- 26.Shahbazi M, Pravica V, Nasreen N, Fakhoury H, Fryer AA, Strange RC, et al. Association between functional polymorphism in EGF +61A/G gene and malignant melanoma. Lancet. 2002;359:397–401. doi: 10.1016/S0140-6736(02)07600-6. [DOI] [PubMed] [Google Scholar]
- 27.Goto Y. No Association between EGF gene polymorphism and gastric cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:2454–56. doi: 10.1158/1055-9965.EPI-05-0401. [DOI] [PubMed] [Google Scholar]
- 28.Gao L-B, Wei Y-S, Zhou B, Wang Y-Y, Liang W-B, Li C, et al. No association between epidermal growth factor and epidermal growth factor receptor polymorphisms and nasopharyngeal carcinoma. Cancer Genet Cytogenet. 2008;185:69–73. doi: 10.1016/j.cancergencyto.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 29.Lin L, Li G, Zhang Z, Wen M, Xu W, Cai J, et al. Association of Epidermal Growth Factor +61 A/G Polymorphism in Chinese Patients with Colon Cancer. Genet Test Mol Biomarkers. 2012;23:23. doi: 10.1089/gtmb.2012.0109. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y-M, Cao C, Liang K. Genetic polymorphism of epidermal growth factor 61A > G and cancer risk: A meta-analysis. Cancer Epidemiol. 2010;34:150–56. doi: 10.1016/j.canep.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 31.Xu X, Xi L, Zeng J, Yao Q. A functional +61G/A polymorphism in epidermal growth factor is associated with glioma risk among Asians. PLoS One. 2012;7:e41470. doi: 10.1371/journal.pone.0041470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe Y, Fukui N, Muratake T, Kaneko N, Someya T. No association of EGF polymorphism with schizophrenia in a Japanese population. Neuroreport. 2005;16:403. doi: 10.1097/00001756-200503150-00019. [DOI] [PubMed] [Google Scholar]
- 33.Lurje G, Nagashima F, Zhang W, Yang D, Chang HM, Gordon MA, et al. Polymorphisms in Cyclooxygenase-2 and Epidermal Growth Factor Receptor Are Associated with Progression-Free Survival Independent of K-ras in Metastatic Colorectal Cancer Patients Treated with Single-Agent Cetuximab. Clin Cancer Res. 2008;14:7884–7895. doi: 10.1158/1078-0432.CCR-07-5165. [DOI] [PubMed] [Google Scholar]
- 34.Li TF, Ren KW, Liu PF. Meta-analysis of epidermal growth factor polymorphisms and cancer risk: involving 9,779 cases and 15,932 controls. 2012;31:568–74. doi: 10.1089/dna.2011.1394. [DOI] [PubMed] [Google Scholar]
- 35.Xu W, Li Y, Wang X, Chen B, Liu S, Wang Y, et al. Association between EGF promoter polymorphisms and cancer risk: a meta-analysis. Med Oncol. 2010;27:1389–1397. doi: 10.1007/s12032-009-9392-8. [DOI] [PubMed] [Google Scholar]