Abstract
Aim
The aim of this study was to investigate the prevalence of GBV-C among Iranian HBV positive patients using PCR-RFLP technique.
Background
GBV-C was a member of flaviviridae family and recently propose to classify as members of a fourth genus in this family, named Pegivirus and suggest that at least one quarter of the world's population has been infected with this virus. GBV-C can be transmitted via the blood-borne route, although vertical and sexual transmission is very well documented.
Patients and methods
100 serum samples were collected from HBsAg positive patients in 2011–2012. RNA was extracted with Qiagene mini kit. cDNA was synthesized by Reverse Transcriptase method and amplified by Semi- nested PCR method. After designing specific primers, the semi nested PCR was optimized, then sequences of PCR products were analyzed with software such as neb cutter, and sites of restriction enzymes were determined and suitable enzymes were selected for RFLP (Restriction Fragment Length Polymorphism).
Results
PCR products were analyzed in 2% agarose gels containing ethidium bromide and were visualized with ultraviolet (UV) light. A 230 bp band was observed in comparison with 100 kb ladder; this band indicates our target gene of GBV-C genome have been isolate from serum samples.
Conclusion
It seems that Co-infection of GBV-C and HBV are common and This method had acceptable sensitivity for detecting GBV-C and determining its genotype, and more affordable than the other techniques; so the results of this study showed the prevalence of GBV-C were 12 serums of 100 serums HBsAg positive in goal population and one sample from 12 GBV-C positive serums was genotype 3 and the others were genotype 2.
Keywords: GBV-C, HBsAg positive, Semi nested –PCR, Genotype
Introduction
The GB virus C (GBV-C) is a single-stranded, positive-sense RNA genome of approximately 9.4 Kb; on a comparison between genome organization and sequence homologies, GBV-C is most closely related to human hepatitis C virus (HCV) However, in contrast to HCV, GBV-C does not appear to be hepatotrophic (1). Based on phylogenetic relationships, genome organization and pathogenic features of the GB viruses, Jack T. Stapleton and colleagues propose to classify GBV-A-like viruses, GBV-C and GBV-D as members of a fourth genus in the family Flaviviridae, named Pegivirus (pe, persistent; g, GB or G)(2).
The clinical significance of human infection with GBV-C/HGV is currently unclear (3). The virus can establish both acute and chronic infection and appears to be sensitive to interferon. GBV-C; can be Transmitted via the blood-borne route, although vertical and sexual transmission is very well documented. GBV-C is distributed globally; a large number of the healthy blood donors in Iran and other countries have been exposed and to be infected with GBV-C. The significantly higher levels of infections seen in the high-risk groups, such as multitransfused patients; hemodialysis patients, hemophilic, HIV, HCV, HBV infected patients, as well as intravenous drug users (3–5).
GBV-C is lymphotropic and not associated with any known Human disease. Co-infection of GBV-C and HBV is common. Recently, it was shown that, co-infection with HIV leads to improved morbidity and mortality for these individuals and slows down progression to acquired immunodeficiency syndrome (AIDS) (6–9).
Most of the studies on GBV-C in Iran have been done on HIV co-infected patients, and the GBV-C infection-rate was reported to be10.97% to 15.5%. In another study, 13% of Iranian hemodialysis patients were reported to be infected with GBV-C.
Also the prevalence of this virus in HCV positive patients, Hemophilic and Thalassemia individuals were 43.6%, 41.4%, and 33.4% respectively. In healthy blood donors infection with this virus was reported up to 8.6%(10–13).
The comparison with new and traditional methods for viruses genotyping such as Sequencing, PCR and Genotype-Specific Primers, Reverse-Phase Hybridization, Mass Spectrometry and other systems, the rapid and sensitive Method was developed based on restriction pattern analysis of the 5’ UTR of the genome (RFLP).
At the time of these study 5 genotypes of the virus with this method, was recognized; hence The aim of the present study was to develop a RFLP analyses of 5’ UTR of 12 GBV-C RNA positive serum isolated from100 serum samples of Iranian HBV infected blood donor.
