Abstract
Aim
The aim of this study is evaluation of molecular assay and the standard staining method.
Background
Cryptosporidium is a protozoon from coccidian subclass, which is one of the most important causes of diarrhea in children and immunocompromised individuals around the world. Diagnosis and treatment are necessary for mentioned cases. Usual diagnostic method for this parasite is fecal smear preparation, modified ziehl-neelsen staining, microscopic consideration and oocyst observation.
Patients and methods
A totally of 2510 stool samples collected from children with diarrhea of 4 pediatric hospitals. Direct smears prepared from fresh fecal samples and from the sediment of formalin-ether method of the same samples. The smears stained with modified ziehl-neelsen method then considered with microscope. The 30 positive samples with staining method considered with DNA extraction and PCR method for cryptosporidiosis infection determination and sensitivity evaluation. 114 random negative samples considered with DNA extraction and PCR method for cryptosporidiosis infection diagnosis and specificity evaluation.
Results
30 positive cases from 2510 fecal samples detected by modified ziehl-neelsen staining and PCR method. We did not have any false positive cases by staining method but 2 cases of negative samples by staining method were positive by PCR technique, which informed us of 2 false negative. The positive samples sequenced for reconfirmation.
Conclusion
Thus, sensitivity of staining method was computed to be 94% and specificity was 100% but sensitivity and specificity of PCR method was calculated to be 100%.
Keywords: Diarrhea, Cryptosporidium, Modified ziehl-neelsen staining, PCR
Introduction
Cryptosporidium is an obligated intracellular protozoan that is able to complete its life cycle intra cellular extra cytoplasmic in the host intestinal epithelial Cells (1). This is a worldwide parasite and it has been reported from different parts of the world. Transmission of these protozoa is fecal–oral and contaminated food and water with oocyst is important factors in its transmission. Therefore, it is classified as a waterborne disease (2.3). Cryptosporidium have been isolated from many types of vertebrates, such as mammals, birds, reptiles and fish. It is a zoonotic disease because of its capability to transmit from animals to human (4). Cryptosporidiosis manifests as a self -imited diarrhea in immunocompetent individuals whereas, in immunocompromised individuals causes severe and chronic diarrhea that if untreated, can lead to death (5). Children due to their low immunity are more susceptible to infection, but most immunocompetent children regarding to nutritional status, health and other environmental factors suffer a self-limited diarrhea. Some outbreaks of cryptosporidiosis revealed the importance of these protozoa (5).
Direct smear preparation of stool or using sediment of formalin ether technique for smear preparation and applying modified Ziehl-neelsen staining (MZN) is ordinary method for Cryptosporidium oocyst diagnosis in microscopic consideration (6.7). Regarding the presence of other acid-fast microorganisms in feces, which are similar in size to Cryptosporidium such as yeast, pollen and other protozoa like Cyclospora, It is not possible to detect these protozoa easily and need to have an expert technician (6, 8). The disadvantage of this technique is a necessity for existence of at least 50,000-500,000 oocyst per gram of stool for detection. It encourages us to look for a new method, which can detect less oocyst per gram of stool and more reliable results (9). Controlling the diseases depend on identifying species and genotyping the agent of the disease (10). Previous reports have shown high sensitivity and specificity for Cryptosporidium molecular detection technique (10). The life cycle pattern of a new genotype helps us to stop the transmission (11). Despite the disadvantages of conventional (MZN) methods, most health care centers are using MZN as a gold standard for Cryptosporidium diagnosis, but there is not enough comparative study for evaluation of molecular method (PCR) with MZN methods in the world especially in Iran. Therefore, it is necessary to compare these two methods with each other, until to declare the best technique for the best results.
Patients and Methods
Fecal specimens
After coordinating with the authorities of different hospitals, 2510 stool samples collected from laboratories from May 2010 until Nov 2012. Samples transferred to the laboratory of parasitology and mycology department, faculty of medicine, Shahid Beheshti University of medical sciences. It should be noted that in this study diarrhea defined as loose or watery stool.
Groups of study
This study has been done on children under 12 years old who have diarrhea and are referred to Shahid Fahmideh hospital, Mofid pediatric hospital, Mahak Medical Center for children with cancer and Medical center of children.
