Abstract
Aim
The aim of this study was to investigate the frequency of betalactamase producing EAEC isolates among young children with diarrhea in Zanjan, Iran.
Background
Entero aggregative Escherichia coli (EAEC) is an emerging enteric pathogen associated with acute and persistent diarrhea and the evolution and spread of acquired extended spectrum betalactamases (ESBLs) among these strains has become a serious problem in the management of infectious diseases in developing countries.
Patients and methods
During the period from March 2011 to January 2012, 140 isolates of E. coli from diarrheal children aged 0–60 months and 90 isolates from age-matched controls without diarrhea were investigated for EAEC using PCR. Antimicrobial susceptibility testing was performed as CLSI guidelines and betalactamase genes, including bla TEM, bla CTX-M, bla IMP, bla VIM and bla NDM-1 investigated in EAEC isolates.
Results
In this study, EAEC was detected with slightly higher frequency in children with (8%) than in children without (4.6%) diarrhea (P > 0.05). Diarrheagenic E. coli exhibited high level resistance to aztreonam (80.7%), amoxicillin (74.4%) and tetracycline (69.3%). Also, 86.4% of E. coli isolates were resistant to at least three different classes of antimicrobial agents and considered as multidrug resistance. Molecular characterization of betalactamase genes showed that bla TEM was the most frequently isolated betalactamase. It was detected in 78.9% of ESBL producing EAEC isolates. Also, the frequency of bla CTX-M was 63.1% (12/19) and 8 (42.1%) isolates carried the bla TEM and bla CTX-M, simultaneously. None MBL producing EAEC was detected in our study.
Conclusion
Our results indicate that ESBLs especially bla TEM and bla CTX-M are widespread among EAEC isolates and appropriate surveillance and control measures are essential to prevent further dissemination of betalactamases in our country.
Keywords: Antibiotic resistance, Diarrhea, EAEC, ESBL
Introduction
Enteroaggregative Escherichia coli (EAEC) is a subgroup of diarrheagenic E. coli (DEC) and has emerged as an important pathogen in travellers’ diarrhea, diarrhea among children and in immunocompromised patients (1, 2). EAEC are associated with watery diarrhea among children younger than five years in developing countries and represent a major public health problem in these areas (3, 4). Nguyen and colleagues, in a study of children younger than 5 years in Vietnam, identified EAEC in 11.6% of diarrheal children, compared with 4.4% of age-matched controls without diarrhea (1). A PCR method based on the presence of essential virulence factors as anti-aggregation protein transporter (CVD432 or the AA probe) would improve the diagnosis of EAEC diseases (5, 6).
The continuous emergence of resistance to antimicrobial agents among the prevalent pathogens is the most dangerous threat for the treatment of infectious disease (6). Compared with other diarrheagenic E. coli, EAEC was found to be highly resistant to many commonly used antimicrobials agents (7). The majority of the EAEC from diarrheal patients in Kolkata, India, exhibited multidrug resistance including resistant to fluoroquinolones (8). Furthermore, E. coli isolates resistant to oxyiminocephalosporins due to the production of extended spectrum betalactamases (ESBL) have emerged worldwide (9). During the next few years, CTX-M has become the predominant ESBL family and CTX-M-producing E. coli has spread globally and has been involved in nosocomial outbreaks and community acquired infections (10, 11). The acquisition of resistance genes by horizontal transfer is currently thought to play a major role in the development of multidrug resistant (MDR) strains (12, 13). Regular surveillance of antibiotic resistance provides information for antibiotic therapy and resistance control (6).
The objectives of the present study were (1) to determine the frequency of EAEC among children younger than 5 years with and without diarrhea and (2) to investigate pattern of antimicrobial resistance and frequency of betalactamase genes including bla CTX-M, bla TEM, bla IMP, bla VIM and bla NDM-1 among EAEC isolates in Zanjan, Iran.
Patients and Methods
This study included 600 stool specimens from children younger than five years of age between March 2012 and February 2013. Patients included 450 children with and 150 without diarrhea attending four major university hospitals in Zanjan, Iran. Control subjects were healthy children with no history of diarrhea and antibiotic therapy for at least 1 month. Stool samples collected in Cary-Blair transport medium were cultured on MacConkey agar (Merck, Germany) and then identified by standard biochemical methods. Verified isolates were preserved at –70 °C in Tripticase soy broth (Merck, Germany) containing 20% (v/v) glycerol for further analysis.
