Abstract
Aim
The aim of this study is to look for the proper methods that would be a major step towards untreated CD diagnosis and seek the metabolic biomarkers causes of CD and compare them to control group.
Background
Celiac disease (CD) is a common autoimmune disorder that is not easily diagnosed using the clinical tests.
Patients and methods
Thirty cases and 30 controls were entered into this study. Metabolic profiling was obtained using proton nuclear magnetic resonance spectroscopy (1HNMR) to seek metabolites that are helpful for the detection of CD. Classification of CD and healthy subject was done using random forest (RF).
Results
The obtained classification model showed an 89% correct classification of CD and healthy subject for the external test set. The metabolites that caused changes in people with CD were identified using RF; these metabolites include lactate, valine and lipid.
Conclusion
The findings of the present study reveal serum lactate, valin and lipid levels in CD patient are lower than healthy cohorts. This metabolite may provide diagnostic tools as well as insight into potential targets for disease therapy.
Keywords: Nuclear magnetic resonance spectroscopy, Celiac disease, Random forest, Metabonomics
Introduction
Celiac disease (CD) is a common systemic disorder, which can have multiple clinical manifestations. It has a multi factorial etiology with a complex genetics and histology. A comparison of recent studies in European and Middle Eastern countries has shown that CD is common in both areas, with an almost similar prevalence (1). Despite advances in investigation techniques, CD remains a challenging problem that often eludes diagnosis and receives sub-optimal attention (2). In this regard, metabonomics can provide powerful techniques for CD diagnosis.
Metabonomics is described as the quantitative measurement of the multi-parametric metabolic response of living systems to pathophysiological stimuli or genetic modification(s) (3, 4). This quantitative measurement can provide multivariate metrics of potential metabolic dysfunction in any living system. There are several available analytical spectroscopic methods to interpret the profiles of metabolism in the biological sample such as urine, plasma or tissue. In the biological systems, proton nuclear magnetic resonance spectroscopy (1HNMR) is a useful method providing valuable data of the metabolites (5, 6). Various metabonomics studies are accomplished on CD. For instance, Bertini, et al. define the metabolic signature of CD through NMR of urine and serum samples of CD patients. In recent study, 1H NMR metabolic profiling of their serum and urine samples examined a cohort of CD patients, before and after gluten free diet (GFD), and healthy controls. The results indicated that altered serum levels of glucose and ketonic bodies suggest alterations of energy metabolism, while the urine data point to alterations of gut microbiota(7). In another review, Bernini and co authors (8) address potential CD patients, defined as subjects who do not have, and have never had, a jejuna biopsy consistent with clear CD, and yet have immunological abnormalities similar to those found in celiac patients. They demonstrated that metabolic alterations may precede the development of small intestinal villous atrophy and provide a further rationale for early institution of GFD in patients with potential CD, as recently suggested by prospective clinical studies (8).
Leo Breiman has recently developed the random forests (RF) (9) that is based on classification and regression methods. RF reduces the variance and improves the prediction accuracy. In this study, we propose to apply RF for discriminating the control and CD subjects. In order to achieve this purpose, we seek the significance of metabolic biomarkers that can lead to the classification of these two groups.
Patients and Methods
Sample population
Thirty blood samples from adult CD patients (14 males and 16 females with mean age (±standard deviation) 34±11 years) and 30 healthy subjects (HS) were collected as described previously (10).
1H NMR spectroscopy
1H NMR experiments were acquired on a Bruker DRX 500 MHz spectrometer equipped with a 5mm NMR tube for analysis. The detail of this technique was presented in our previous study (11–13). Metabolites present in serum samples were identified on the basis of several previous studies (14–16).
Random forest (RF)
Random Forest (RF) classifiers are used to classify the serum samples of healthy and CD subjects and seeking the fundamental metabolites for desecrating (9). RF is a modified non-linear classification and regression trees (CART) method providing an importance ranking for the effectiveness of each metabolite. CART maximize the difference of heterogeneity, but the over fitting problem causes the classifier to have a high error of prediction in the test set whereas the bagging mechanism in RF algorithm can improve over fitting problem (18). The RF algorithm is illustrated in Ref (9). This algorithm builds every tree that is different owing to two factors. In first step, a best split is chosen at each node. This selection occurs from a random subset of the predictors rather than all of them. In next step, a bootstrap sample of the observations builds every tree. The out-of-bag (OOB) data are one-third of the observations. They can be used to estimate the prediction accuracy. Finally, based on averaging over all the trees is calculated overall prediction.