Patients and Methods
One hundred serum samples of HBV infected patients were collected from Iranian blood transfusion organization and stored at −70 °C; were analyzed for the study during the period October 2010 to November 2011. The study and sampling was approved by Tarbiat Modares University's Ethics Approval Committee.
RNA Extraction and RT-PCR
The PCR primers were designed based on the most conserved regions derived from the known sequences available in GenBank. All so-called complete GBV-C sequences deposited in GenBank and partial sequences from the 5’ UTR were analyses for primer synthesis (Table 1).
Table 1.
Primer sequences for round 1
| Primer name | Primer Sequence | Attach site | Product length |
|---|---|---|---|
| sense, outer/S1 | 5'GGTCGTAAATCCCGGTCACC3’ | 139–400 | 262 bp |
| antisense, outer/AS1 | 5'CCCACTGGTCCTTGTCAAC3’ |
These primers were confirmed to be specific for GBV-C with Gene Runner, Mega4, Oligoanalyser and NCBI Blast software.
The primers were prepared with an Alfa sequencing Company (Canada) (Table 1). Standards precautions for avoiding contamination for PCR were observed. A negative control serum was also included in each run to ensure specificity. All of the samples were analyzed for RNA extraction with Qiagene mini Kit (Germany). cDNA was synthesized by Reverse Transcriptase method and amplified by Semi- nested PCR by specific primers.
For RFLP analysis, the 237 bp DNA fragment of the viral genome was amplified by RT-PCR with specific primers for conserved 5’ UTR of the GBV-C.
RNA was initially obtained from 200 ml of a serum samples using the method described by QIAamp RNA Viral Mini Kit (Qiagene, Germany). 10 µl of extracted RNA was used for cDNA synthesis in a final volume of 21 µl. cDNA was synthesized by incubation at 70 °C with Specific antisense primer (AS1) of GBV-C for 5 min and M-MuLV reverse transcriptase at 42°C for 60 min. Each 21 µl RT master mixture contained 0.2 mM of dNTPmix, 0.16 pmol Primer, 20unit M-MuLV-reverse transcriptase, 2 units RNase inhibitor and 2 µl 1 X RT buffer. The cDNA was stored at −20 °C for using in Semi nested- PCR method.
Semi Nested-PCR conditions
After sequence alignment, primers were designed for 5’ UTR of GBV-C genome and were used in Semi nested- PCR and procedures of this method was carefully optimized.
The 5 µl cDNA was used for 25 ul volume first round PCR reaction which contain: 2.5 µl of polymerase buffer1x, 0.75 ul of 1.5 mmol/l MgCl2,0.5 µl of 0.2 mmol/l dNTPmix, 0.3 ul of Taq (1.25Unit) and 0.16 pmol Sense primer S1 and 0.4 µl of 0.16 pmols AS1(Table 1). After a denaturation step (5min at 95°C), 32 cycles of PCR, with 1 cycle consisting of denaturation (50s at 94°C), annealing (40s at 55°C), primer extension (50 s at 72°C) and the end for inactivation of Taq DNA polymerase followed (3 min at 72°C).
Semi-Nested PCR was performed in a volume of 25 µl after transfer of 1 µl from the first round of PCR product to a mix with 0.4 µl of 0.16 pmol/ µl sense primer; S2, and 0.4 µl of 0.16 pmol AS1, and other components was similar to first round(Table 2).
Table 2.