Antimicrobial susceptibility testing and phenotypic characterization
Susceptibility of isolates to the following antibiotics was examined using the disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (14): Amoxicilin (25µg), Aztreonam (30µg), Amikacin (30µg), Cefotaxime (30µg), Cefoxitine (30µg), Ceftazidime (30µg), Ciprofloxacin (5µg), Co-amoxiclav (30µg), Co-trimoxazole (25µg), Erythromycin (25µg), Gentamicin (10µg), Imipenem (10µg) and Tetracycline (30µg) (MAST, UK). Isolates shown to be resistant to at least three different classes of antimicrobial agents were determined to be multidrug resistant (MDR). Escherichia coli ATCC 25922 was used as control for antibiotic resistance. Phenotypic characterization of extended spectrum betalactamases (ESBLs) and metallo betalactamase (MBLs) was determined using Double Disk Synergy Test (DDST) according to CLSI criteria.
Detection of Enteroaggregative E. coli and betalactamases by PCR
Enteroaggregative E. coli was detected by virulence markers pCVD432 (the nucleotide sequence of the EcoRI-PstI DNA fragment of pCVD432) and astA (enteroaggregative heat stable toxin). Furthermore, ESBL or MBL producing isolates were tested for bla genes including bla TEM, bla CTX-M, bla IMP, bla VIM and bla NDM-1 using the primers listed in Table 1.
Table 1.
Primers used in this study
| Target | Primer sequence (5’→3’) | Amplicon size (bp) | Ref. |
|---|---|---|---|
| bla TEM | TCCGCTCATGAG ACA ATA ACCTTCGTCTGACAGTTACCAATGC | 931 | Kiratisin et al. 2008 |
| bla CTX-M | GGTTAAAAAATCACTGCGTCTTGGTGACGATTTTAGCCGC | 909 | Kiratisin et al. 2008 |
| bla IMP | GGAATAGAGTGGCTTAATTCTCCCAAACCACTACGTTATCT | 188 | Ellington et al. 2007 |
| bla VIM | GATGGTGTTTGGTCGCATACGAATGCGCAGCACCAG | 390 | Ellington et al. 2007 |
| pCVD432 | CTGGCGAAAGACTGTATCATAAATGTATAGAAATCCGCTGTT | 630 | Aslani et al. 2011 |
| astA | CCATCAACACAGTATATCCGAGGTCGCGAGTGACGGCTTTGT | 111 | Aslani et al. 2011 |
| NDM-1 | ACCGCCTGGACCGATGACCAGCCAAAGTTGGGCGCGGTTG | 263 | Shahcheraghi et al. 2013 |
The total DNA was extracted from whole organisms by boiling. The PCR mixtures with a final volume of 25 µl consisted of 5 µl template DNA; 0.2 mM of each deoxynucleoside triphosphate; 10 pmol of each primers; 10 mM Tris- HCl; 1.5 mM MgCl2; 50 mM KCl; 1.5 U of Taq DNA polymerase. PCR was performed with the Gene Atlas 322 system (ASTEC, Japan). Amplification involved an initial denaturation at 94°C, 5 min followed by 35 cycles of denaturation (94°C, 50 s), annealing (50°C, 1 min for blaTEM, blaCTX-M, blaVIM and blaIMP, 52°C, 40 s for astA, 55°C, 40 s for pCVD432 and 58°C, 50 s for blaNDM-1) and extension (72°C, 1 min), with a final extension step (72°C, 8 min). The amplified DNA was separated by submarine gel electrophoresis on 1% agarose, stained with ethidium bromide, and visualized under UV transillumination. The following reference strains were used as positive and negative controls: EAEC 97R (pCVD432), E. coli ATCC 35218 (blaTEM), K. pneumoniae 7881 (blaCTX-M), A. baumannii AC54/97 (blaIMP), E. coli K12 DH5α (no virulence gene) and E. coli ATCC 25922 (non betalactamase produser).
Statistical analysis
The data were analyzed with SSPS version 17.0 software (SPSS, Inc., Chicago, IL). The chi-square test was used to determine the statistical significance of the data. A P value of < 0.05 was considered significant.
Results
Detection and characterization of Enteroaggregative E. coli from clinical stool samples
A total of 450 children with diarrhea and 150 control children without diarrhea were studied. Among the total children, 133 (22.1%) children were younger than 12 months, 180 (30%) were 13–24 months and 287 (47.9%) were 25–60 months. The mean of age in patient and control groups was 24 and 20 month, respectively. The sex distribution was 364 (60.7%) male and 236 (39.3%) female.