RF package is a readily accessible implementation of the RF algorithm and can be downloaded from the website. The data preprocessing and the modeling was executed utilizing MATLAB (version 6.5.1, The Math works, Cambridge, UK). RF has been applied on mean-centered data set and validated by predicting the classes of test set not used in the training set (17).
Results
A total of 42 different training and 18 test sets were built by random splitting for NMR spectra. Test set contain about 1/3 of the samples. In the classification model, these descriptive variables are the integral at the difference of the chemical shift in NMR spectra while the class numbers of the different samples were employed as response. According to RF model, we concluded that using three descriptors, CD and control groups could be classified. These metabolites include lactate, valine and lipid. Table 1 presents the summaries of the metabolite level distributions which are considerably (P-value < 0.001) different between CD patients and control group. Table 1 depicts serum lipid, valine and lactate levels in CD patient are lower than healthy group.
Table 1.
Metabolites present in serum samples of celiac patients and control α
| Metabolite | Assignment | lH chemical shift (ppm) | CD group |
|---|---|---|---|
| Lactate | βCH3 | 1.32 | ↓ |
| Valine | dCH3 | 1.03 | ↓ |
| Lipid | CH2CH2CO | 1.56 | ↓ |
α The arrows (↓) indicate decrease of metabolites levels in CD group.
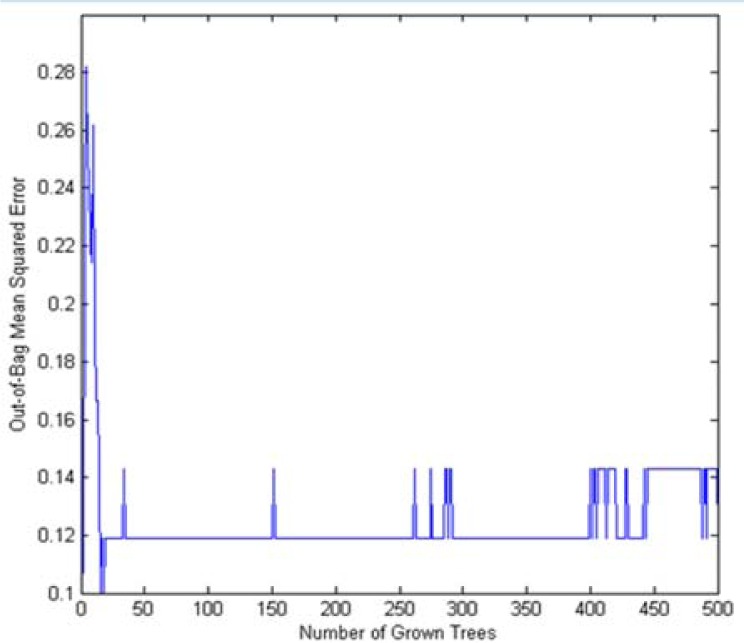
Samples of the training set were classified using RF in which 500 trees were grown. The OOB data was used to estimate the prediction accuracy of classification. Figure 1 presents the OOB error rate. The RF algorithm considers how much prediction error increases. At this time OOB data for that variable is permuted while all other variables are left unchanged. By this method the significance of a variable can be estimated. Confusion matrix is a tool to illustrate the relations between real class and predicted classes. Table 2 presents confusion matrix of the RF model for the training and test set. In detecting CD patients of test set, as it is clear from Table 2, RF model has an accuracy of 0.89. With respect to these results, RF model has great chance in diagnosis of CD.
Figure 1.
Plot of OOB error for RF classification of CD and control group
Table 2.
Confusion matrix for training and test set.
| Predicted | |||
|---|---|---|---|
| Training set | Observed | CD class | Healthy class |
| CD class | 20 | 1 | |
| Healthy class | 2 | 19 | |
| Test set | CD class | 8 | 1 |
| Healthy class | 1 | 8 | |
Table 3 reports the classification specificity and other classification parameters for each individual in the training and test set. Another evidence for capability of RF model in CD diagnosis is the high non-error rate in the external test set.
Table 3.