Statistical sample volume
To evaluate and compare the sensitivity and specificity between MZN and PCR respectively, it requires a certain positive and negative samples of Cryptosporidium. According to the guidance of statistic advisor and use of statistical formulas Number 1 & 2, 30 positive samples were needed to evaluate the sensitivity, 114 negative samples were needed to evaluate the specificity. To collect these 30 positive and 114 negative samples, with the prevalence of 1%, the maximum confidence level of 95% and 0.1 error, required samples were calculated to be 2510 (formula number 3).
All samples were numbered, after transferring to the laboratory, then smear prepared from all samples by the direct method and by concentration method. In direct smear method a drop of fecal suspension placed on a glass slide and spread to form a thin smear. In formalin-ether method, fecal suspension was made by 2 grams of soft and loose stool or with 2 ml of watery stool, then 10 ml of 10% formalin added to the suspension and mixed well then filtered with No. 10 mesh screen and poured to conical tubes. Then 3ml ether was added to tube and strongly was shaken then centrifuged at 500g for 15 minutes, supernatants were disposed and the smear prepared from the sediment. Smears were stained by modified Ziehl Neelsen method and dried by the air then fixed by ethanol, later alkaline fushin was poured on the slides and heated until it brought up to steam, but not boiled, after 5 minute slides were washed by water and decolorized by 2.5% sulfuric acid for 1 minute, depending on the film thickness then counter stained with 1% methylene blue for 1 min then washed and air dried and examined with 100X objective. One slide was reviewed per case at a rate of 5 min per slide.
DNA extraction and PCR were done on the samples, which were positive by MZN for detection of Cryptosporidium infection and sensitivity. Also, evaluation was done on 114 samples, which were negative by MZN, for specificity and Cryptosporidium infection detection.
DNA extraction
DNA was extracted from fecal samples using QIAamp DNA stool Mini Kit (QIAGEN, Hilden, Germany). Following manufacturer's instructions with a modification of five times for 5-minute freeze-thaw cycle to rupture the Cryptosporidium oocyst wall for better DNA extraction. The concentration of extracted DNA was measured by nanodrap (Thermo, ND1000).
Amplification
A nested PCR targeting the small subunit of 18s-rRNAgene was performed with use of earlier reported primers (Xiao et al., 2001). Initial primers Cr18PA: (5′ -TTC TAG AGC TAA TAC ATG CG-3′) and Cr18PB: (5′-CCC ATT TCC TTC GAA ACA GGA-3′) amplified a 1.3 kb fragment of 18S-rRNA gene. The inner primer Cr18NA: 5′-GGA AGG GTT GTA TTT ATT AGA TAA AG-3′ and Cr18NB: 5′-CTC ATA AGG TGC TGA AGG AGT A-3′ amplified a 826-864 bp fragment of former amplified sequence (12). The reaction mixture of the PCR consisted of 3µl, 1X PCR buffer (50 mM KCL, 20 mM Tris-HCL, 2.5 mM Mgcl2, pH 8.4) and 1µl, of 0.1 µg/ml BSA, 2µl of the initial primers, and 1µl of 0.3 mM concentration of each of dNTPs, 0.5µl of recombinant Taq polymerase and 2 µl of the purified DNA and 2µl of 1.5 mM mgcl2. For the second round of amplification, the reaction mixture was prepared as described above, except the primers and DNA template. We used 1µl of first PCR product as a source of DNA and the inner primers. Primary amplification was carried out in 30 cycles, each consisting of 94°C for 35s, 50°C for 35s, and 72°C for 45s, with an initial denaturation at 94°C for 8 min and a final extension at 72°C for 5 min. For secondary amplification, 35 cycles were used, with identical temperatures and times. All PCRs were run in a Thechne PCR thermocycler, PCR products were analyzed by electrophoresis on 1.5% agarose gels, stained with ethidium bromide, and visualized on a UV transilluminator (IN genius from SYNGEN company). Positive controls including PCR mixture reaction with the DNA extract of known Cryptosporidium positive stool samples were used in both PCR respectively. PCR mixture reaction without DNA, were used as negative controls in both PCR too.
Statistical analysis
The maximum likelihood method was used to estimate the sensitivity and specificity of MZN & PCR methods. After setting up the PCR protocol, it was used for all samples.