Overall, 230 (38.3%) E. coli isolates were identified in the 600 stool samples: 140 isolates from diarrheal patients and 90 isolates from control group. The frequency of EAEC in the diarrheal patients and healthy controls was 36 (8%) and 7 (4.6%) isolates, respectively. EAEC isolates were identified with slightly higher frequencies in diarrheal patients than in control group (P > 0.05). Out of the 36 EAEC isolates harboring the pCVD432, 12 (2.7%) also had the astA gene.
Antimicrobial susceptibility testing
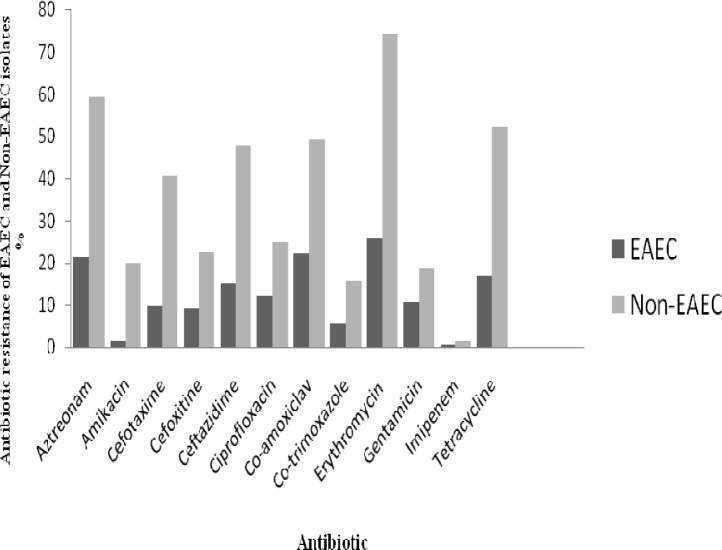
Antimicrobial susceptibility of EAEC and non-EAEC strains isolated from diarrheal patients is presented in Table 2 and Fig. 1. The highest rate of resistance among diarrheagenic E. coli showed against to Erythromycin (100%) followed by aztreonam (80.7%) and amoxicillin (74.4%). Although, imipenem resistance rate was 2.1% (3 isolates), but the intermediate resistant isolates (25%) should be concerned. A total of 121 (86.4%) isolates of E. coli were multidrug resistant (MDR). Out of the 36 EAEC isolates, 19 (52.7%) were ESBL positive, whilst none imipenem resistant and MBL producing EAEC was detected.
Table 2.
Antimicrobial susceptibility of EAEC and Non EAEC isolates
| Antimicrobial agents | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| EAEC (n= 36) (%) | Non-EAEC (n= 104)(%) | Total DGEC (n= 140) (%) | |||||||
| R | I | S | R | I | S | R | I | S | |
| Erythromycin | 36(25.7) | 0 | 0 | 104(74.3) | 0 | 0 | 140 (100) | 0 | 0 |
| Amoxicillin | 26(18.6) | 1(0.7) | 9(6.4) | 78(55.7) | 3(2.7) | 23(16.4) | 104(74.4) | 4(2.8) | 32(22.8) |
| Aztreonam | 3(04) | 2(1.4) | 4(2.8) | 83(59.3) | 17(12.1) | 4(2.8) | 113(80.7) | 19(13.6) | 8(5.7) |
| Co-amoxiclav | 31(22.1) | 1(0.7) | 4(2.8) | 69 (49.3) | 5(3.6) | 30(21.4) | 100(71.4) | 6(4.3) | 34 (24.3) |
| Cefotaxime | 14(10) | 10(7.1) | 12(8.6) | 57(40.7) | 9(6.4) | 38(27.1) | 71(50.7) | 19(13.6) | 50 (35.7) |
| Ceftazidime | 21(15) | 9(6.4) | 6(4.3) | 67(47.8) | 18(12.8) | 19(13.6) | 88(62.8) | 27(19.4) | 25 (17.8) |
| Cefoxitine | 13(9.3) | 1(0.7) | 22(15.7) | 32(22.8) | 6(4.3) | 66(47.1) | 45(32.1) | 7(5) | 88 (62.9) |
| Co-trimoxazole | 8 (5.7) | 14(10) | 14 (10) | 22(15.7) | 8(5.7) | 74(52.8) | 30(21.5) | 22(15.7) | 88(62.9) |
| Ciprofloxacin | 17(12.1) | 5(3.6) | 14(10) | 35(25) | 22(15.7) | 47(33.6) | 52(37.1) | 27(19.3) | 61 (43.6) |
| Tetracycline | 24(17.1) | 9(6.4) | 3 (9.3) | 73(52.1) | 24(17.1) | 7 (5) | 97(69.3) | 33(23.6) | 10 (7.1) |
| Amikacin | 2(1.4) | 15(10.7) | 19(13.6) | 28(20) | 20(14.3) | 56 (40) | 30(21.4) | 35(25) | 75 (53.6) |
| Gentamicin | 15(10.7) | 12(8.6) | 9 (6.4) | 26(18.6) | 38(27.1) | 40(28.6) | 41(29.3) | 50(35.7) | 49 (35) |
| Imipenem | 1(0.7) | 5(3.6) | 30(21.4) | 2(1.4) | 30(21.4) | 72(51.4) | 3(2.1) | 35(25) | 102 (72.9) |
Figure 1.