Primer sequences for round 2
| Primer name | Primer Sequence | Attach site | Product length |
|---|---|---|---|
| sense, inner/S2 | 5'TAGCCACTATAGGTGGGTCT3’ | 163–400 | 237 bp |
| anti-sense, outer/AS1 | 5'CCCACTGGTCCTTGTCAACT3’ |
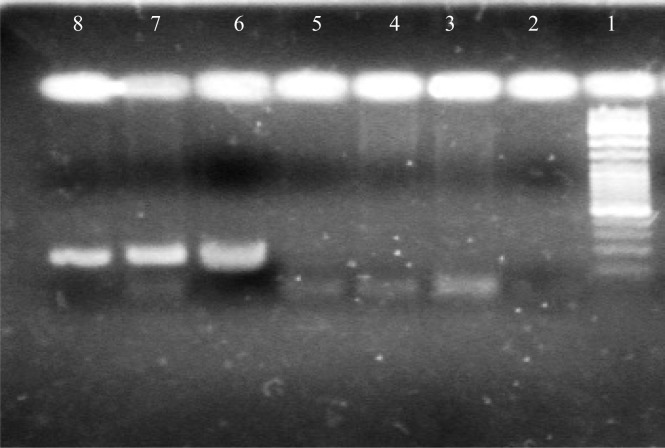
The second PCR was performed after a denaturation step as given above (5min at 95°C) and 32 cycles of PCR followed with 1 cycle consisting of denaturation (5min at 95°C), annealing (50s at 64°C), and extension (40 s at 72°C) and a final extension (3 min at 72°C) follow by a hold at 4°C. the second-round PCR products were analyzed by electrophoresis in 2% agarose gels and stained with ethidium bromide, then visualized under ultraviolet(UV) light. The PCR product, 237bp from the 5’ UTR of GBV-C was expected (Figure 1).
Figure 1.
1, 2 and 3: 237 bp band (positive sample); 4, 5 and 6: negative sample; 7: negative control; 8: size marker100 bp
Determination of restriction enzymes for RFLP
In order to design a RFLP method, an attempt was made to identify specific restriction sites that are unique for each genotype. To reach this goal, all partial sequences from the 5’ UTR and complete GBV-C sequences deposited in GeneBank at the time of this study, were used for analysis and determined restriction enzymes pattern with Neb cutter and web cutter software. In the beginning 3 restriction enzymes were selected: Ban II (FriOI), Dra II (ECO0109I) and Ava II (Bme18I) which allows the determination of all known genotypes of GBV-C (Table 3).
Table 3.
Selected Restriction Enzymes and their cutting site sequences
| Location identified | Enzyme name | |
|---|---|---|
| 5’ RG↓GNCCY3’3'YCCNG↑GY5’ | Dra II | (ECO 0109I) |
| 5’ G↓GWCC3’3'CCWG↑G5’ | Ava II | (Bme18I) |
| 5'GRGCY↓C3’3'C↑YCGRG5’ | FriOI | (Ban II) |
RFLP Analysis
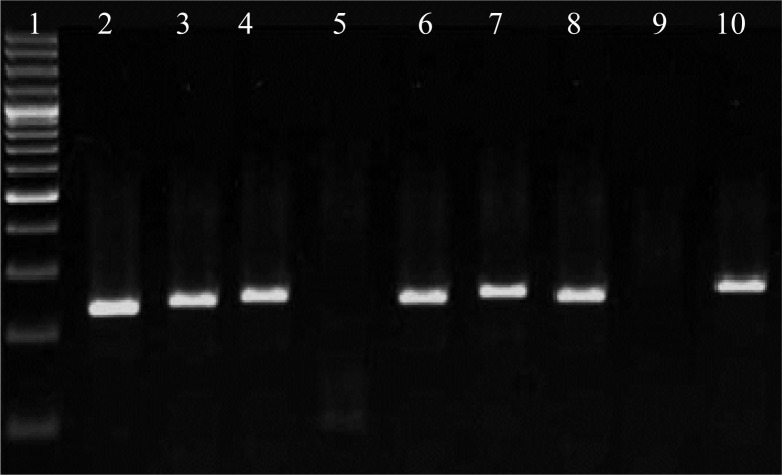
RFLP analysis was performed after the genotype specific restriction sites were identified. Restriction cleavage was carried out with 0.1–0.5 µg of second PCR product mixed with 1x reaction buffer, restriction enzymes and incubated at 37°C for 16 to 24 hour according to the manufacturer's recommendations. After incubation samples were run on a 2% agarose gel and the RFLP pattern was then evaluated and visualized by ethidium bromide staining under ultraviolet light (Figure 2). 12% acrylamide and 2% agarose gel were used for increasing sensitivity detection and band differentiating.