The calculated error and non-error rates of the classification index and the classification performances of training and test sets
| Set | Error rate | Non-error rate | specificity | sensitivity | accuracy |
|---|---|---|---|---|---|
| Training | 0.07 | 0.93 | 0.95 | 0.91 | 0.93 |
| Test | 0.11 | 0.89 | 0.89 | 0.89 | 0.89 |
Discussion
Lipids are a group of naturally occurring molecules such as types of vitamins, monoglycerides, and others. Lipids act as structural components of cell membranes. The majority of lipids in biological systems include energy storage (18, 19). Krums and et.al stated lipid metabolism was evaluated in patients with CD (20). The reason of this function in patients with CD is disorders of lipid metabolism in the small intestine. Lewis and co-authors investigated cholesterol profile in people with newly diagnosed CD (21). They suggested that untreated CD is associated with lower total cholesterol than in the general population. Bertini, et al. (7) expressed that lipid oxidation should be increased in CD. In these cases intake of lipids is reduced because of malabsorption in CD patients. They explained that the lower levels of lipids in sera to be due to an enhanced lipids oxidation and malabsorption. Also they found valine and lactate levels in the serum of healthy individuals to be less than those found in CD patients. Our classification results are better than results of Bertini and coauthors (7). They found the classification accuracy of CD and healthy control groups was 79.7-83.4% for serum and 69.3% for urine. We applied the RF method for classification that has optimization parameters which are less than support vector machines (SVM) (applied method in Ref (7)).
An essential amino acid is valine that must be ingested. Valine both in structure and function is closely related to leucine and isoleucine. These amino acids are important for supplying energy to muscles and increase endurance and aid in muscle tissue recovery and repair. Hernanz, et.al analyzed amino acid concentrations in plasma from control and treated and untreated patients with CD (22). They found both treated and untreated cohorts had significantly decreased plasma concentrations of citrulline, tyrosine, valine, isoleucine, and leucine compared with control cohorts. In another review, Bernini et.al stated that glycolysis is somehow impaired in CD explaining both a lowering of lactate levels and an increase of glucose levels in blood (8). The body produces lactate in throughout the day. It is actually an important fuel used by the muscles during prolonged exercise. Also Bertini and coworkers defined metabonomics for CD in three components including malabsorption, energy metabolism and the third related to alterations of gut microflora (7).
In Conclusion, metabonomics and analysis of the important above mentioned metabolites in serum is applied widely in early stage of CD disease. Due to the results RF proved to be quite powerful in discriminating between CD and healthy subjects.
Acknowledgements
We gratefully acknowledge financial support from Iran National Science Foundation (INSF), Sharif University of Technology and Shahid Beheshti University of Medical Sciences.
(Please cite as: Fathi F, Ektefa F, Arefi Oskouie A, Rostami K, Rezaei-Tavirani M, Mohammad-Alizadeh AH, et al. NMR based metabonomics study on celiac disease in the blood serum. Gastroenterol Hepatol Bed Bench 2013;6(4):190-194).
References
- 1.Rostami K, Malekzadeh R, Shahbazkhani B, Akbari M, Catassi C. Celiac disease in Middle Eastern countries: a challenge for the evolutionary history of this complex disorder? Dig Liver Dis. 2004;36:694–97. doi: 10.1016/j.dld.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Rostami Nejad M, Rostami K, Pourhoseingholi M, Nazemalhosseini E, Dabiri H, Habibi M. Atypical presentation is dominant and typical for Celiac Disease. J Gastrointestin Liver Dis. 2009;18:285–91. [PubMed] [Google Scholar]