Thus, PCR performed on 30 samples, which were positive by staining, to evaluate the sensitivity and 114 samples, which were negative by the same method to assess the specificity of PCR. In comparative studies to evaluate and compare the two methods, sensitivity and specificity of them were determinate and compared. Thus, PCR performed on 30 positive and 114 negative samples to determine the true and false, positive and negative samples.
To examine the sensitivity, the formula No. 5-2 and 6-2 were used to assess the specificity and the data from this study was analyzed by using the software SPSS16.
Results
Microscopic assay
A total of 2510 diarrheic fecal samples were screened by microscope after MZN, 30 positive samples for cryptosporidium oocyst and 114 randomized negative samples from all collected samples were preserved in 70% ethanol.
PCR method
All 144 samples spiked with positive control, amplification of Cryptosporidium DNA indicates that PCR inhibition was not a factor in this trial.
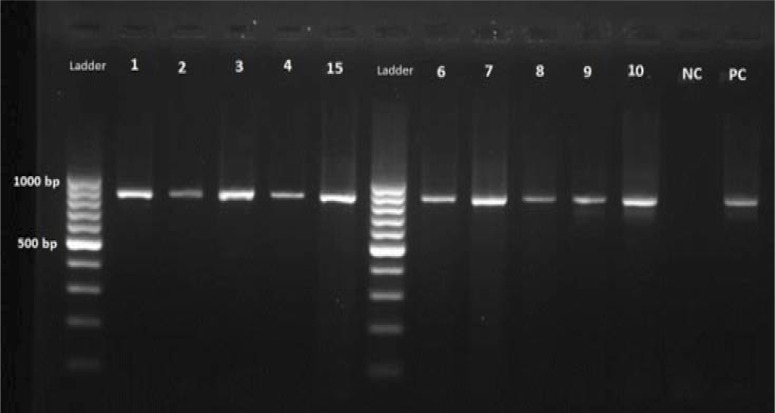
Microscopy detected a total of 30 positive (1.1%). PCR analysis detected a total of 32 (1.2%) positive, amplification of the samples, which previously were positive by MZN, identified 30 positive samples as well, which indicated no false positive by MZN method. All 30 positive samples had an 840bp band on electrophoresis gel after PCR reaction (Figure 1), so there were no false positive results with MZN method.
Figure 1.
Positive samples by PCR method, which also were positive by MZN
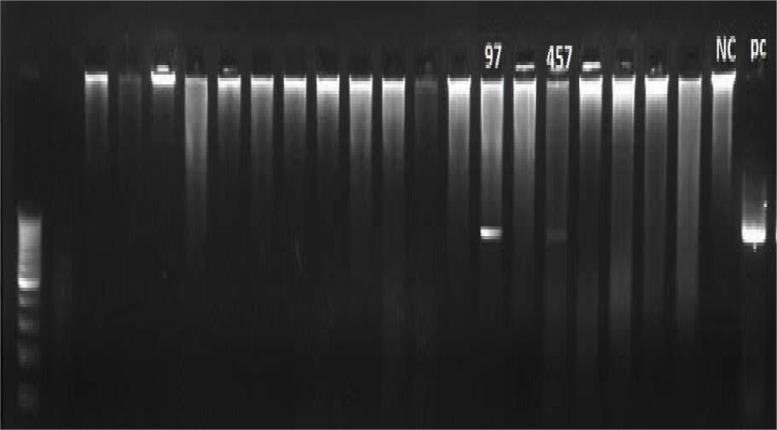
On the other hand, 114 negative samples with MZN method which collected randomly for specificity evaluation were tested by PCR reaction and 2 positive cases were identified in them (Figure 2), and the results indicated that 2 false negative by MZN method and the rest of 112 samples were true negative.
Figure 2.
Revealed two positive samples by PCR method, which previously were negative by MZN method
Comparison of MZN & PCR results for sensitivity and specificity informed us 94% sensitivity for MZN and 100% for PCR and also 100% specificity for both methods. (See Table 1).
Table 1.