Figure 2.
1: size marker (100bp); 2, 3, 4, 6, 7 and 8: 225 bp band (genotype 2); 5: 105& 106 bp band (genotype 3); 9: without digest fragment as negative control; 10: 225 bp band as positive control
Results
In the current study we developed Semi Nested-PCR for the sensitive and specific detection of GBV-C RNA and RFLP method to precise determination of 6 genotypes of this virus in study population.
This study showed GBV-C RNA was detected in 12 out of the 100 serum samples of Iranian blood donors with hepatitis B infection. In the beginning, the specific restriction site for DraII at nucleotide position 225, induced 15 and 225 bp fragments was found only in genotype2. While genotypes 1, 4 and 5 did not possess a DraII cleavage site in the selected region of the 5’ UTR (nucleotide position163–400) and in the gel 237 bp fragment were seen.
The specific restriction site for DraII at nucleotide position35 and 102 induced 35,102 and 100 bp fragments was found only in genotypes3 or 6. Then restriction enzyme AvaII did not have cleavage site for genotype 5 but the specific restriction site for genotype 1 at nucleotide position 35 and 68 was found and induced 35, 33 and 172 bp fragments. The digestion site in nucleotide position 169 and 68 for genotype 4 was found and induce d 68,101 and 71bp fragments. Finally Ban II was selected, which restriction site for genotype 6, was absent but at nucleotide position135 and 106 restriction site were seen that produced 105,29 and 106 bp fragments was found only in genotypes3. The pattern of restriction enzyme cutting by Ban II (FriOI), Dra II (ECO0109I) and Ava II (Bme18I) allowed the assignment of all known specific genotypes of GBV-C in this sample.
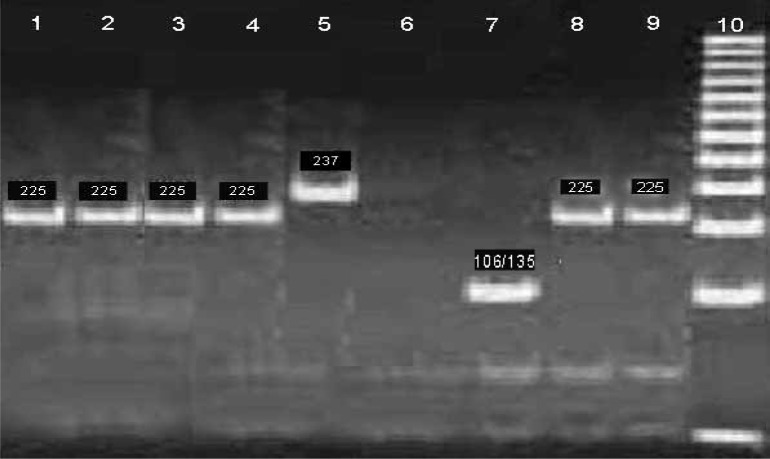
Agarose gel 2% visualized by ethidium bromide staining under ultraviolet light. By using of 12% Acrylamide gel electrophoresis method, increased band differentiating (Figure 3).
Figure 3.
Digested with enzymes DraII I and Ban on 12% polyacrylamide gels; 1, 2, 3, 4, 8, and 9: cut with DraII and 225 bp bands representing genotype 2; 5: 237 bp band (no digestion enzyme reaction); 6: negative control; 7: Cut with Ban II and 106 and 135 bands representing genotype 3.
This study showed genotype 2 in 11 (91.6%) and genotype 3 in 1 (8.3%) of the 12 positive GBV-C serum samples of Iranian blood donors with hepatitis B infection was detected, and Based on the nucleotide sequence of the 5’-UTR region (139 –400) Genotype 2,was prevalence in this population.
Discussion
Studies of GBV-C have reported a relatively high prevalence in high risk groups such as hemophilic and hemodialysis patients, intravenous drug users as well as HIV and HCV patients (15).