Antibiotic resistance of EAEC and non EAEC isolates
Molecular characterization of ESBL and MBL genes
EAEC isolates were subjected to PCR experiments to detect betalactamase genes, including bla TEM, bla CTX-M, bla IMP, bla VIM and bla NDM-1. Bla TEM was the most frequently isolated betalactamase. It was detected in 78.9% (15/19) of ESBL producing EAEC isolates. The frequency of bla CTX-M was 63.1% (12/19) and 8 (42.1%) isolates of EAEC producing ESBL carried the bla TEM and bla CTX-M, simultaneously. None MBL producing EAEC was detected in our study.
Discussion
EAEC is an emerging enteric pathogen associated with acute and persistent diarrhea (≥ 14 days) and may cause malnutrition and growth defects in children. It has been identified in traveller's diarrhea in both developing and developed countries and has been isolated in immunocompromised patients (15, 16). An increasing number of studies support the association of EAEC with diarrhea in populations in developing countries, most prominently in association with persistent diarrhea. In several studies, culture of EAEC from the stool during the first few days of diarrhea is predictive of a longer duration of illness. The association of EAEC with diarrhea appears to vary geographically, and many studies have demonstrated the importance of EAEC in pediatric diarrhea. In studies carried out in Vietnam and the USA, EAEC was isolated at higher prevalence in children with diarrhea (11.6 and 4.5%, respectively) than in controls (7.2 and 1.7%, respectively) (1, 17, 18). Our findings, however, were in contrast to these studies. According to results, EAEC pathotype isolated in both children with and without diarrhea and there was no significant association between EAEC and diarrhea. The prevalence of this pathotype in diarrheal and healthy children's was 8% and 4.6%, respectively. This lack of association with diarrhea has been observed in another study. In a study carried out in Nicaragua, EAEC strains were the most frequently isolated pathotype of E. coli; however, the isolation rate as a total was slightly higher in the healthy group than in the diarrheal group. It has also been shown that EAEC is a heterogeneous group of E. coli and not all strains are capable of causing diarrhea (19). Among the few published studies in Iran, only Salmanzadeh-Ahrabi et al has reported the association of EAEC isolates with diarrhea. They reported EAEC isolates in 24% of childrens with diarrhea and 8% of controls (p < 0.0001) (20, 21).
Antimicrobial resistance and the spread of betalactamases among human pathogens has become a major public health problem in developing countries. In a study carried out in Kolkata, India, majority of EAEC isolates from diarrheal patients exhibited multidrug resistance including resistant to fluoroquinolones (8). Peruvian EAEC isolates were also multidrug resistant, especially to ampicillin, cotrimoxazole, tetracycline and nalidixic acid (22). According to results, EAEC isolates exhibited high level resistance to various antibiotics. The high frequency of antibiotic resistant isolates of EAEC may be due to the widespread use of numerous antimicrobial agents in our country. Furthermore, a majority of isolates were resistant to aztreonam, amoxicillin and tetracycline, resulting in a high percentage of multidrug resistance.