- 3.Nicholson J, Connelly J, Lindon J, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nature Rev Drug Disc. 2002;1:153–61. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 4.Nicholson J, Lindon J, Holmes E. Metabonomics’: understanding the metabolic responses of living systems to athophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–89. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 5.Lindon JC, Nicholson JK, Holmes E, Everett JR. Metabonomic: metabolic processes studied by NMR spectroscopy of biofluids. Concepts in Magnetic Resonance. 2000;12:289–320. [Google Scholar]
- 6.Bollard ME, Stanley EG, Lindon JC, Nicholson JK, Holmes E. NMR-based metabonomic approaches for evaluating physiological influences on biofluid composition. NMR Biomed. 2005;18:143–62. doi: 10.1002/nbm.935. [DOI] [PubMed] [Google Scholar]
- 7.Bertini I, Calabro A, Carli VD, Luchinat C, Nepi S, Porfirio B, et al. The metabonomic signature of celiac disease. J Proteome Res. 2009;8:170–77. doi: 10.1021/pr800548z. [DOI] [PubMed] [Google Scholar]
- 8.Bernini P, Bertini I, Calabro A, Marca Gl, Gabriele Lami, Luchinat C. Are patients with potential celiac disease really potential? The answer of metabonomics. J Proteome Res. 2011;10:714–21. doi: 10.1021/pr100896s. [DOI] [PubMed] [Google Scholar]
- 9.Steffens M, Lamina C, Illig T, Bettecken T, Vogler R, Entz P, et al. SNP-based analysis of genetic substructure in the German population. Hum Hered. 2006;62:20–29. doi: 10.1159/000095850. [DOI] [PubMed] [Google Scholar]
- 10.Fathi F, Ektefa F, Tafazzoli M, Rostami K, Rostami Nejad M, Fathi M, et al. A concentration of serum zinc in celiac patients compare to healthy subject in Tehran. Gastroenterol Hepatol Bed Bench. 2013;6:92–95. [PMC free article] [PubMed] [Google Scholar]
- 11.Fathi F, Kyani A, Darvizeh F, Mehrpour M, Tafazzoli M, Shahidi G. Relationship between serum level of selenium and metabolites using 1 HNMR-based metabonomics in Parkinson's disease. Appl Magn Reson. 2013;44:721–34. [Google Scholar]
- 12.Mortazavi-Tabatabaei SAR, Fathi F, Ektefa F, Tafazzoli M, Arefi Oskouie A, Rezaie-Tavirani M, et al. Investigation of metabonomics technique by analyze of NMR data, which method is better?, Mean center or auto scale? Journal of Paramedical Sciences (JPS) 2013;4:2–9. [Google Scholar]
- 13.Mehrpour M, Kyani A, Tafazzoli M, Fathi F, Joghataie M-T. A metabonomics investigation of multiple sclerosis by nuclear magnetic resonance. Magn Reson Chem. 2013;51:102–9. doi: 10.1002/mrc.3915. [DOI] [PubMed] [Google Scholar]
- 14.Viant MR. Improved methods for the acquisition and interpretation of NMR metabolomic data. Biochem Biophys Res Commun. 2003;310:943–48. doi: 10.1016/j.bbrc.2003.09.092. [DOI] [PubMed] [Google Scholar]
- 15.Pang H, Lin A, Holford M, Enerson BE, Lu B, Lawton MP, et al. Pathway analysis using random forests classification and regression. Bioinformatics. 2006;22:2028–36. doi: 10.1093/bioinformatics/btl344. [DOI] [PubMed] [Google Scholar]
- 16.Nicholson JK, Foxall PJ, Spraul M, Farrant RD, Lindon JC. 750 MHz 1H and 1H-13C NMR spectroscopy of human blood plasma. Anal Chem. 1995;67:793–811. doi: 10.1021/ac00101a004. [DOI] [PubMed] [Google Scholar]
- 17.Verwaest KA, Vu TN, Laukens K, Clemens LE, Nguyen HP, Gasse BV, et al. 1H NMR based metabolomics of CSF and blood serum: A metabolic profile for a transgenic rat model of Huntington disease. Biochim Biophys Acta. 2011;1812:1371–79. doi: 10.1016/j.bbadis.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Fan M. Metabolite profiling by one-and two-dimensional NMR analysis of complex mixtures. Prog Nucl Magn Reson Spectrosc. 1996;28:161–219. [Google Scholar]
- 19.Lindon JC. NMR Spectroscopy on biofluids. Annu Rep NMR Spectrosc. 1999;38:1–88. [Google Scholar]
- 20.Fahy E, Subramaniam S, Murphy R, Nishijima M, Raetz C, Shimizu T, et al. Update of the LIPID MAPS comprehensive classification system for lipids. J Lipid Res. 2009;50:S9–14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramaniam S, Fahy E, Gupta S, Sud M, Byrnes RW, Cotter D, et al. Bioinformatics and Systems Biology of the Lipidome. Chem Rev. 2011;111:6452–90. doi: 10.1021/cr200295k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hernanz A, Polanco I. Plasma precursor amino acids of central nervousvsystem monoamines in children with coeliac disease. Gut. 1991;32:1478–81. doi: 10.1136/gut.32.12.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]