Different studies in countries showed, the prevalence of GBV-C exposure was 6.8% in the dialysis patients, 35.2% in hemophilic patients; while the prevalence in HCV and HIV was 24.2% and 18.2%, respectively and, between 1% to 4% of healthy blood donors have active infection and could detect viral RNA in their sera. (14, 6)
In Iranian studies, Several researchers evaluate the effect of GBV-C coinfection in HIV and some of them investigated on GBV-C infection in HCV positive individuals and high risk group such as hemodialysis. The prevalence of GBV-C in HIV and HCV patients was 15.5% and 43.6%, respectively and in the new study 8.6% of healthy blood donors infected which did not correlate with gender, age, or history of blood transfusions (10–13).
GB virus type C is a non-pathogenic flavivirus associated with prolonged survival in HIV-infected individuals and correlation between GBV-C and other disease were not seen but in coinfection with other viruses might affected response to therapy or clinical course in patients.
By itself, GBV-C infection has not been associated with any specific disease. No association has been found between GBV-C and such conditions as hepatocellular carcinoma, lichen planus, cryoglobulinaemia, Sjogren's disease or various malignant or non-malignant hematological disorders (16).
To date, several methods have been made in order to provide a rapid method for detect and genotyping of viruses such as, Sequencing, PCR and Genotype-Specific Primers, Reverse-Phase Hybridization, Mass Spectrometry (17). In this study because of the increasing number of GBV- C sequences deposited in the GenBank database RFLP of the initial methods, to be used. RFLP analysis was: less time consuming, Rapid and simple procedures, sensitive method without computer processing and Affordable.
This method of genotyping is still an important and reliable tool; even in the presence of automated methods. These methods have been advantages such as Good qualities compared with genotyping based on sequencing and comparative alignment and Capable to detect all known GBV-C genotypes/isolates in just a serum sample. Ability to detect, distinguish of virus, in mixed viral populations. Good for transmission and epidemic studies, Determination of all genotypes of GBV-C in large number of samples (18–20). By using of RFLP method, we differentiated six GBV-C genotypes by restriction endonuclease enzymes that digest of the amplified cDNA from 5’ UTR by Semi- Nested PCR.
We have utilized simple genotyping method by Using three restriction enzymes: Dra II (ECO 0109I) and Ava II (Bme18I) and Ban II (FriO), which digest genotype-specific site(s) in PCR products, by RFLP, rapidly and easily. With this method (similar to other studies and methods) based on the 5’- UTR, six genotypes (1 to 6) of GBV-C was classified.
As previously reported in Iran, ghanbari and et al determine of genotypes of this virus in HCV positive patients by using of sequencing and phylogenic analysis (21).
In other country studies, genotyping of GBV-C had done with sequencing, phylogenic analysis and RFLP methods (16). In OUBINA study in 1999, three genotypes of GBV-C were recognized and another research, Sabine B. Schleicher and et al in 2003, RFLP and phylogenetic analysis were designed; which five genotypes of this virus were determined. In study by Mizokamia and et al. by using of two restriction enzymes (ScrFI and BsmFI), recognized three genotypes of GBV-C/HGV. With this method in these studies, a simple restriction fragment polymorphism analysis was developed for genotyping (19, 20, 22–25).
In this study we developed Semi-Nested PCR in 237 bp region of GBV-C genome with specific primers for 5'UTR of virus and utilized of 3 restriction enzymes for RFLP method. The results of this study shown that Co-infection of GBV-C and HBV are common and majority of circulating genotype in goal population were genotype 2 and 11 out of 12 positive serum confected with GBV-C was genotype 2 and 1 out of 12 samples was genotype 3 that digested by Dra II and Ban II. So we utilized RFLP method is useful for differentiating the infections with variant genotype of GBV-C in co-infected serum with blood-borne viruses such as HIV, HBV and HCV positive patients; and PCR-RFLP method is accurate and inexpensive to detect genome of GBV-C and circulated genotypes in HBV infected patients. Based in the results, this method has acceptable for detection and diagnosis of GBV-C infection.