Comparison of PCR versus microscopic detection of Cryptosporidium
| Method | Examined samples | Positive samples | Negative samples | Sensitivity (%) | Specificity (%) | Genotyping ability |
|---|---|---|---|---|---|---|
| PCR | 144 | 32 | 112 | 100 | 100 | Yes |
| MZN | 144 | 30 | 114 | 94 | 100 | No |
Discussion
Cryptosporidium is one of the most important causes of diarrhea in immunocompromised individuals and children. The significance of cryptosporidiosis depends on number of immunocompromised persons such as AIDS patients or consumers of immunosuppressive drugs. This study was done for evaluation of PCR as an accurate method for cryptosporidium diagnosis.
Morgan UM, Pallant L et al. in 1998 compared PCR with staining method in Cryptosporidium parvum diagnosis in diarrheic samples of patients. From 511 fecal samples, 36 samples were positive by PCR and 29 samples were positive by staining and 400X power. As a result the number of false negative calculated was 7. After performing the PCR on the samples, which they were positive by MZN, 5 samples considered as false positive. Their study showed 83.7% sensitivity and 98.9% specificity for staining method. According to the results, PCR technique is a useful method in cryptosporidium diagnosis (11). Our results showed the similar sensitivity to Morgan results for number of false negative by MZN, which depends on the number of oocyst in stool samples. On the other hand, our results revealed 100% specificity for MZN, which is different to Morgan‘s results. In our study, we had no false positive while they had detected 5 false positive, which is due to different study methods. The use of concentration methods, MZN staining, and 1000X power decreases the amount of false positives and detects the oocysts more accurately in our study. Kaushik et al. in 2008, in India, evaluated the four methods including: Ziehl neelsen staining, safranin methylen blue staining, Ag detection and Nested-PCR for Cryptosporidium detection. This experiment was done on 359 diarrheic stool samples of which 206 individual were HIV seropositive and 153 were HIV seronegative. Their results showed 100% sensitivity for PCR and 41.5% sensitivity for MZN staining, which are similar to ours. In addition to sensitivity the results of specificity is the same and is 100%, which is similar to our results (13). Zeihda in Malaysia compared the PCR and MZN for Cryptosporidium detection in AIDS patients. 8 out of 59 were detected by PCR and just 2 were detected by MZN and 1 was detected by both methods. Their results indicated high sensitivity for PCR and less for MZN and also specificity for both methods are equal and are 100%, which are similar to our results. The low sensitivity of MZN is because of not using concentration method (14). Results of Bialek revealed that sensitivity of immunologic methods is sufficient for the cryptosporidium screening in feces samples and showed that PCR didn't lead to any significant increase in sensitivity (15). However, Zeigler announced the increase in sensitivity by PCR in Cryptosporidium diagnosis, which is similar to our results (16). Another scientist revealed that PCR is the most sensitive method for the Cryptosporidium diagnosis, but in developing countries, the concentration methods are reliable in case of lacking PCR in medical laboratories (17).
Conclusion
According to the importance of Cryptosporidium especially in high risks groups such as AIDS patients and transplant patients who use immunocompromised drugs whom are very sensitive and susceptible to infection and are required to have diagnosis and treatment immediately suggest that laboratory of pediatric hospitals and general hospital with aids section, to use PCR for Cryptosporidium diagnosis, but regarding to the cost of PCR and its unavailability in all laboratories and hospitals, MZN staining on smears which is prepared by formalin-ether method seems to be useful and has enough accuracy for Cryptosporidium diagnosis for low risk patients.
Acknowledgements
The present research was supported by research grant of Pediatric Infectious Research Center of Shahid Beheshti University of Medical sciences Tehran, Iran. We appreciate the cooperation of the lab directors of Mofid hospital and this paper is access and utilization of postgraduate dissertation.
(Please cite as: Tahvildar-Biderouni F, Salehi N. Detection of Cryptosporidium infection by modified zeihl-neelsen and PCR methods in children with diarrheal samples in pediatric hospitals in Tehran. Gastroenterol Hepatol Bed Bench 2014;7(2):125-130).