In Asia, the frequency of ESBL-positive Enterobacteriaceae has been shown to vary in different countries. National survey data have indicated the prevalence of ESBLs in 5 to 8% of E. coli isolates from Korea, Japan, Malaysia and Singapore but in 12-24% in Thailand, Taiwan, the Philippines, Indonesia, Hong Kong and China (23). In the present study, the frequency of ESBL positive isolates of EAEC was 52.7%. The frequency of bla TEM and bla CTX-M in EAEC isolates was 78.9% and 63.1%, respectively. Metallo betalactamases have been reported from many countries, particularly in multidrug resistance pathogens like Pseudomonas aeruginosa and Acinetobacter species. None MBL producing EAEC was detected in our study. Our results indicate that ESBLs especially bla TEM and bla CTX-M are widespread among EAEC isolates and appropriate surveillance and control measures are essential to prevent further dissemination of betalactamases in our country.
Acknowledgements
The authors are grateful to the Department of Medical Microbiology, Zanjan University of Medical Sciences.
(Please cite as: Khoshvaght H, Haghi F, Zeighami H. Extended spectrum betalactamase producing Enteroaggregative Escherichia coli from young children in Iran. Gastroenterol Hepatol Bed Bench 2014;7(2):131-136).
References
- 1.Nguyen TV, Le Van P, Le Huy C, Gia KN, Weintraub A. Detection and characterization of diarrheagenic Escherichia coli from young children in Hanoi, Vietnam. J Clin Microbiol. 2005;43:755–60. doi: 10.1128/JCM.43.2.755-760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nataro JP. Enteroaggregative Escherichia coli pathogenesis. Curr Opin Gastroenterol. 2005;21:4–8. [PubMed] [Google Scholar]
- 3.Tobias J, Vutukuru SR. Simple and rapid multiplex PCR for identification of the main human diarrheagenic Escherichia coli . Microbiological Research. 2012;167:564–70. doi: 10.1016/j.micres.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 4.Taniuchi M, Walters CC, Gratz J, Maro A, Kumburu H, Serichantalergs O, et al. Development of a multiplex polymerase chain reaction assay for diarrheagenic Escherichia coli and Shigella spp. and its evaluation on colonies, culture broths, and stool. Diagn Microbiol Infect Dis. 2012;73:121–28. doi: 10.1016/j.diagmicrobio.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aranda KRS, Fagundes-Neto U, Scaletsky ICA. Evaluation of multiplex PCRs for diagnosis of infection with diarrheagenic Escherichia coli and Shigella spp. J Clin Microbiol. 2004;42:5849–53. doi: 10.1128/JCM.42.12.5849-5853.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aslani MM, Alikhani MY, Zavari A, Yousefi R, Zamani AR. Characterization of enteroaggregative Escherichia coli (EAEC) clinical isolates and their antibiotic resistance pattern. Int J Infect Dis. 2011;15:136–39. doi: 10.1016/j.ijid.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Arancibia EM, Pitart C, Ruiz J, Marco F, Gasco′n J, Vila J. Evolution of antimicrobial resistance in enteroaggregative Escherichia coli and enterotoxigenic Escherichia coli causing traveller's diarrhea. J Antimicrob Chemother. 2009;64:343–47. doi: 10.1093/jac/dkp178. [DOI] [PubMed] [Google Scholar]
- 8.Kahali S, Sarkar B, Rajendran K, Khanam J, Yamasaki Sh, Nandy RK, et al. Virulence characteristics and molecular epidemiology of enteroaggregative Escherichia coli isolates from hospitalized diarrheal patients in Kolkata, India. J Clin Microbiol. 2004;42:4111–20. doi: 10.1128/JCM.42.9.4111-4120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghafourian S, bin Sekawi Z, Sadeghifard N, Mohebi Neela VK, Maleki A, et al. The Prevalence of ESBLs producing Klebsiella pneumoniae isolates in some major hospitals, Iran. Open Microbiol J. 2011;5:91–95. doi: 10.2174/1874285801105010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiratisin P, Apisarnthanarak A, Laesripa Ch, Saifon P. Molecular characterization and epidemiology of extended-spectrum-ß-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates causing health care-associated infection in Thailand, where the CTX-M family is endemic. Antimicrob Agents Chemother. 2008;52:2818–24. doi: 10.1128/AAC.00171-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bandekar N, Vinodkumar CS, Basavarajappa KG, Prabhakar PJ, Nagaraj P. Betalactamases mediated resistance amongst gram negative bacilli in burn infection. Int J Biol Med Res. 2011;2:766–70. [Google Scholar]