There remaining many questions which require further studies about viral course of infection GBV-C and coinfection with other viruses such as HBV that must answer. In addition, a better Understanding of any role different genotypes of GBV-C may play in HIV progression and course of hepatitis disease such as hepatocellular carcinoma, response and resistant to viral therapy is required. Although GBV-C has been shown that no correlation with human diseases but so far a little work has been done on coinfection of GBV-C with HBV.
Acknowledgements
The authors acknowledge the support of Tarbiat Modares University for this study.
(Please cite as: Yazdani L, Ravanshad M, Khanlari Z, Mousavi Nasab SD, Ahmadi NA, Imanzad M. Prevalence of GBV-C among Iranian HBV positive patients using PCR-RFLP technique. Gastroenterol Hepatol Bed Bench 2013;6(Suppl.):S70-S76).
References
- 1.Sehgal R, Sharma A. Hepatitis G virus (HGV): current perspectives. Indian J Pathol Microbiol. 2002;45:123–28. [PubMed] [Google Scholar]
- 2.Stapleton Jack T, Muerhoff Scott A. The GB viruses: a review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae. J Gen Virol. 2011:233–46. doi: 10.1099/vir.0.027490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramezani A, Gachkar L. Prevalence of Hepatitis G virus Exposure in Blood Donors. Iranian J Infect Dis Trop Med. 2007;12:41–46. [Google Scholar]
- 4.Vanesa Ruiz, Mirta Giordano, Minassian ML, Cuestas ML, Trinks J, et al. GB virus C quasispecies detected in plasma and lymphocyte subsets in a natural human infection. J Gen Virol. 2010;91:1687–92. doi: 10.1099/vir.0.019877-0. [DOI] [PubMed] [Google Scholar]
- 5.Wiwanitkit Viroj. Hepatitis G virus RNA positivity among the voluntary blood donors: a summary. Ann Hepatol. 2005;4:43–46. [PubMed] [Google Scholar]
- 6.Stapleton JT. GB virus type C/hepatitis G virus. Semin Liver Dis. 2003;23:137–48. doi: 10.1055/s-2003-39943. [DOI] [PubMed] [Google Scholar]
- 7.Xiang J, Wünschmann S, Diekema DJ, Klinzman D, Patrick KD, George SL, et al. Effect of coinfection with GB virus C on survival among patients with HIV infection. N Engl J Med. 2001;345:707–14. doi: 10.1056/NEJMoa003364. [DOI] [PubMed] [Google Scholar]
- 8.Antonucci G, Girardi E, Cozzi-Lepri A, Capobianchi MR, Morsica G, Pizzaferri P, et al. Response to HAART and GB virus type C coinfection in cohort of antiretroviral-naive HIV-infected individuals. Antiviral Therapy. 2005;10:109–11. [PubMed] [Google Scholar]
- 9.Keyvani H, Mohammadi A, Haji-abdolbaghi M. Prevalence of GBV-C RNA in HIV infected individuals in Tehran Iran. Iranian J Publ Health. 2010;39:22–27. [PMC free article] [PubMed] [Google Scholar]
- 10.Fallahian F, Alavian SM, Rasoulinejad M. Epidemiology and transmission of Hepatitis G virus infection in dialysis patients. Saudi J Kidney Dis Transpl. 2010;21:831–4. [PubMed] [Google Scholar]
- 11.Khanizadeh S, Ravanshad M, Mohebbi SR, Naghoosi H, Abrahim Tahaei M, Mousavi Nasab SD, et al. Hepatitis G virus in Iranian Blood donors and High-Risk Groups. Hepat Mon. 2009;9:282–86. [Google Scholar]
- 12.Ziaee M, Zarban A, Malekinejad P, Akhbary H. Evaluation of HGV Viremia Prevalence and Its Co-Infection with HBV, HCV, HIV and HTLV-1 in Hemophilic Patients of Southern Khorassan Iran. Hepat Mon. 2007;7:11–14. [Google Scholar]