References
- 1.Fayer R, Xiao L, editors. New York: Clearance Center; 2007. Cryptosporidium and cryptosporidiosis. [Google Scholar]
- 2.Unger BLP, Dubey JP, Speer CA, Fayer R, editors. Boca Rotan. FL: CRC Press; 1990. Cryptosporidiosis in human (homo sapiens)Cryptosporidiosis of man and animals. [Google Scholar]
- 3.Sunnotel O, Lowery CJ, Moore JE, Dooley JSG, Xiao L, Millar BC, et al. Cryptosporidium. Let Appl Microbiol. 2006;43:7–16. doi: 10.1111/j.1472-765X.2006.01936.x. [DOI] [PubMed] [Google Scholar]
- 4.Sanford SE, Josephson GKA. Bovine Cryptosporidiosis Clinical and Pathological Findings in Forty-two Scouring Neonatal Calves. Can Vet J. 1982;23:343–47. [PMC free article] [PubMed] [Google Scholar]
- 5.Mc Donald V. Welcome Trust illustrated history of tropical disease. Cryptosporidiosis. In: Cox FEG, editor. London, England: The Welcome Trust; 1996. pp. 256–63. [Google Scholar]
- 6.Neva F, Brown HW, editors. 6th ed. New York: WB Saunders; 1994. Basic clinical parasitology. [Google Scholar]
- 7.Fayer R, Morgan U, Upton SJ. Epidemiology of Cryptosporidium: transmission, detection and identification. Int J Parasitol. 2000;30:1305–22. doi: 10.1016/s0020-7519(00)00135-1. [DOI] [PubMed] [Google Scholar]
- 8.Garcia LS, Brewer TC, Bruckner DA. Fluorescence detection of Cryptosporidium oocysts in human fecal specimens by using monoclonal antibodies. Clin Microbiol J. 1987;25:119–21. doi: 10.1128/jcm.25.1.119-121.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anonymous. Cryptosporidium outbreak in British Colombia, Cryptosporidium Capsule 1; 1996. pp. 1–3. [Google Scholar]
- 10.Morgan U, Thompson RCA. PCR Detection of Cryptosporidium: Parasitol Today. 1998;14:241–45. doi: 10.1016/s0169-4758(98)01247-2. [DOI] [PubMed] [Google Scholar]
- 11.Morgan UM, Pallant L, Dwyer BW, Forbes DA, Rich G, Thompson CA. Comparison of PCR and microscopy for detection of Cryptosporidium parvum in human fecal specimens. Clinical Trial Microbiol. 1998;36:995–98. doi: 10.1128/jcm.36.4.995-998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao L, Bern C, Limor J, Sulaiman I, Roberts J, Checkley W, et al. Identification of 5 Types of Cryptosporidium Parasites in Children in Lima, Peru. J Infect Dis. 2001;183:492–97. doi: 10.1086/318090. [DOI] [PubMed] [Google Scholar]
- 13.Kaushik K, Khurana S, Wanchu A, Malla N. Evaluation of staining techniques, antigen detection and nested PCR for the diagnosis of cryptosporidiosis in HIV seropositive and seronegative patients. Parasitol Res. 2008;107(1):1–7. doi: 10.1016/j.actatropica.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Zaidah AR, Chan YY, SitiAsma H, Shukri A, Nurhaslindawati A, Salleh M, et al. Detection of Cryptosporidium parvum in HIV-infected patients in Malaysia using a molecular approach. South ASI J Trop Med. 2008;39:140–51. [PubMed] [Google Scholar]
- 15.Bialeka R, Bindera N, Dietzb K, Joachimc A, Knobloch J, Ulrike E, et al. Comparison of fluorescence, antigen and PCR assays to detect Cryptosporidium parvum in fecal specimens. Diagnostic Microbiol Inf Dis. 2002;43:283–88. doi: 10.1016/s0732-8893(02)00408-x. [DOI] [PubMed] [Google Scholar]
- 16.Ziegler PE, Santucci F, Lindergard G, Nydam DV, Wade SE, Schaaf SL, et al. Evaluation of polymerase chain reaction diagnosis of Cryptosporidium spp in dairy cattle and wildlife. Vet Ther. 2007;8:148–59. [PubMed] [Google Scholar]
- 17.Paul S, Chandra D, Tewari AK, Banerjee PS, Ray DD, Boral R. Comparative evaluation and economic assessment of coprological diagnostic methods and PCR for detection of Cryptosporidium spp. in bovines. Vet Parasitol Vol. 2009;164:295–91. doi: 10.1016/j.vetpar.2009.06.015. [DOI] [PubMed] [Google Scholar]