- 12.Rodriguez-Martinez JM, Nordmann P, Fortineau N, Poirel L. VIM-19, a metallo-ß-lactamase with increased carbapenemase activity from Escherichia coli and Klebsiella pneumoniae. Antimicrob Agents Chemother. 2010;54:471–76. doi: 10.1128/AAC.00458-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gundogdu A, Beverley Long Y, Vollmerhausen TL, Katouli M. Antimicrobial resistance and distribution of sul genes and integron-associated intI genes among uropathogenic Escherichia coli in Queensland, Australia. J Med Microbiol. 2011;60:1633–42. doi: 10.1099/jmm.0.034140-0. [DOI] [PubMed] [Google Scholar]
- 14.Clinical and laboratory standards institute. Performance standards for antimicrobial susceptibility testing; Twenty third informational supplement. CLSI document M100-S23. 2013;33(1) [Google Scholar]
- 15.Weintraub Andrej. Enteroaggregative Escherichia coli: epidemiology, virulence and detection. J Med Microbiol. 2007;56:4–8. doi: 10.1099/jmm.0.46930-0. [DOI] [PubMed] [Google Scholar]
- 16.Amaya E, Reyes D, Vilchez S, Paniagua M, Mollby R, Nord KE, et al. Antibiotic resistance patterns of intestinal Escherichia coli isolates from Nicaraguan children. J Med Microbiol. 2011;60:216–22. doi: 10.1099/jmm.0.020842-0. [DOI] [PubMed] [Google Scholar]
- 17.Nataro JP, Mai V, Johnson J, Blackwelder WC, Heimer R, Tirrell S, et al. Diarrheagenic Escherichia coli infection in Baltimore, Maryland, and New Haven, Connecticut. Clin Infect Dis. 2006;43:402–407. doi: 10.1086/505867. [DOI] [PubMed] [Google Scholar]
- 18.Usein CR, Tatu-Chitoiu D, Ciontea S, Condei M, Damian M. Escherichia pathotypes associated with diarrhea in Romanian children younger than five years of age. Jpn J Infect Dis. 2009;62:289–93. [PubMed] [Google Scholar]
- 19.Vilchez S, Bucardo F, Reyes D, Paniagua M, Mollby R, Weintraub A. Prevalence of diarrhoeagenic Escherichia coli in children from Leon, Nicaragua. J Med Microbiol. 2009;58:630–637. doi: 10.1099/jmm.0.007369-0. [DOI] [PubMed] [Google Scholar]
- 20.Jafari A, Aslani MM, Bouzari S. Enteroaggregative Escherichia coli, a heterogenous, underestimated and under-diagnosed E. coli pathotype in Iran. Gastroenterol Hepatol Bed Bench. 2013;6:71–79. [PMC free article] [PubMed] [Google Scholar]
- 21.Salmanzadeh Ahrabi S, Habibi E, Jaafari F, Zali MR. Molecular epidemiology of Escherichia coli diarrhoea in children in Tehran. Ann Trop Paediatr. 2005;25:35–39. doi: 10.1179/146532805X23335. [DOI] [PubMed] [Google Scholar]
- 22.Ochoa TJ, Ecker L, Barletta F, Mispireta ML, Gil AI, Contreras C, et al. Age-related susceptibility to infection with diarrheagenic Escherichia coli among infants from Periurban areas in Lima, Peru. Clin Infect Dis. 2009;49:1694–702. doi: 10.1086/648069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phongpaichit S, Tunyapanit W, Pruekprasert P. Antimicrobial resistance, class 1 integrons and extended spectrum betalactamases in E. coli clinical isolates from patients in south Thailand. J Health Sci. 2011;57:281–88. [Google Scholar]
- 24.Ellington MJ, Kistler J, Livermore DM, Woodford N. Multiplex PCR for rapid detection of genes encoding acquired metallo- β -lactamases. J Antimicrob Chemother. 2007;59:321–22. doi: 10.1093/jac/dkl481. [DOI] [PubMed] [Google Scholar]
- 25.Shahcheraghi F, Nobari S, Rahmati Ghezelgeh F, Nasiri S, Owlia P, Nikbin VS, et al. First report of New Delhi Metallo-Beta-Lactamase-1-producing Klebsiella pneumoniae in Iran. Microb Drug Resis. 2013;19:30–36. doi: 10.1089/mdr.2012.0078. [DOI] [PubMed] [Google Scholar]