- 13.Rezvan H, Sharafi M. A Preliminary Report on Prevalence of Antibody Response to GBV-C E2 Protein in Iranian Blood Donors and Multitransfused Patients. Iranian J Publ Health. 2007;36:46–11. [Google Scholar]
- 14.Hekmat S, Mohraz M, Vahabpour R, Jam S, Bahramali G, Banifazl M, et al. Frequency and Genotyping of GB Virus C Among Iranian Patients Infected With HIV. J Med Virol. 2008;80:1941–46. doi: 10.1002/jmv.21314. [DOI] [PubMed] [Google Scholar]
- 15.Mohr EL, Stapleton JT. GB virus type C interactions with HIV: the role of envelope glycoproteins. J Viral Hepat. 2009;16:757–68. doi: 10.1111/j.1365-2893.2009.01194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polgreen PM, Xiang J, Stapleton JT, Chang Q. GB virus type C/hepatitis G virus: a non-pathogenic flavivirus associated with prolonged survival in HIV-infected individuals. Microbes Infect. 2003;5:1255–61. doi: 10.1016/j.micinf.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe MA, Milanezi CM, Silva WA, Jr, de Lucena Angulo, I, Santis G, Kashima S, et al. Molecular investigation of GB virus C RNA in hemodialysis and thalassemics patients from Brazil. Ren Fail. 2003;25:67–75. doi: 10.1081/jdi-120017469. [DOI] [PubMed] [Google Scholar]
- 18.Gauthier J, DesLauriers M, Bourne EJ, Carrick KM, Baldanti F, et al. Two sensitive PCR-based methods for detection of hepatitis B virus variants associated with reduced susceptibility to lamivudine. J Clin microbiol. 1999;37:3338–47. doi: 10.1128/jcm.37.10.3338-3347.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabine B. Schleicher, Bertram F. Genotyping of GB Virus C by Restriction pattern Analysis of the 5’ Untranslated Region. J Med Virol. 2003;71:226–32. doi: 10.1002/jmv.10474. [DOI] [PubMed] [Google Scholar]
- 20.Quarleri JF, Mathet VL, Feld M, Ferrario D. GB virus C /hepatitis G virus groups and subgroups: classification by a restriction fragment length polymorphism method based on phylogenetic analysis of 5-untranslated region. J Clin microbiol. 1999;37:1340–47. doi: 10.1128/jcm.37.5.1340-1347.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghanbari R, Ravanshad M, Hosseini SY, Yaghobi R, Shahzamani K. Genotyping and Infection Rate of GBV-C among Iranian HCV-Infected Patients. Hepat Mon. 2010;27:80–87. [PMC free article] [PubMed] [Google Scholar]
- 22.Muerhoff AS, Dawson GJ, Desai SM. A Previously Unrecognized Sixth Genotype of GB Virus C Revealed by Analysis of 5. Untranslated Region Sequences. J Med Virol. 2006;78:105–11. doi: 10.1002/jmv.20510. [DOI] [PubMed] [Google Scholar]
- 23.Naito H, Abe K. Genotyping system of GBV-C/HGV type 1 to type 4 by the polymerase chain reaction using type-specific primers and geographical distribution of viral genotypes. J Virol Methods. 2001;91:3–9. doi: 10.1016/s0166-0934(00)00207-x. [DOI] [PubMed] [Google Scholar]
- 24.Smith DB, Cuceanu N, Davidson F. Discrimination of hepatitis G virus/GBV-C geographical variants by analysis of the 5, non-coding region. J Gen Virol. 1997;78:1533–42. doi: 10.1099/0022-1317-78-7-1533. [DOI] [PubMed] [Google Scholar]
- 25.Mukaide M, Mizokami M, Orito E, Ohba K, Nakano T, Ueda R, et al. Three different GB virus C/hepatitis G virus genotypes: Phylogenetic analysis and a genotyping assay based on restriction fragment length polymorphism. FEBS Letters. 1997;407:51–58. doi: 10.1016/s0014-5793(97)00136-1. [DOI] [PubMed] [Google Scholar]